Synthesis and Characterization of Edaravone Analogues as Remyelinating Agents and Putative Mechanistic Probes
Abstract
1. Introduction
2. Results
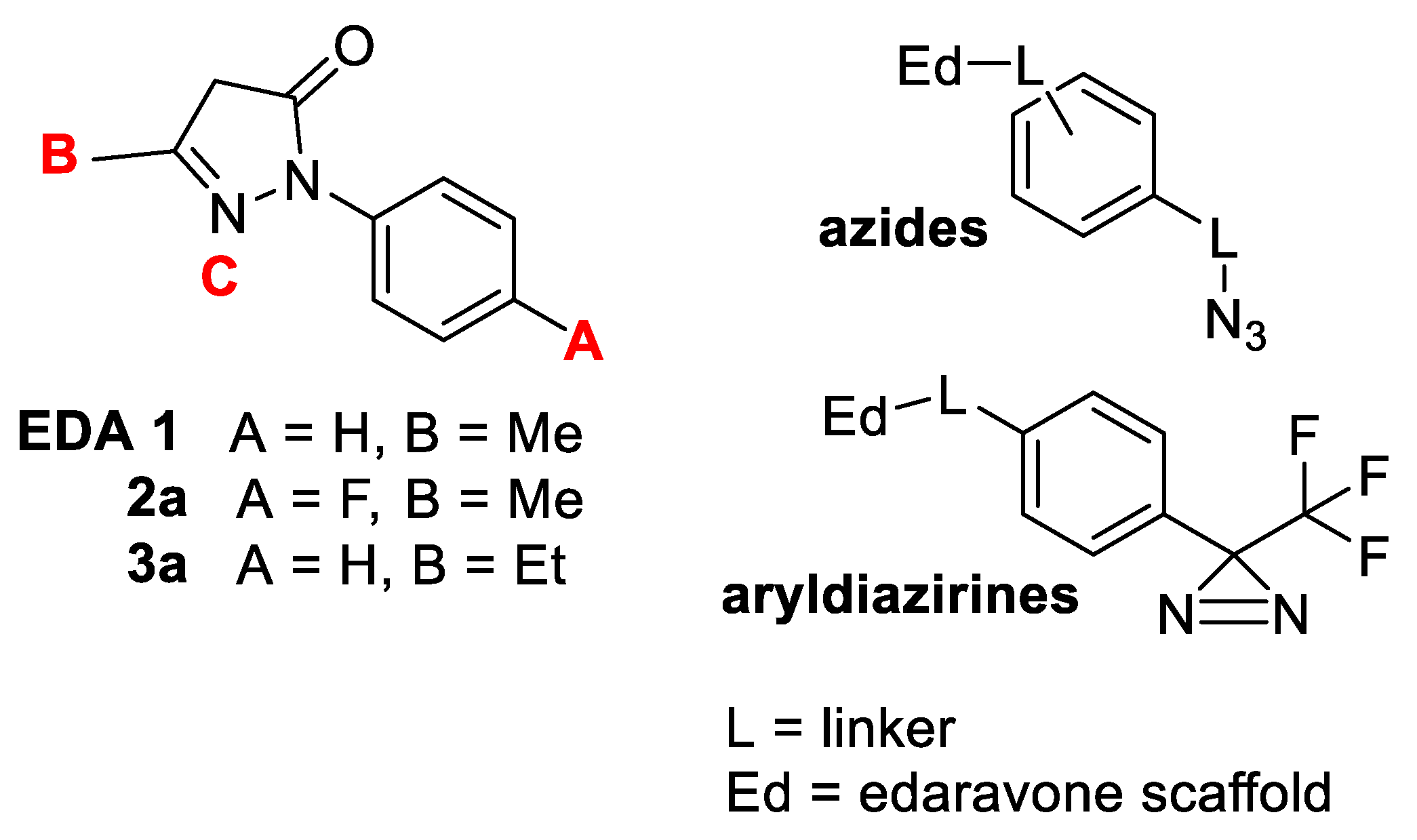
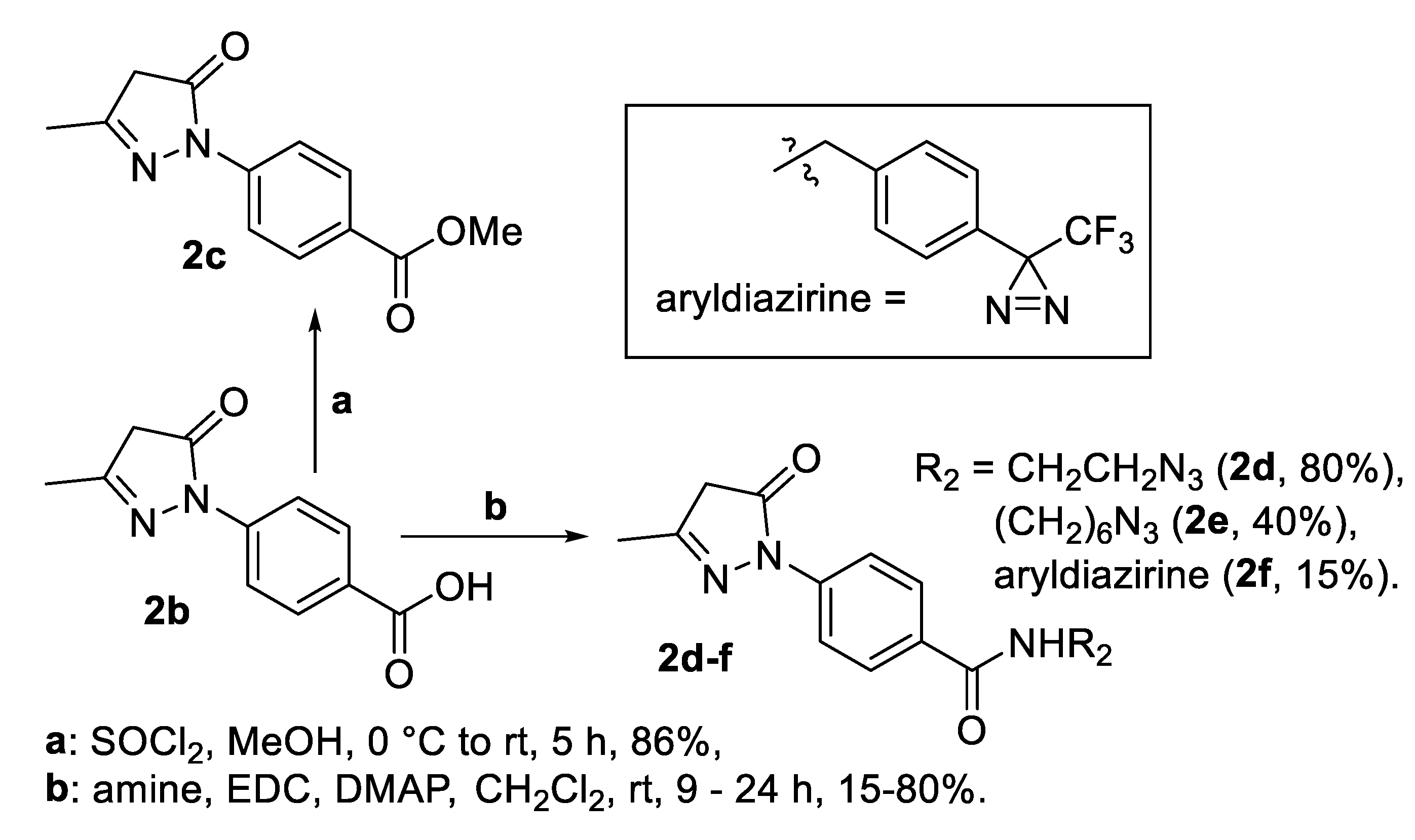
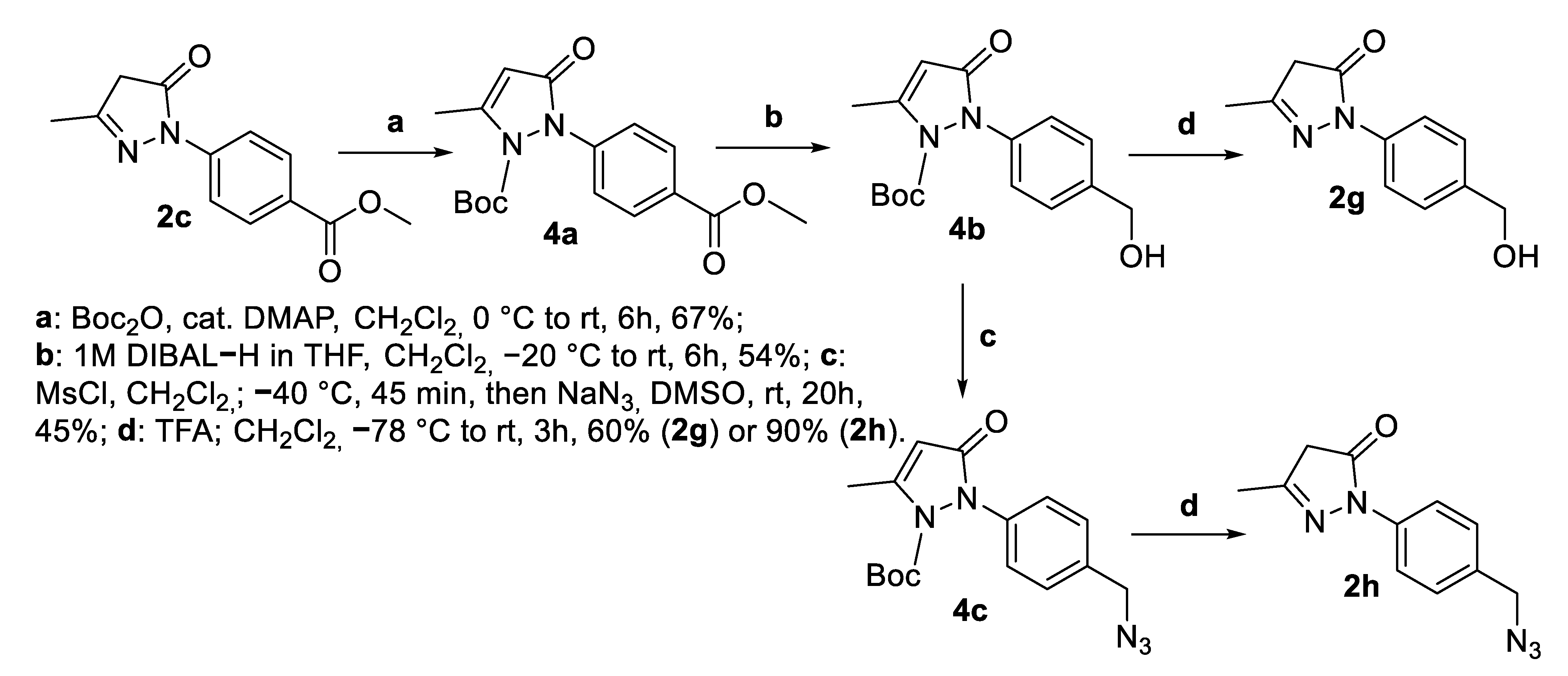
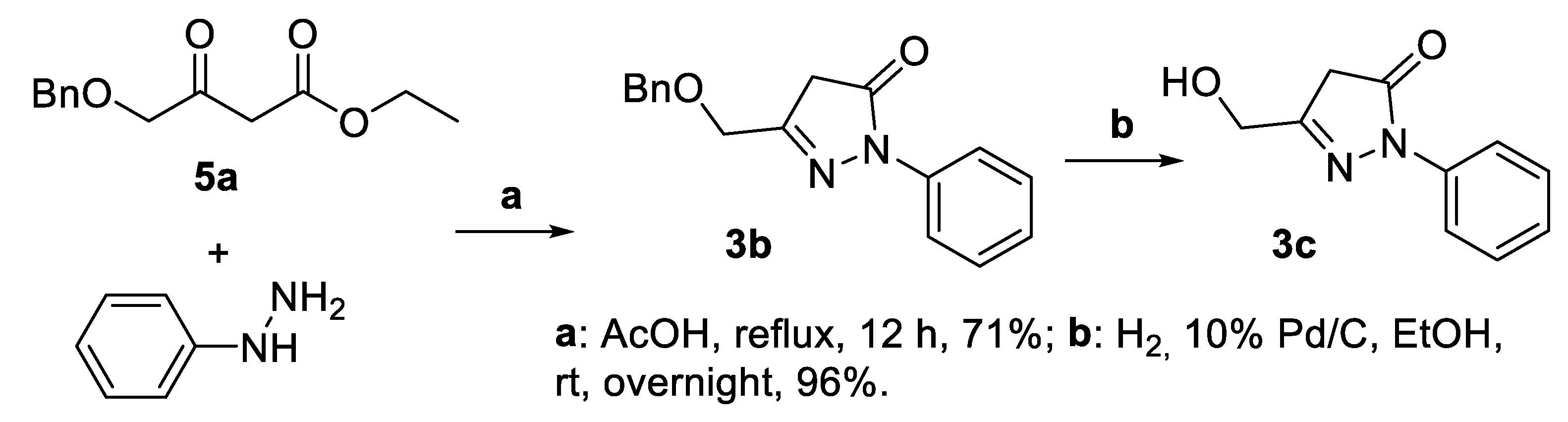
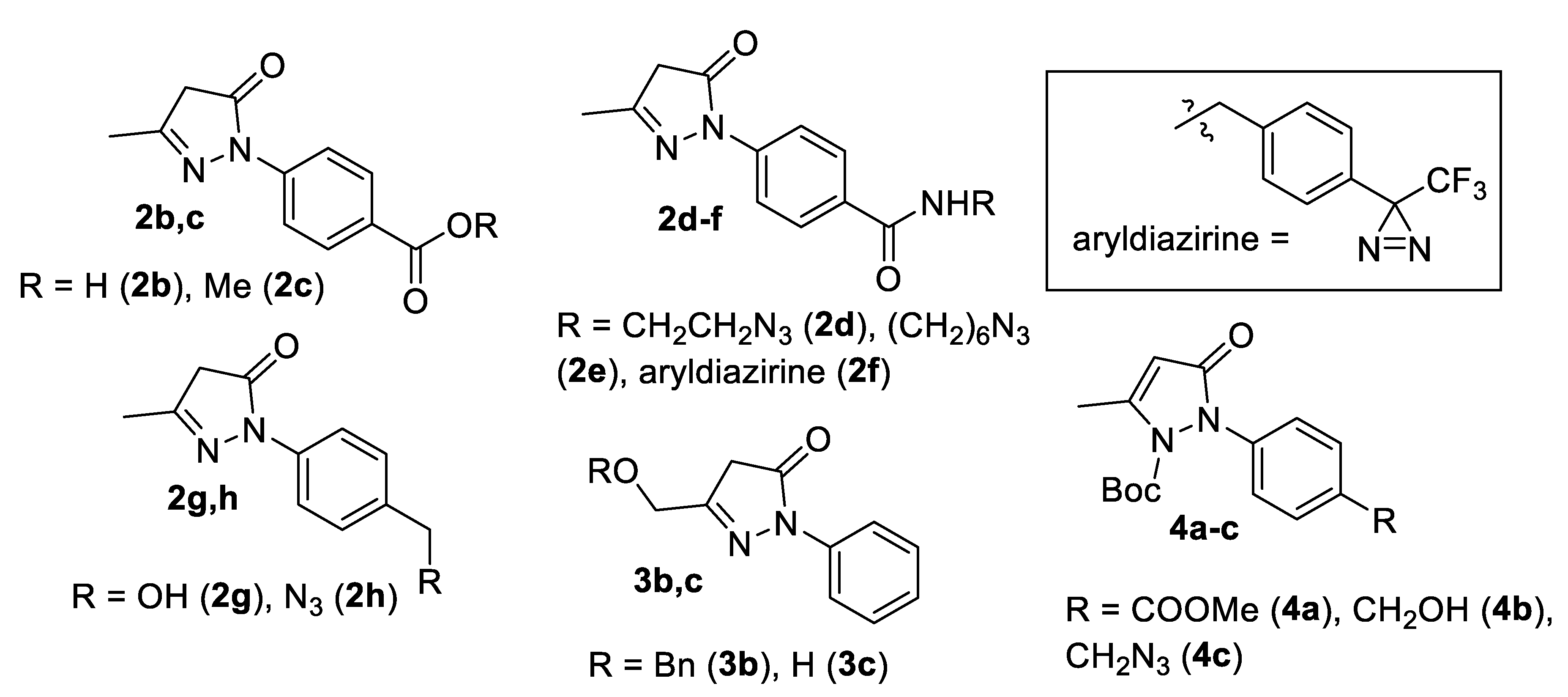
2.1. Synthesis
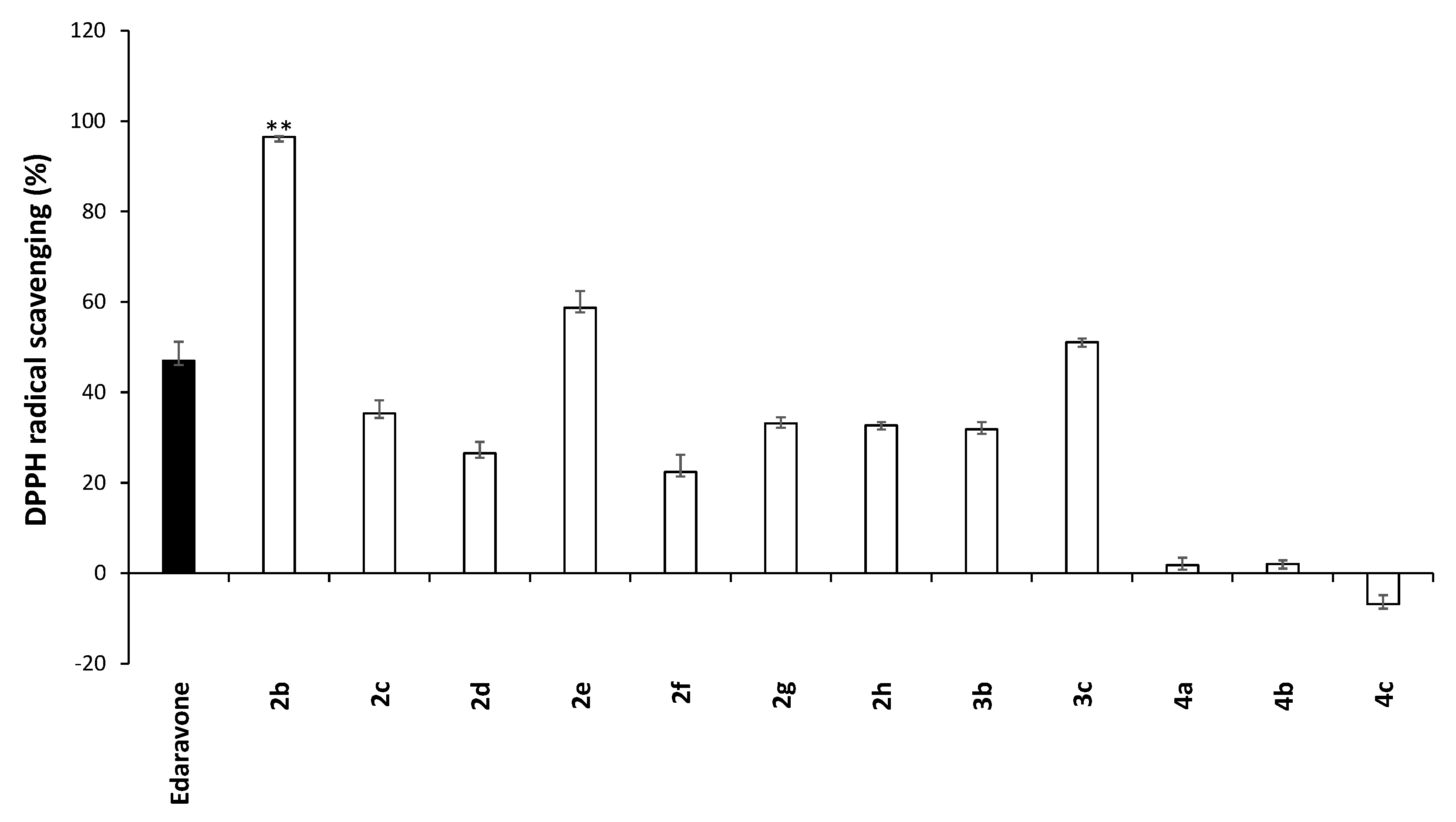
2.2. Primary Biological/ADME Profiling of EDA Analogues 2b–4c
2.3. Detailed Characterization—EDA Analogues 2b, 2c, 4a
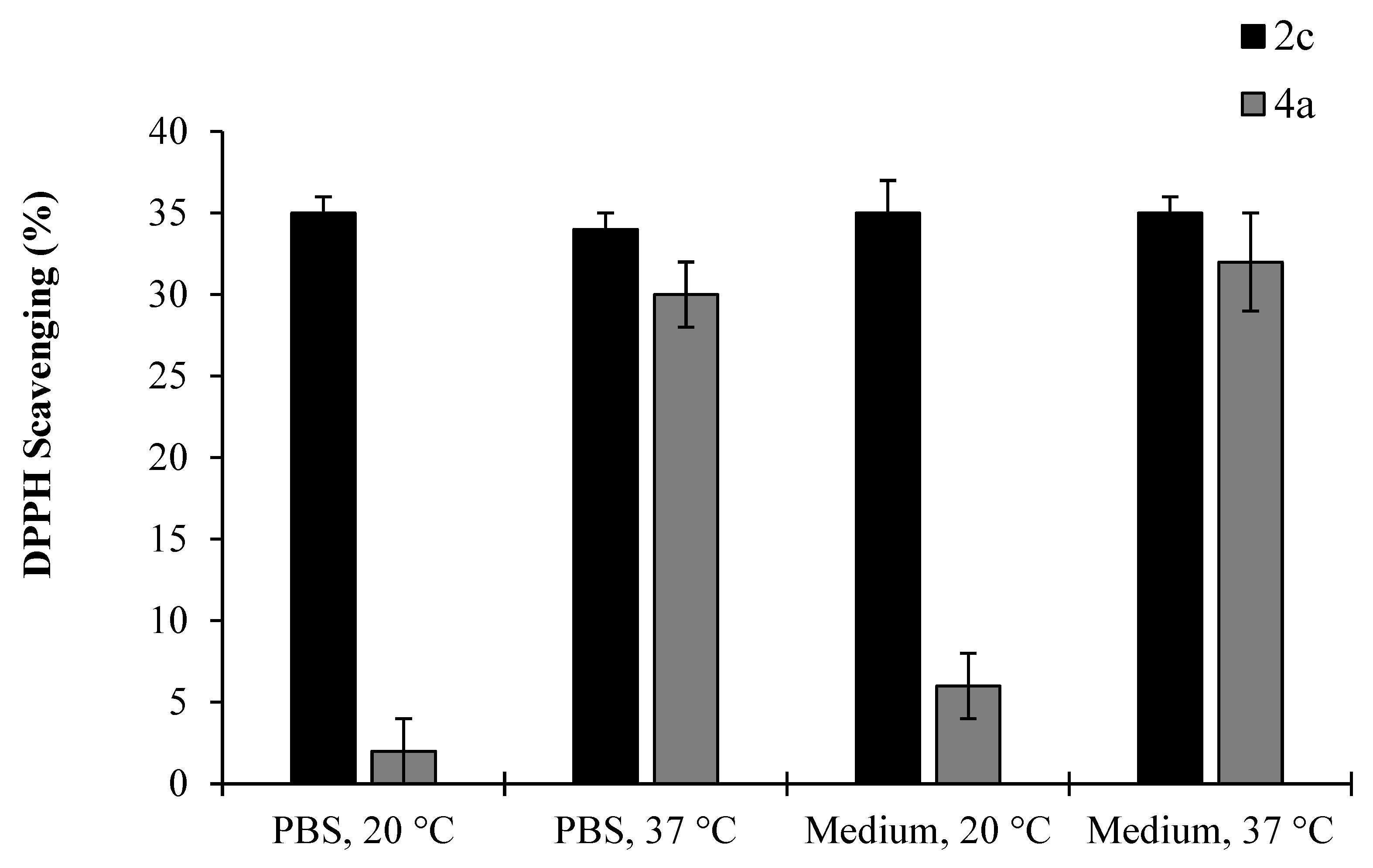
2.4. Stability Studies—EDA Analogues 2c and 4a
3. Discussion and Conclusions
4. Materials and Methods
4.1. Synthesis
4.1.1. General
4.1.2. Synthesis of Methyl 4-(3-methyl-5-oxo-4,5-dihydro-1H-pyrazol-1-yl)benzoate 2c
4.1.3. Synthesis of Tert-butyl 2-(4-(methoxycarbonyl)phenyl)-5-methyl-3-oxo-2,3-dihydro-1H-pyrazole-1-carboxylate 4a
4.2. Biology
4.2.1. Animals
4.2.2. OPC Cultures
4.2.3. 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Reduction Assay
4.2.4. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) Radical Scavenging Assay
4.2.5. Chemical Stability—DPPH Radical Scavenging Assay
4.2.6. Determination of Distribution Coefficient (Log D7.4)
4.2.7. Interaction Studies with Liposome Cell Membrane Models
4.3. Computational Studies
4.3.1. In Silico ADME Predictions
4.3.2. Quantum Chemical Simulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- McGinley, M.P.; Goldschmidt, C.H.; Rae-Grant, A.D. Diagnosis and treatment of multiple sclerosis. JAMA 2021, 325, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, C.H.; McGinley, M.P. Advances in the treatment of multiple sclerosis. Neurol. Clin. 2021, 39, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Tepavčević, V.; Lubetzki, C. Oligodendrocyte progenitor cell recruitment and remyelination in multiple sclerosis: The more, the merrier? Brain 2022, 145, 4178–4192. [Google Scholar] [CrossRef]
- Eleuteri, C.; Olla, S.; Veroni, C.; Umeton, R.; Mechelli, R.; Romano, S.; Buscarinu, M.; Ferrari, F.; Calò, G.; Ristori, G.; et al. A Staged Screening of Registered Drugs Highlights Remyelinating Drug Candidates for Clinical Trials. Sci. Rep. 2017, 7, 45780. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Xu, H.; Xu, L.; Jiang, W.; Chen, C.; Chang, Y.; Liu, C.; Tian, Z.; Qiu, X.; Xie, C.; et al. Remyelination in neuromyelitis optica spectrum disorder is promoted by edaravone through mTORC1 signaling activation. Glia 2023, 71, 284–304. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiari, M.; Ghasemi, N.; Salehi, H.; Amirpour, N.; Kazemi, M.; Mardani, M. Evaluation of Edaravone effects on the differentiation of human adipose derived stem cells into oligodendrocyte cells in multiple sclerosis disease in rats. Life Sci. 2021, 282, 119812. [Google Scholar] [CrossRef]
- Cho, H.; Shukla, S. Role of Edaravone as a Treatment Option for Patients with Amyotrophic Lateral Sclerosis. Pharmaceuticals 2021, 14, 29. [Google Scholar] [CrossRef]
- Ziegler, S.; Pries, V.; Hedberg, C.; Waldmann, H. Target Identification for Small Bioactive Molecules: Finding the Needle in the Haystack. Angew. Chem. Int. Ed. 2013, 52, 2744–2792. [Google Scholar] [CrossRef]
- Kumar, B.; Verma, R.K. Esterification at Room Temperature: A Mixing Affair Only. Synth. Commun. 1984, 14, 1359–1363. [Google Scholar] [CrossRef]
- Queiroz, A.N.; Martins, C.C.; Santos, K.L.B.; Carvalho, E.S.; Owiti, A.O.; Oliveira, K.R.M.; Herculano, A.M.; da Silva, A.B.F.; Borges, R.S. Experimental and Theoretical Study on Structure-Tautomerism among Edaravone, Isoxazolone, and Their Heterocycles Derivatives as Antioxidants. Saudi Pharm. J. 2020, 28, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Smith, R.; Vivoli, M.; Ema, M.; Goos, N.; Gehrke, S.; Harmer, N.J.; Wagner, G.K. Covalent inhibitors of LgtC: A blueprint for the discovery of non-substrate-like inhibitors for bacterial glycosyltransferases. Bioorg. Med. Chem. 2017, 25, 3182–3194. [Google Scholar] [CrossRef][Green Version]
- Jiang, J.; Sigua, L.H.; Chan, A.; Kalra, P.; Pomerantz, W.C.K.; Schönbrunn, E.; Qi, J.; Georg, G.I. Dihydropyridine lactam analogs targeting BET bromodomains. ChemMedChem 2022, 17, e202100407. [Google Scholar] [CrossRef]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12827. [Google Scholar] [CrossRef]
- Minnelli, C.; Laudadio, E.; Galeazzi, R.; Rusciano, D.; Armeni, T.; Stipa, P.; Cantarini, M.; Mobbili, G. Synthesis, Characterization and Antioxidant Properties of a New Lipophilic Derivative of Edaravone. Antioxidants 2019, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- QikProp. Small-Molecule Drug Discovery Suite 2021-1; Schrodinger, LLC: New York, NY, USA, 2021; Available online: https://www.schrodinger.com/products/qikprop (accessed on 8 September 2020).
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Islam, N. Investigation of comparative shielding of Morin against oxidative damage by radicals: A DFT study. Cogent Chem. 2015, 1, 1078272. [Google Scholar] [CrossRef]
- Kremer, D.; Akkermann, R.; Küry, P.; Dutta, R. Current advancements in promoting remyelination in multiple sclerosis. Mult. Scler. J. 2019, 25, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Morinaka, Y.; Iseki, K.; Watanabe, T.; Yuki, S.; Nishi, H. Structure–activity relationship of 3-methyl-1-phenyl-2-pyrazolin-5-one (edaravone). Redox Rep. 2003, 8, 151–155. [Google Scholar] [CrossRef]
- Nevitt, T.; Ohrvik, H.; Thiele, D.J. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 2012, 182, 1580–1593. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Holzer, W.; Plagens, B.; Lorenz, K. Alkylation of Pyrazolones via the Mitsunobu Reaction. Heterocycles 1997, 45, 309–314. [Google Scholar] [CrossRef]
- Haessner, R.; Hennig, L.; Gaca, J. 13C NMR Data for Chlorine- or Nitro-Substituted Azomethine Dyes. Magn. Reson. Chem. 1990, 28, 817–819. [Google Scholar] [CrossRef]
- Fan, W.; Li, W.; Ma, X.; Tao, X.; Li, X.; Yao, Y.; Xie, X.; Zhang, Z. Ru-Catalyzed Asymmetric Hydrogenation of γ-Heteroatom Substituted β-Keto Esters. J. Org. Chem. 2011, 76, 9444–9451. [Google Scholar] [CrossRef] [PubMed]
- Kotha, S.; Shirbhate, M.E. Diversity-Oriented Approach to Macrocyclic Cyclophane Derivatives via RingClosing Metathesis. Synlett 2012, 23, 2183–2188. [Google Scholar] [CrossRef]

| Compound a | LogP, n-octanol/Water Ratio | Apparent Caco-2 Permeability nm/sec | Log BB, Brain/Blood Ratio | Apparent MDCK Permeability nm/sec | Predicted CNS Access b | Lipinski Rule of 5 Violations |
|---|---|---|---|---|---|---|
| 1 to 3 good | <25 poor, >500 good | −2.7 to −1.5 good | <25 poor, >500 good | −2 to +2 b | <2 good | |
| 1 (MTT, DPPH) | 1.320 | 2693 | 0.065 | 1443 | 1 | 0 |
| 2b (DPPH) | 1.091 | 62 | −0.974 | 31 | −1 | 0 |
| 2c (MTT) | 1.151 | 791 | −0.561 | 384 | 0 | 0 |
| 2d | 0.260 | 95 | −1.815 | 38 | −2 | 0 |
| 2e | 1.634 | 91 | −2.292 | 37 | −2 | 0 |
| 2f | 3.172 | 217 | −1.261 | 376 | −2 | 0 |
| 2g | 1.063 | 761 | −0.591 | 368 | 0 | 0 |
| 2h | 0.319 | 224 | −1.174 | 98 | −2 | 0 |
| 4a (MTT) | 2.227 | 726 | −0.691 | 350 | 0 | 0 |
| 4b | 2.167 | 664 | −0.770 | 318 | −1 | 0 |
| 4c | 1.416 | 194 | −1.383 | 84 | −2 | 0 |
| 3b | 3.184 | 2568 | −0.293 | 1371 | 0 | 0 |
| 3c | 0.800 | 838 | −0.511 | 408 | 0 | 0 |
| Compound | Log D7.4 a | Affinity for a Model Lipid Bilayer b | Cu-Induced ROS Inhibition c,a | |
|---|---|---|---|---|
| 20 °C | 37 °C | |||
| 1 | 1.1 ± 0.1 | 37 ± 3 | 53 ± 2 | 14 ± 2 |
| 2b | −0.74 ± 0.19 | 38 ± 4 | 45 ± 2 | 38 ± 5 |
| 2c | 1.0 ± 0.1 | 52 ± 2 | 71 ± 2 | 53 ± 7 |
| 4a | 2.3 ± 0.4 | 88 ± 6 | 77 ± 4 | 65 ± 6 |
| Compound | HOMO (eV) | LUMO (eV) | IP (eV) | EA (eV) | |
|---|---|---|---|---|---|
| 1 | −6.13 | −1.01 | 5.12 | 6.13 | 1.01 |
| 2b | −6.34 | −1.71 | 4.63 | 6.34 | 1.71 |
| 2c | −6.31 | −1.66 | 4.65 | 6.31 | 1.66 |
| 4a | −6.45 | −1.71 | 4.74 | 6.45 | 1.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colombo, E.; Olla, S.; Minnelli, C.; Formato, A.; Veroni, C.; Corbisiero, S.; Pericolo, M.; Siguri, C.; Mobbili, G.; Agresti, C.; et al. Synthesis and Characterization of Edaravone Analogues as Remyelinating Agents and Putative Mechanistic Probes. Molecules 2023, 28, 6928. https://doi.org/10.3390/molecules28196928
Colombo E, Olla S, Minnelli C, Formato A, Veroni C, Corbisiero S, Pericolo M, Siguri C, Mobbili G, Agresti C, et al. Synthesis and Characterization of Edaravone Analogues as Remyelinating Agents and Putative Mechanistic Probes. Molecules. 2023; 28(19):6928. https://doi.org/10.3390/molecules28196928
Chicago/Turabian StyleColombo, Eleonora, Stefania Olla, Cristina Minnelli, Alessia Formato, Caterina Veroni, Silvia Corbisiero, Mattia Pericolo, Chiara Siguri, Giovanna Mobbili, Cristina Agresti, and et al. 2023. "Synthesis and Characterization of Edaravone Analogues as Remyelinating Agents and Putative Mechanistic Probes" Molecules 28, no. 19: 6928. https://doi.org/10.3390/molecules28196928
APA StyleColombo, E., Olla, S., Minnelli, C., Formato, A., Veroni, C., Corbisiero, S., Pericolo, M., Siguri, C., Mobbili, G., Agresti, C., & Seneci, P. (2023). Synthesis and Characterization of Edaravone Analogues as Remyelinating Agents and Putative Mechanistic Probes. Molecules, 28(19), 6928. https://doi.org/10.3390/molecules28196928











