The Antitumour Activity of a Curcumin and Piperine Loaded iRGD-Modified Liposome: In Vitro and In Vivo Evaluation
Abstract
:1. Introduction
2. Results and Discussion
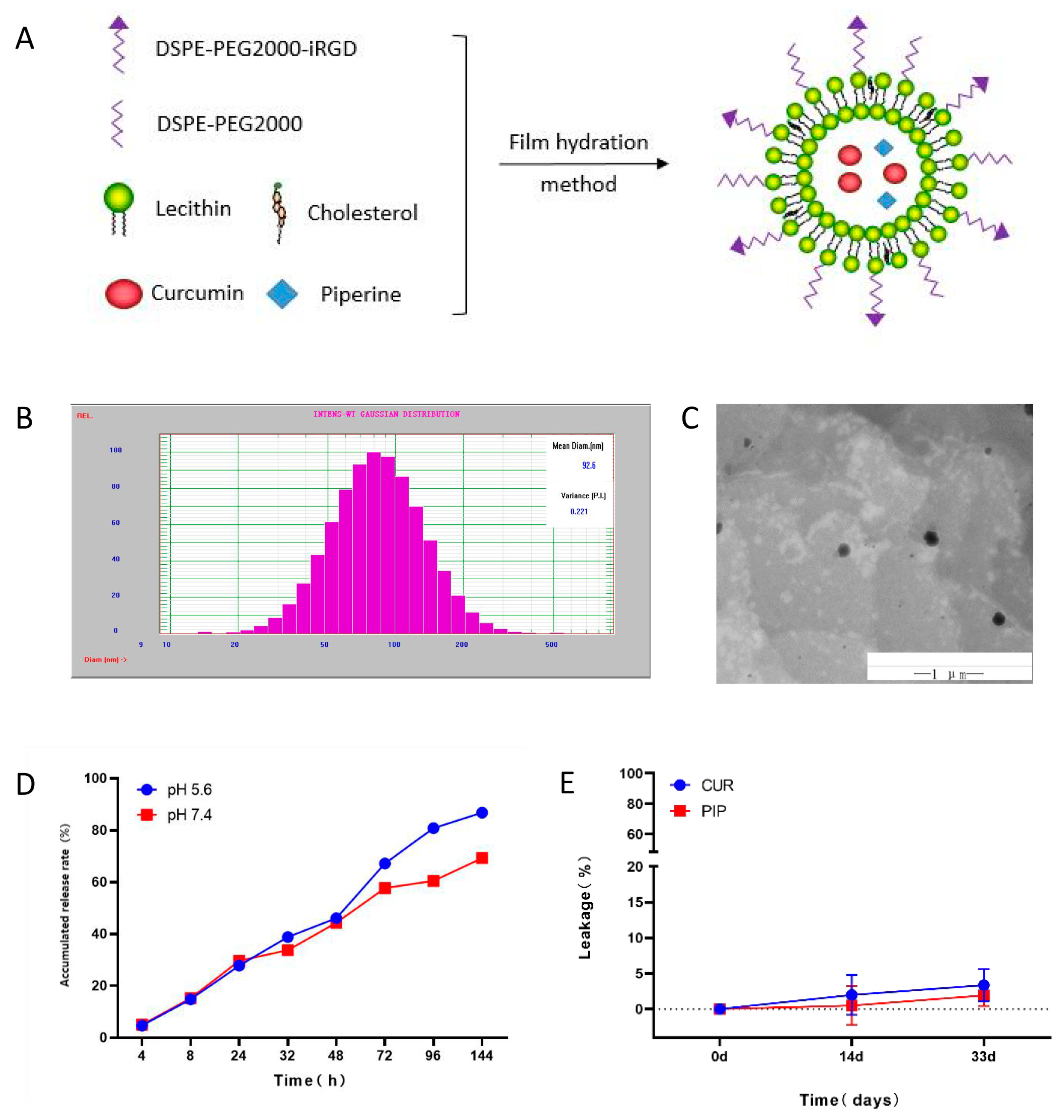
2.1. Characterisation of iRGD-LP-CUR-PIP
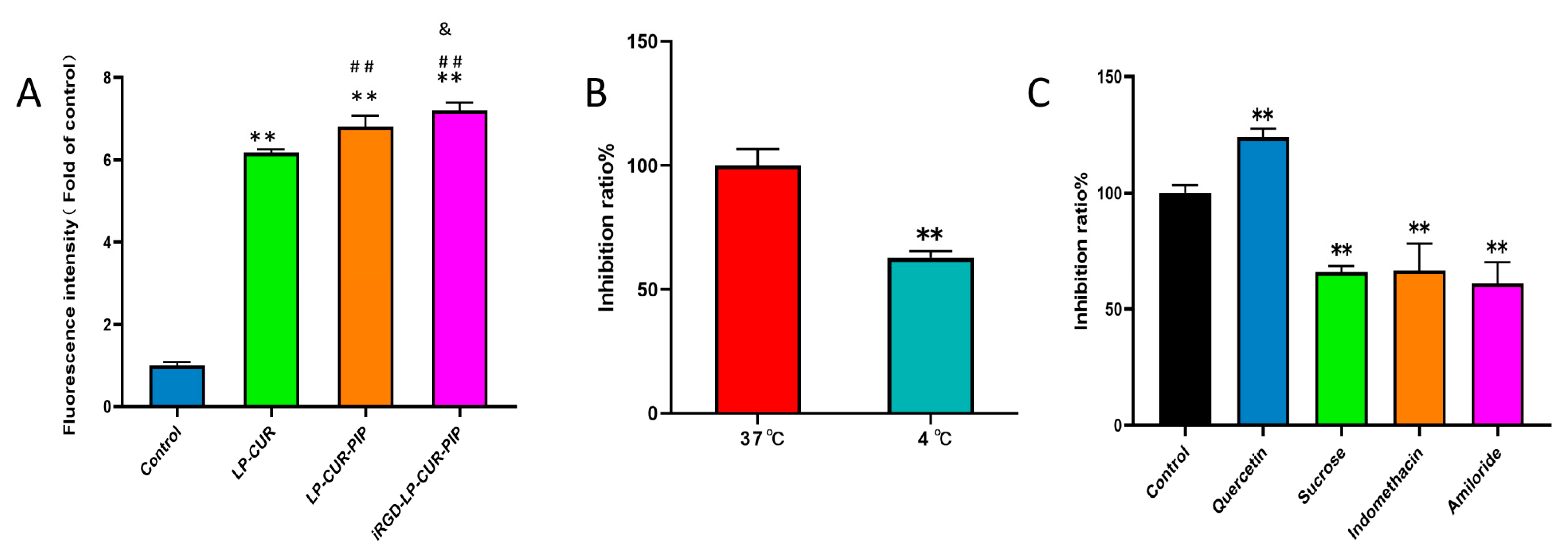
2.2. In Vitro Cellular Uptake
2.3. In Vitro Cytotoxicity
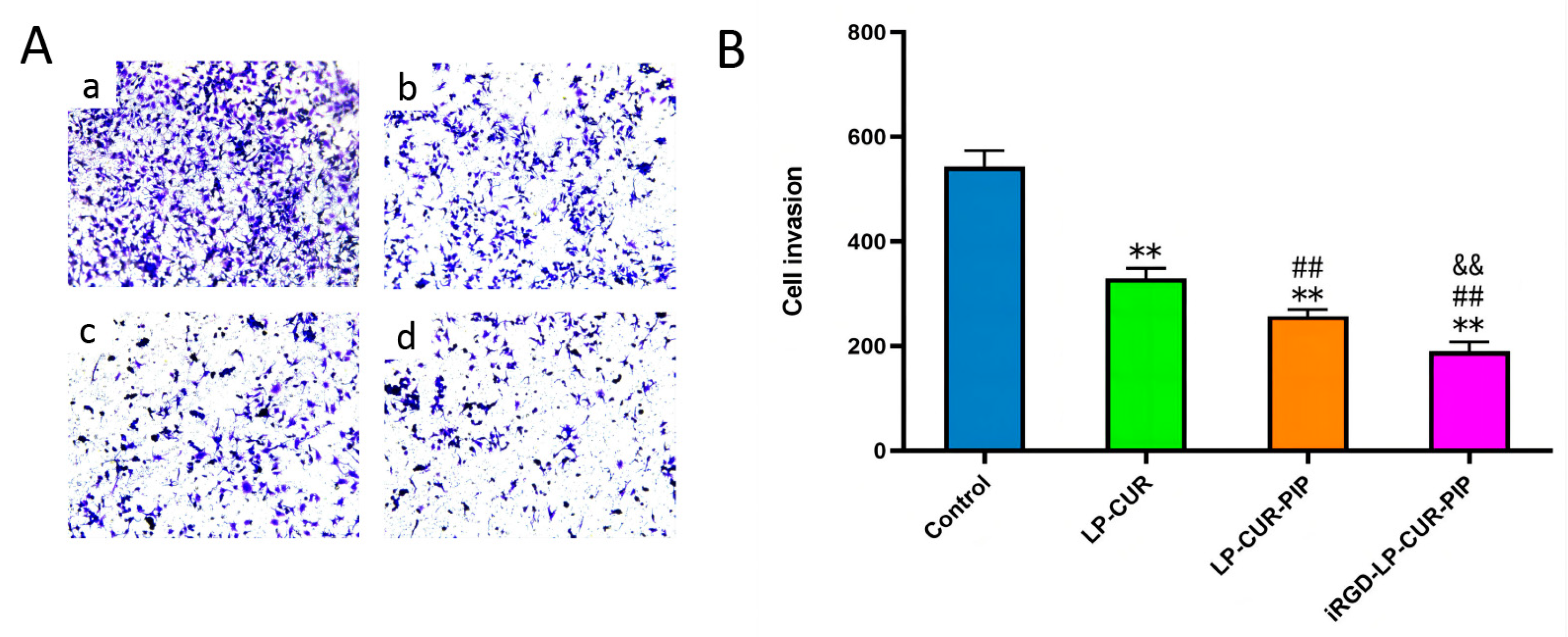
2.4. Inhibition of In Vitro Invasion
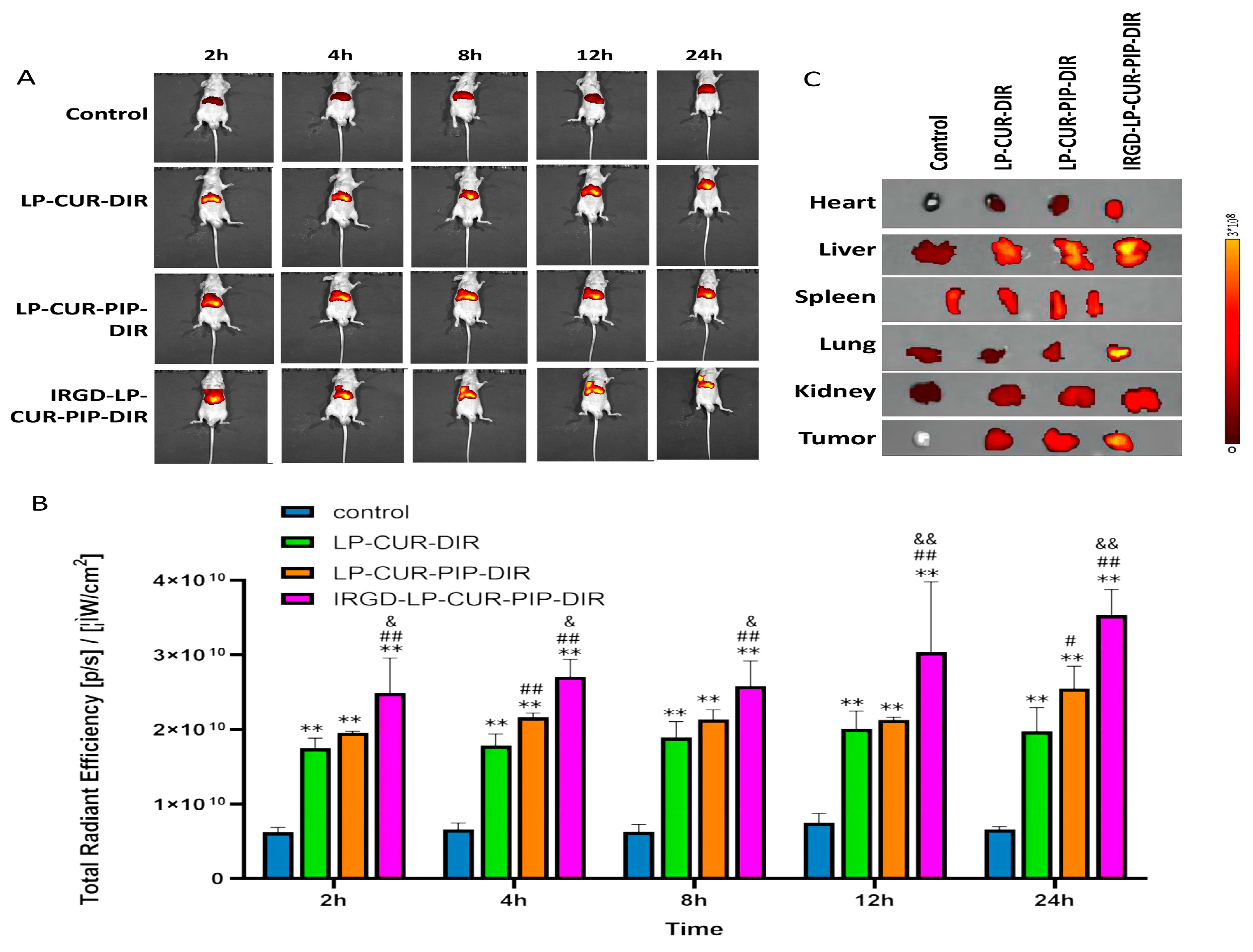
2.5. Biodistribution Characteristics of DiR-Loaded Liposomes
2.6. In Vivo Antitumour Activity of iRGD-LP-CUR-PIP
3. Materials and Methods
3.1. Materials
3.2. Method
3.2.1. Preparation of iRGD-LP-CUR-PIP
3.2.2. Characterisation of iRGD-LP-CUR-PIP
3.2.3. Stability of iRGD-LP-CUR-PIP
3.2.4. In Vitro Cellular Uptake
3.2.5. In Vitro Cell Invasion Inhibition
3.2.6. In Vitro Cytotoxicity
3.2.7. Imaging of Biodistribution In Vivo
3.2.8. In Vivo Antitumour Activity of iRGD-LP-CUR-PIP
3.2.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Arriagada, R.; Dunant, A.; Pignon, J.P.; Bergman, B.; Chabowski, M.; Grunenwald, D.; Kozlowski, M.; Le Pechoux, C.; Pirker, R.; Pinel, M.I.; et al. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant cisplatin-based chemotherapy in resected lung cancer. J. Clin. Oncol. 2010, 28, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Panda, A.K.; Chakraborty, D.; Sarkar, I.; Khan, T.; Sa, G. New insights into therapeutic activity and anticancer properties of curcumin. J. Exp. Pharmacol. 2017, 9, 31–45. [Google Scholar] [CrossRef]
- Kao, N.J.; Hu, J.Y.; Wu, C.S.; Kong, Z.L. Curcumin represses the activity of inhibitor-kappab kinase in dextran sulfate sodium-induced colitis by s-nitrosylation. Int. Immunopharmacol. 2016, 38, 1–7. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Daglia, M.; Moghaddam, A.H.; Habtemariam, S.; Nabavi, S.M. Curcumin and liver disease: From chemistry to medicine. Compr. Rev. Food Sci. Food Saf. 2014, 13, 62–77. [Google Scholar] [CrossRef]
- Hussain, H.; Ahmad, S.; Shah, S.; Ghias, M.; Ullah, A.; Rahman, S.U.; Kamal, Z.; Khan, F.A.; Khan, N.M.; Muhammad, J.; et al. Neuroprotective potential of synthetic mono-carbonyl curcumin analogs assessed by molecular docking studies. Molecules 2021, 26, 7168. [Google Scholar] [CrossRef]
- Hussain, H.; Ahmad, S.; Shah, S.; Ullah, A.; Ali, N.; Almehmadi, M.; Ahmad, M.; Khalil, A.; Jamal, S.B.; Ahmad, H.; et al. Attenuation of scopolamine-induced amnesia via cholinergic modulation in mice by synthetic curcumin analogs. Molecules 2022, 27, 2468. [Google Scholar] [CrossRef]
- Hussain, H.; Ahmad, S.; Shah, S.; Ullah, A.; Almehmadi, M.; Abdulaziz, O.; Allahyani, M.; Alsaiari, A.A.; Halawi, M.; Alamer, E. Investigation of antistress and antidepressant activities of synthetic curcumin analogues: Behavioral and biomarker approach. Biomedicines 2022, 10, 2385. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Zhou, L.; Yu, F.; Ding, H.; Li, P.; Zhou, M.; Wang, K. Potential mechanisms of action of curcumin for cancer prevention: Focus on cellular signaling pathways and mirnas. Int. J. Biol. Sci. 2019, 15, 1200–1214. [Google Scholar] [CrossRef]
- Hegde, M.; Girisa, S.; Bharathwajchetty, B.; Vishwa, R.; Kunnumakkara, A.B. Curcumin formulations for better bioavailability: What we learned from clinical trials thus far? ACS Omega 2023, 8, 10713–10746. [Google Scholar] [CrossRef] [PubMed]
- Karaboga, A.A.; Uzunhisarcikli, E.; Yerer, M.B.; Bishayee, A. The golden spice curcumin in cancer: A perspective on finalized clinical trials during the last 10 years. J. Cancer Res. Ther. 2022, 18, 19–26. [Google Scholar] [CrossRef]
- Wan, M.T.W.; Lajis, N.H.; Abas, F.; Othman, I.; Naidu, R. Mechanistic understanding of curcumin’s therapeutic effects in lung cancer. Nutrients 2019, 11, 2989. [Google Scholar] [CrossRef]
- Zhang, L.; Tao, X.; Fu, Q.; Ge, C.; Li, R.; Li, Z.; Zhu, Y.; Tian, H.; Li, Q.; Liu, M.; et al. Curcumin inhibits cell proliferation and migration in nsclc through a synergistic effect on the tlr4/myd88 and egfr pathways. Oncol. Rep. 2019, 42, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Liu, Y.; Xiao, Y.; Yang, X.; Su, W.; Zhang, M.; Liao, Y.; Kuang, H.; Wang, X. High drug payload curcumin nanosuspensions stabilized by mpeg-dspe and spc: In vitro and in vivo evaluation. Drug Deliv. 2017, 24, 109–120. [Google Scholar] [CrossRef]
- Chen, X.P.; Li, Y.; Zhang, Y.; Li, G.W. Formulation, characterization and evaluation of curcumin- loaded plga- tpgs nanoparticles for liver cancer treatment. Drug Des. Dev. Ther. 2019, 13, 3569–3578. [Google Scholar] [CrossRef]
- Hu, Y.; He, Y.; Ji, J.; Zheng, S.; Cheng, Y. Tumor targeted curcumin delivery by folate-modified mpeg-pcl self-assembly micelles for colorectal cancer therapy. Int. J. Nanomed. 2020, 15, 1239–1252. [Google Scholar] [CrossRef]
- Xie, X.; Li, Y.; Zhao, D.; Fang, C.; He, D.; Yang, Q.; Yang, L.; Chen, R.; Tan, Q.; Zhang, J. Oral administration of natural polyphenol-loaded natural polysaccharide-cloaked lipidic nanocarriers to improve efficacy against small-cell lung cancer. Nanomedicine 2020, 29, 102261. [Google Scholar] [CrossRef]
- Baspinar, Y.; Ustundas, M.; Bayraktar, O.; Sezgin, C. Curcumin and piperine loaded zein-chitosan nanoparticles: Development and in-vitro characterisation. Saudi Pharm. J. 2018, 26, 323–334. [Google Scholar] [CrossRef]
- Suresh, D.; Srinivasan, K. Tissue distribution & elimination of capsaicin, piperine & curcumin following oral intake in rats. Indian J. Med. Res. 2010, 131, 682–691. [Google Scholar] [PubMed]
- Zheng, J.; Zhou, Y.; Li, Y.; Xu, D.P.; Li, S.; Li, H.B. Spices for prevention and treatment of cancers. Nutrients 2016, 8, 495. [Google Scholar] [CrossRef]
- Patial, V.; Mahesh, S.; Sharma, S.; Pratap, K.; Singh, D.; Padwad, Y.S. Synergistic effect of curcumin and piperine in suppression of dena-induced hepatocellular carcinoma in rats. Environ. Toxicol. Pharmacol. 2015, 40, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H. Irgd: A promising peptide for cancer imaging and a potential therapeutic agent for various cancers. J. Oncol. 2019, 2019, 9367845. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Greenwald, D.R.; Ruoslahti, E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science 2010, 328, 1031–1035. [Google Scholar] [CrossRef]
- Kang, S.; Lee, S.; Park, S. Irgd peptide as a tumor-penetrating enhancer for tumor-targeted drug delivery. Polymers 2020, 12, 1906. [Google Scholar] [CrossRef]
- Patel, S.S.; Acharya, A.; Ray, R.S.; Agrawal, R.; Raghuwanshi, R.; Jain, P. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit. Rev. Food Sci. Nutr. 2020, 60, 887–939. [Google Scholar] [CrossRef]
- Ashby, G.; Keng, K.E.; Hayden, C.C.; Gollapudi, S.; Houser, J.R.; Jamal, S.; Stachowiak, J.C. Selective endocytic uptake of targeted liposomes occurs within a narrow range of liposome diameter. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, Q.; Meng, F.; Ding, N.; Yan, J.; Liu, B. Modification of erythrocytes by internalizing arg-gly-asp (irgd) in boosting the curative effect of radiotherapy for gastric carcinoma. J. Gastrointest. Oncol. 2022, 13, 2249–2258. [Google Scholar] [CrossRef]
- Erel-Akbaba, G.; Carvalho, L.A.; Tian, T.; Zinter, M.; Akbaba, H.; Obeid, P.J.; Chiocca, E.A.; Weissleder, R.; Kantarci, A.G.; Tannous, B.A. Radiation-induced targeted nanoparticle-based gene delivery for brain tumor therapy. ACS Nano 2019, 13, 4028–4040. [Google Scholar] [CrossRef]
- Le, T.D.; Nakagawa, O.; Fisher, M.; Juliano, R.L.; Yoo, H. Rgd conjugated dendritic polylysine for cellular delivery of antisense oligonucleotide. J. Nanosci. Nanotechnol. 2017, 17, 2353–2357. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Girard, O.M.; Hanahan, D.; Mattrey, R.F.; Ruoslahti, E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 2009, 16, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Yagolovich, A.V.; Isakova, A.A.; Artykov, A.A.; Vorontsova, Y.V.; Mazur, D.V.; Antipova, N.V.; Pavlyukov, M.S.; Shakhparonov, M.I.; Gileva, A.M.; Markvicheva, E.A.; et al. Dr5-selective trail variant dr5-b functionalized with tumor-penetrating irgd peptide for enhanced antitumor activity against glioblastoma. Int. J. Mol. Sci. 2022, 23, 12687. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Sun, S.Q.; Wu, W.Z.; Yang, S.L.; Tan, J.M. Characterization of a water-soluble polysaccharide from boletus edulis and its antitumor and immunomodulatory activities on renal cancer in mice. Carbohydr. Polym. 2014, 105, 127–134. [Google Scholar] [CrossRef]
- Bai, L.; Zhu, L.Y.; Yang, B.S.; Shi, L.J.; Liu, Y.; Jiang, A.M.; Zhao, L.L.; Song, G.; Liu, T.F. Antitumor and immunomodulating activity of a polysaccharide from sophora flavescens ait. Int. J. Biol. Macromol. 2012, 51, 705–709. [Google Scholar] [CrossRef]
- Xie, J.H.; Jin, M.L.; Morris, G.A.; Zha, X.Q.; Chen, H.Q.; Yi, Y.; Li, J.E.; Wang, Z.J.; Gao, J.; Nie, S.P.; et al. Advances on bioactive polysaccharides from medicinal plants. Crit. Rev. Food Sci. Nutr. 2016, 56 (Suppl. S1), S60–S84. [Google Scholar] [CrossRef]
- Zhao, Y.; Yan, B.; Wang, Z.; Li, M.; Zhao, W. Natural polysaccharides with immunomodulatory activities. Mini-Rev. Med. Chem. 2020, 20, 96–106. [Google Scholar] [CrossRef]
- Kuo, I.M.; Lee, J.J.; Wang, Y.S.; Chiang, H.C.; Huang, C.C.; Hsieh, P.J.; Han, W.; Ke, C.H.; Liao, A.; Lin, C.S. Potential enhancement of host immunity and anti-tumor efficacy of nanoscale curcumin and resveratrol in colorectal cancers by modulated electro-hyperthermia. BMC Cancer 2020, 20, 603. [Google Scholar] [CrossRef]
- Zhan, Q.Q.; Liu, Q.Y.; Yang, X.; Ge, Y.H.; Xu, L.; Ding, G.Y.; Guo, S.; Zhu, B.; Xu, W.G. Effects of silencing neuropilin-2 on proliferation, migration, and invasion of colorectal cancer ht-29. Bioengineered 2022, 13, 11042–11049. [Google Scholar] [CrossRef]
- Balakrishna, A.; Kumar, M.H. Evaluation of synergetic anticancer activity of berberine and curcumin on different models of a549, hep-g2, mcf-7, jurkat, and k562 cell lines. BioMed Res. Int. 2015, 2015, 354614. [Google Scholar] [CrossRef]


| Average Particle Size (nm) | Polydispersity | Zeta Potential (mV) | Entrapment Efficiency (%) | ||
|---|---|---|---|---|---|
| CUR | PIP | ||||
| LP-CUR mean ± SD | 86.0 ± 2.5 | 0.341 ± 0.009 | −0.4 ± 0.1 | 94.26% ± 1.82 | - |
| LP-CUR-PIP mean ± SD | 95.5 ± 1.8 | 0.337 ± 0.021 | −0.3 ± 0.1 | 90.30% ± 0.62 | 92.46% ± 0.76 |
| iRGD-LP-CUR-PIP mean ± SD | 93.9 ± 0.9 | 0.200 ± 0.015 | 0.3 ± 0.1 | 95.23% ± 1.08 | 97.04% ± 1.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Huang, X.; Chen, H.; Wu, Q.; Zhao, Q.; Fu, D.; Liu, Q.; Wang, Y. The Antitumour Activity of a Curcumin and Piperine Loaded iRGD-Modified Liposome: In Vitro and In Vivo Evaluation. Molecules 2023, 28, 6532. https://doi.org/10.3390/molecules28186532
Wang Y, Huang X, Chen H, Wu Q, Zhao Q, Fu D, Liu Q, Wang Y. The Antitumour Activity of a Curcumin and Piperine Loaded iRGD-Modified Liposome: In Vitro and In Vivo Evaluation. Molecules. 2023; 28(18):6532. https://doi.org/10.3390/molecules28186532
Chicago/Turabian StyleWang, Yingzheng, Xunhua Huang, Hanzhi Chen, Qianyuan Wu, Qianqian Zhao, Dezhuang Fu, Qinghua Liu, and Yinghao Wang. 2023. "The Antitumour Activity of a Curcumin and Piperine Loaded iRGD-Modified Liposome: In Vitro and In Vivo Evaluation" Molecules 28, no. 18: 6532. https://doi.org/10.3390/molecules28186532
APA StyleWang, Y., Huang, X., Chen, H., Wu, Q., Zhao, Q., Fu, D., Liu, Q., & Wang, Y. (2023). The Antitumour Activity of a Curcumin and Piperine Loaded iRGD-Modified Liposome: In Vitro and In Vivo Evaluation. Molecules, 28(18), 6532. https://doi.org/10.3390/molecules28186532





