Nucleoside Analog Reverse-Transcriptase Inhibitors in Membrane Environment: Molecular Dynamics Simulations
Abstract
1. Introduction
2. Results and Discussion
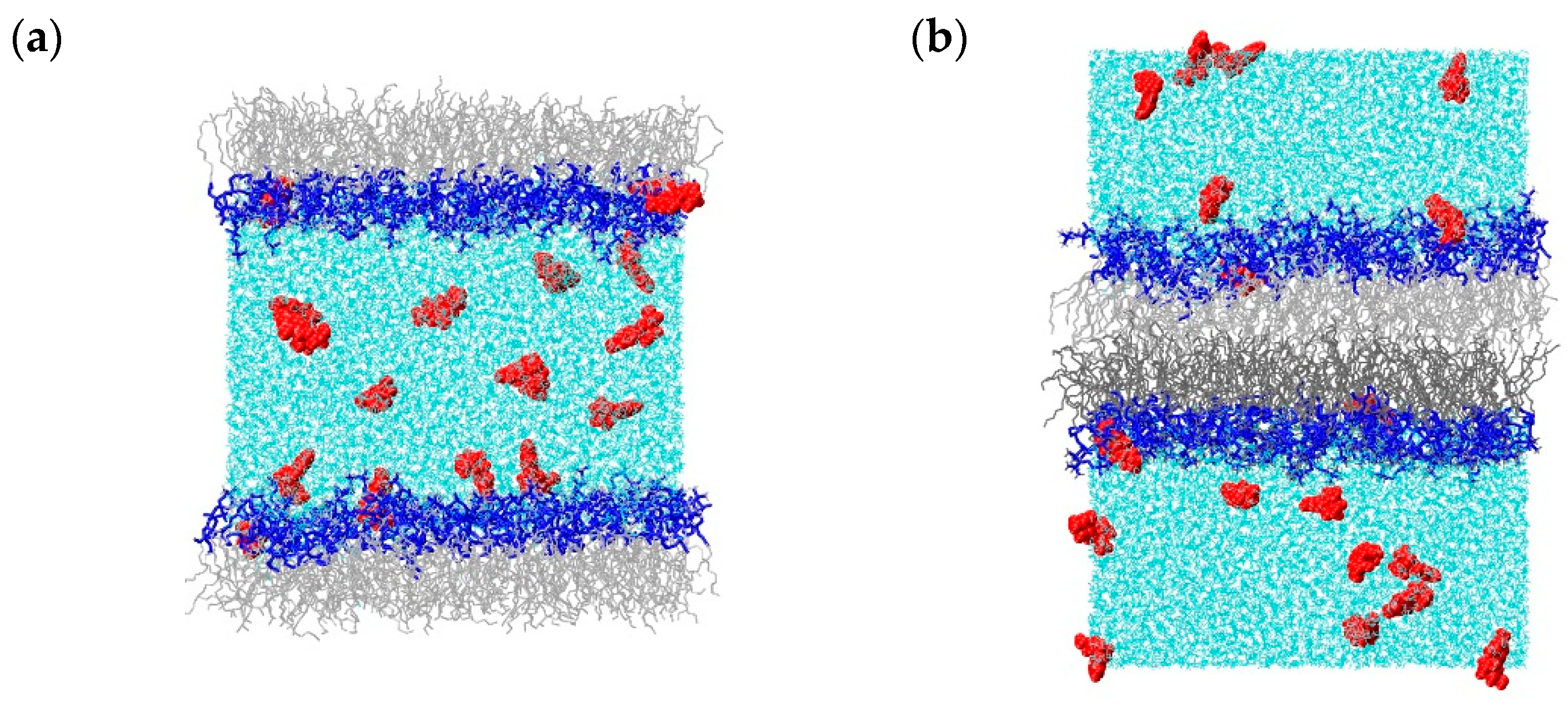
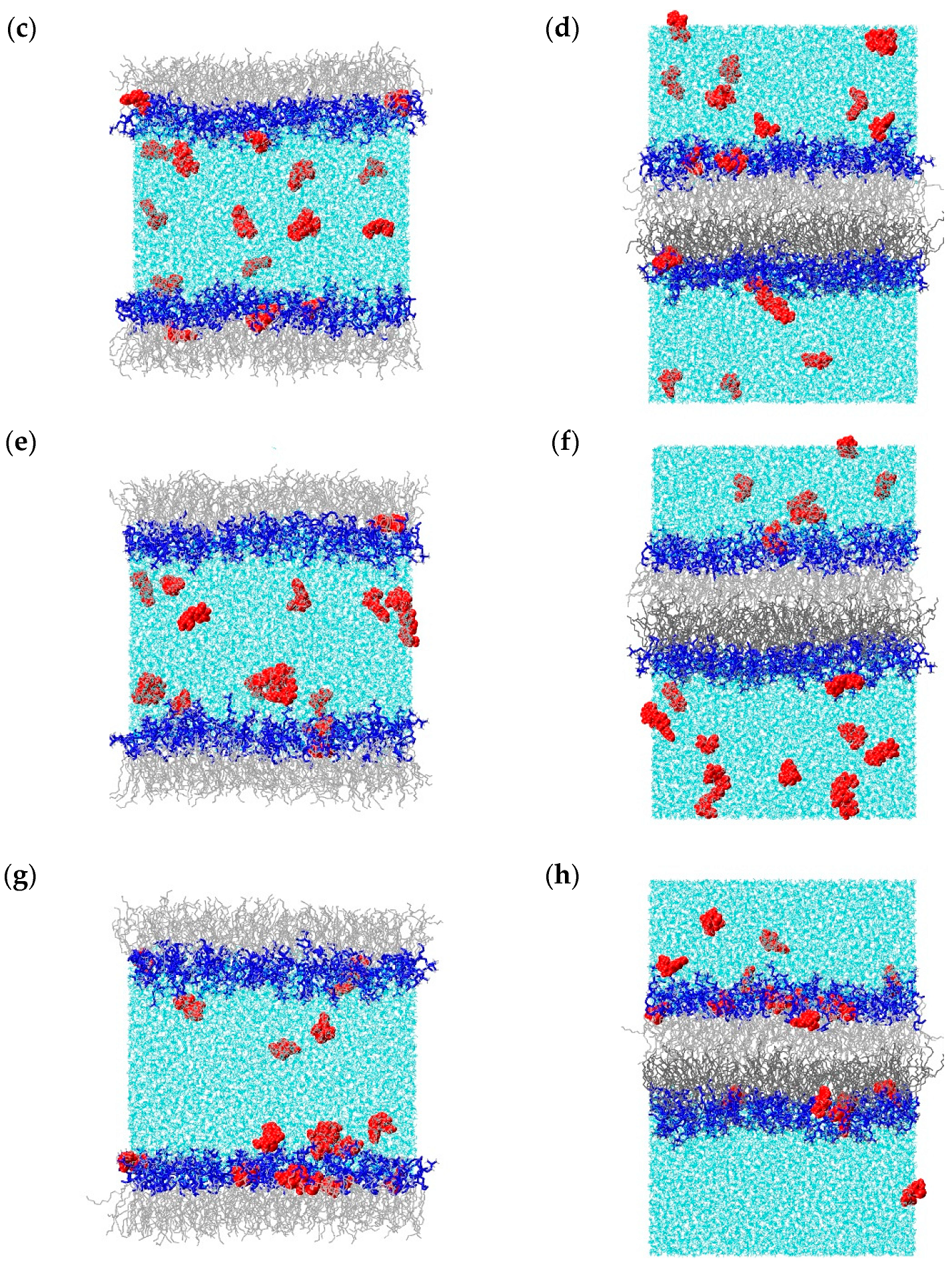
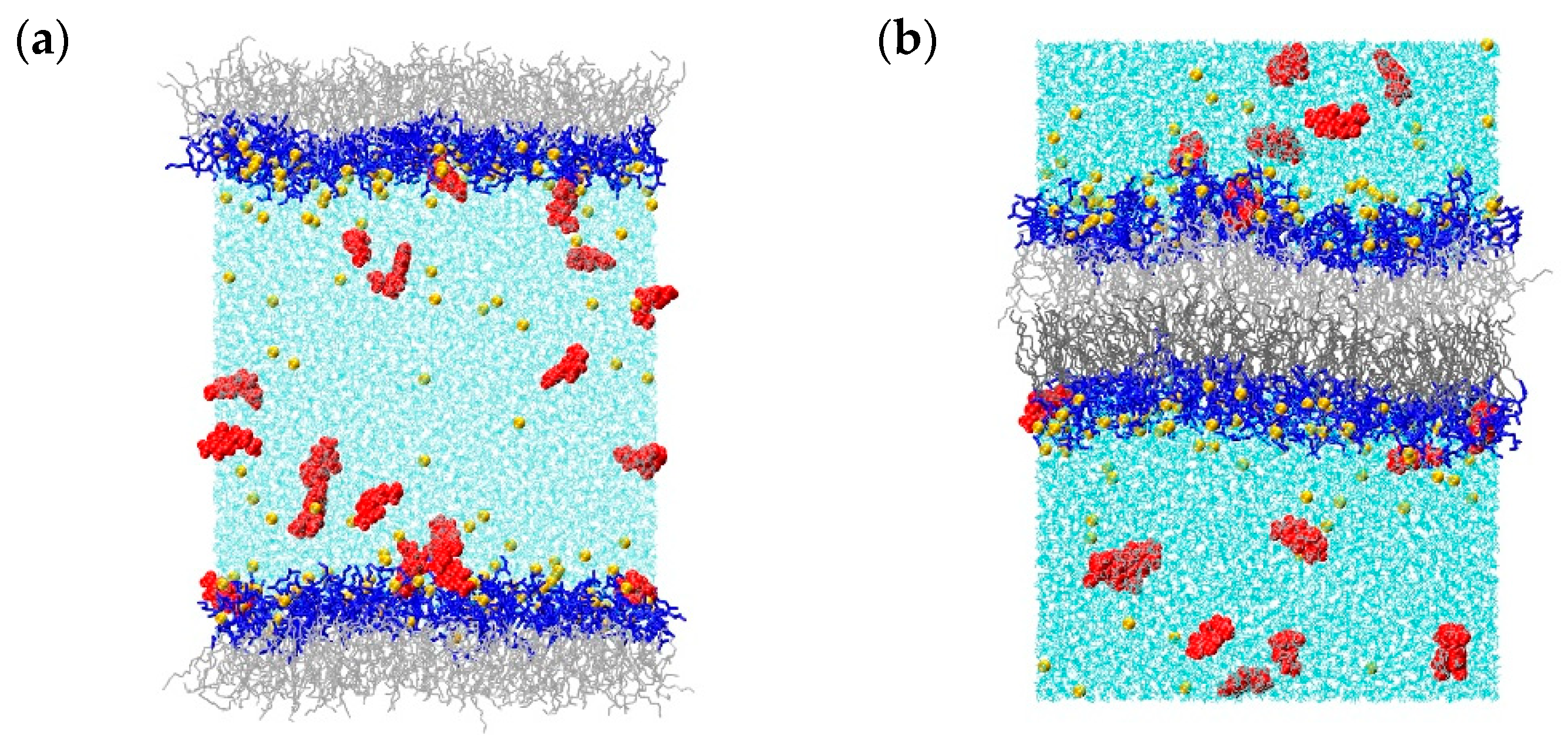
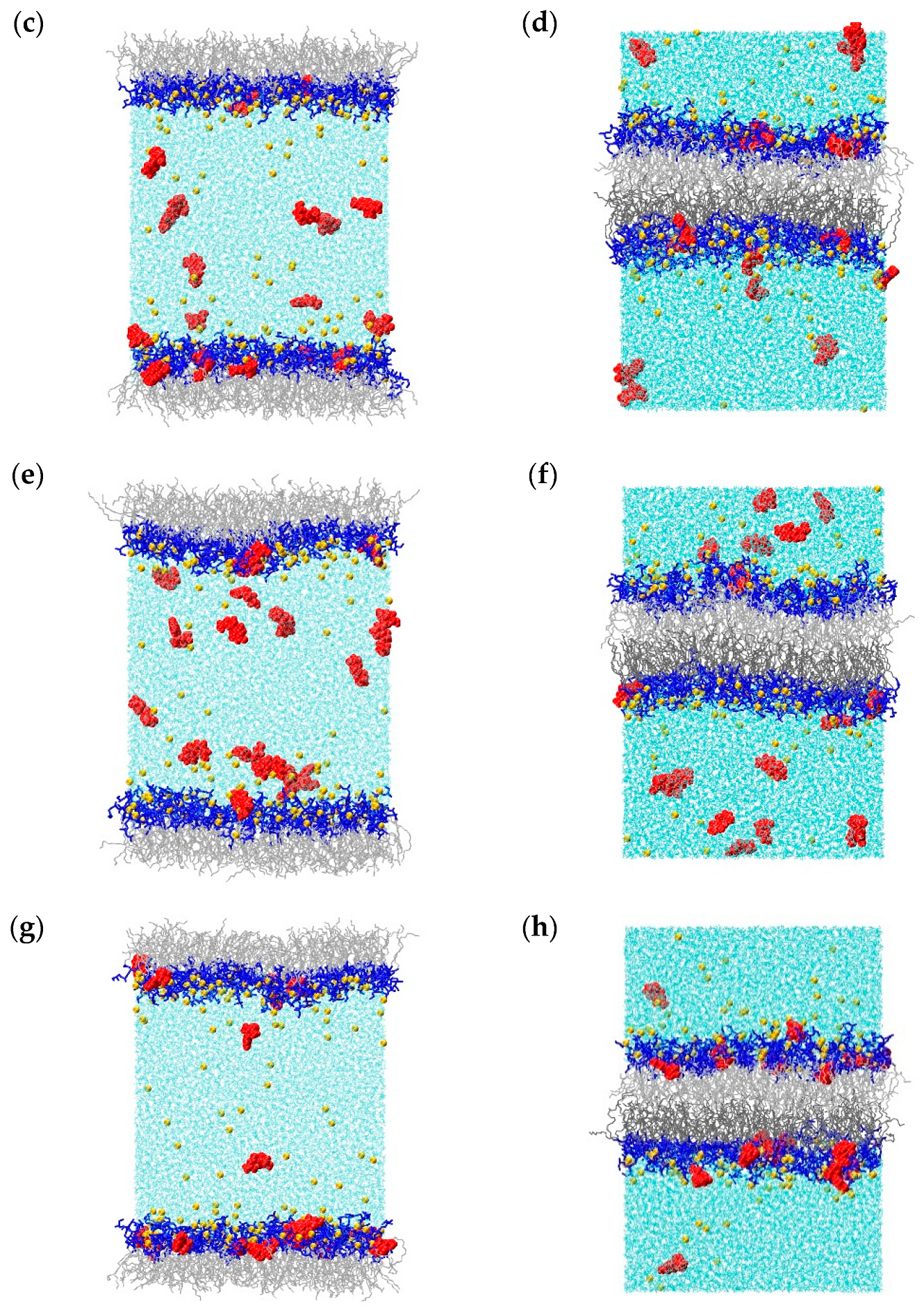
2.1. Molecular Dynamics Simulations
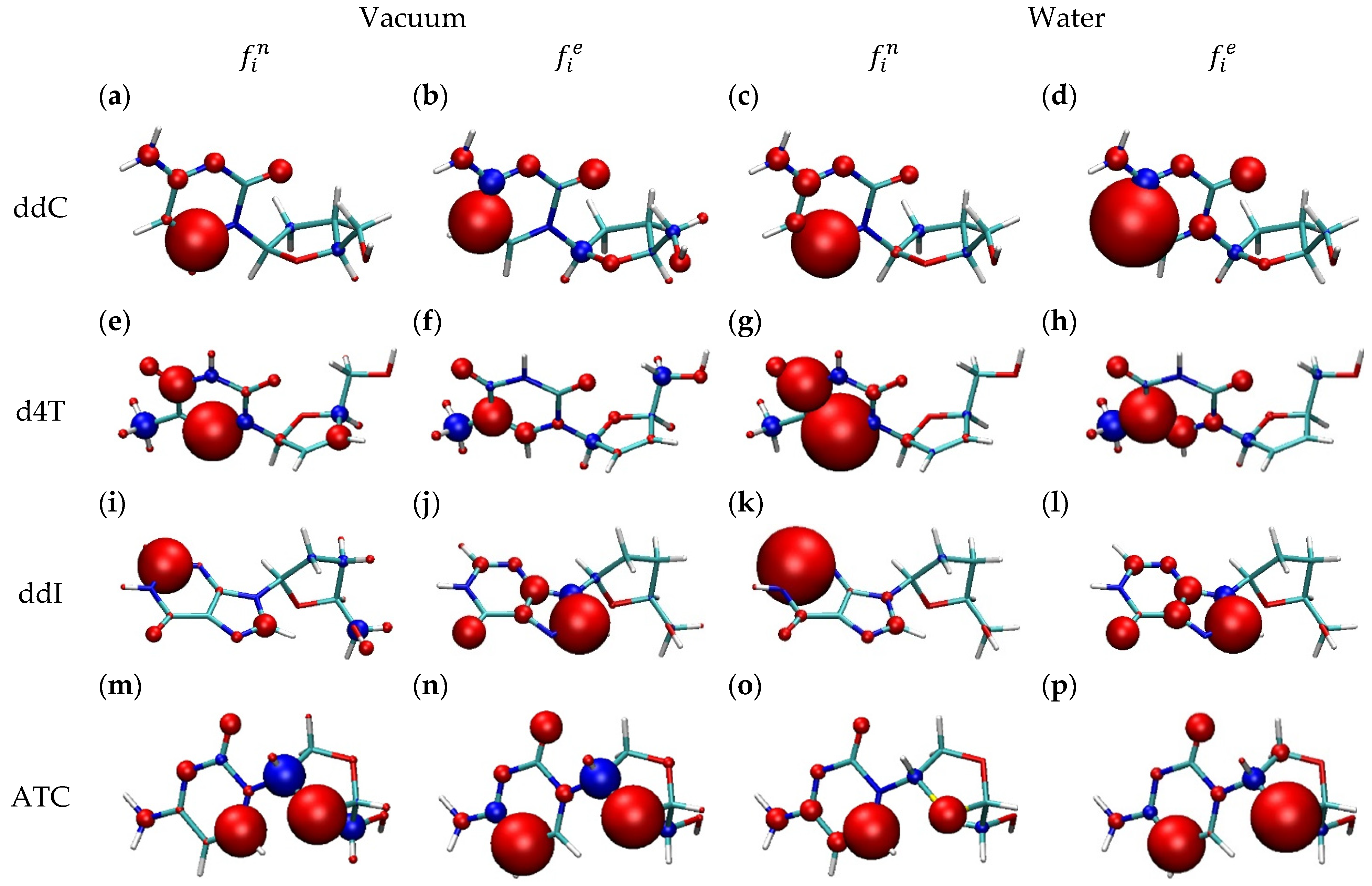
2.2. Drug Donor/Acceptor Properties
2.3. NRTI–Lipid Interaction Energies
3. Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cihlar, T.; Ray, A.S. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antivir. Res. 2010, 85, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, C.; Pannecouque, C.; De Clercq, E.; Chen, F. Development of non-nucleoside reverse transcriptase inhibitors (NNRTIs): Our past twenty years. Acta Pharm. Sin. B 2020, 10, 961–978. [Google Scholar] [CrossRef]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV/AIDS-Res. Palliat. Care 2015, 7, 95–104. [Google Scholar] [CrossRef]
- Shafer, R.; Vuitton, D. Highly active antiretroviral therapy (Haart) for the treatment of infection with human immunodeficiency virus type 1. Biomed. Pharmacother. 1999, 53, 73–86. [Google Scholar] [CrossRef]
- Warnke, D.; Barreto, J.; Temesgen, Z. Therapeutic review: Antiretroviral drugs. J. Clin. Pharmacol. 2007, 47, 1570–1579. [Google Scholar] [CrossRef]
- Samji, H.; Cescon, A.; Hogg, R.S.; Modur, S.P.; Althoff, K.N.; Buchacz, K.; Burchell, A.N.; Cohen, M.; Gebo, K.A.; Gill, M.J.; et al. Closing the Gap: Increases in Life Expectancy among Treated HIV-Positive Individuals in the United States and Canada. PLoS ONE 2013, 8, e81355. [Google Scholar] [CrossRef] [PubMed]
- Lohse, N.; Obel, N. Update of Survival for Persons with HIV Infection in Denmark. Ann. Intern. Med. 2016, 165, 749–750. [Google Scholar] [CrossRef]
- Grant, R.M.; Lama, J.R.; Anderson, P.L.; McMahan, V.; Liu, A.Y.; Vargas, L.; Goicochea, P.; Casapía, M.; Guanira-Carranza, J.V.; Ramirez-Cardich, M.E.; et al. Preexposure Chemoprophylaxis for HIV Prevention in Men Who Have Sex with Men. N. Engl. J. Med. 2010, 363, 2587–2599. [Google Scholar] [CrossRef]
- Fonner, V.A.; Dalglish, S.L.; Kennedy, C.E.; Baggaley, R.; O’reilly, K.R.; Koechlin, F.M.; Rodolph, M.; Hodges-Mameletzis, I.; Grant, R.M. Effectiveness and safety of oral HIV preexposure prophylaxis for all populations. AIDS 2016, 30, 1973–1983. [Google Scholar] [CrossRef] [PubMed]
- Townsend, C.L.; Cortina-Borja, M.; Peckham, C.S.; de Ruiter, A.; Lyall, H.; Tookey, P.A. Low rates of mother-to-child transmission of HIV following effective pregnancy interventions in the United Kingdom and Ireland, 2000–2006. AIDS 2008, 22, 973–981. [Google Scholar] [CrossRef]
- Hurwitz, S.J.; Schinazi, R.F. Practical considerations for developing nucleoside reverse transcriptase inhibitors. Drug Discov. Today Technol. 2012, 9, e183–e193. [Google Scholar] [CrossRef]
- Gubernick, S.I.; Félix, N.; Lee, D.; Xu, J.J.; Hamad, B. The HIV therapy market. Nat. Rev. Drug Discov. 2016, 15, 451–452. [Google Scholar] [CrossRef]
- Bertoletti, N.; Chan, A.H.; Schinazi, R.F.; Anderson, K.S. Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance. Molecules 2020, 25, 4868. [Google Scholar] [CrossRef] [PubMed]
- Vaccaro, J.A.; Parnell, K.M.; Terezakis, S.A.; Anderson, K.S. Mechanism of Inhibition of the Human Immunodeficiency Virus Type 1 Reverse Transcriptase by d4TTP: An Equivalent Incorporation Efficiency Relative to the Natural Substrate dTTP. Antimicrob. Agents Chemother. 2000, 44, 217–221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- de Béthune, M.P. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: A review of the last 20 years (1989–2009). Antivir. Res. 2010, 85, 75–90. [Google Scholar] [CrossRef]
- Spence, R.A.; Kati, W.M.; Anderson, K.S.; Johnson, K.A. Mechanism of Inhibition of HIV-1 Reverse Transcriptase by Nonnucleoside Inhibitors. Science 1995, 267, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.A. Ibalizumab: A Review in Multidrug-Resistant HIV-1 Infection. Drugs 2020, 80, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.R.; Savjani, J.K. Recent updates for designing CCR5 antagonists as anti-retroviral agents. Eur. J. Med. Chem. 2018, 147, 115–129. [Google Scholar] [CrossRef]
- Bangsberg, D.R.; Acosta, E.P.; Gupta, R.; Guzman, D.; Riley, E.D.; Harrigan, P.R.; Parkin, N.; Deeks, S.G. Adherence–resistance relationships for protease and non-nucleoside reverse transcriptase inhibitors explained by virological fitness. AIDS 2006, 20, 223–231. [Google Scholar] [CrossRef]
- Whyte-Allman, S.-K.; Bendayan, R. HIV-1 Sanctuary Sites—The Role of Membrane-Associated Drug Transporters and Drug Metabolic Enzymes. AAPS J. 2020, 22, 118. [Google Scholar] [CrossRef]
- Kulpa, D.A.; Chomont, N. HIV persistence in the setting of antiretroviral therapy: When, where and how does HIV hide? J. Virus Erad. 2015, 1, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Dahl, V.; Josefsson, L.; Palmer, S. HIV reservoirs, latency, and reactivation: Prospects for eradication. Antivir. Res. 2010, 85, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Redondo, R.; Fryer, H.R.; Bedford, T.; Kim, E.Y.; Archer, J.; Kosakovsky Pond, S.L.K.; Chung, Y.S.; Penugonda, S.; Chipman, J.G.; Fletcher, C.V.; et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016, 530, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Estes, J.D.; Kityo, C.; Ssali, F.; Swainson, L.; Makamdop, K.N.; Del Prete, G.Q.; Deeks, S.G.; Luciw, P.A.; Chipman, J.G.; Beilman, G.J.; et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 2017, 23, 1271–1276. [Google Scholar] [CrossRef]
- Fletcher, C.V.; Staskus, K.; Wietgrefe, S.W.; Rothenberger, M.; Reilly, C.; Chipman, J.G.; Beilman, G.J.; Khoruts, A.; Thorkelson, A.; Schmidt, T.E.; et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. USA 2014, 111, 2307–2312. [Google Scholar] [CrossRef]
- Smit, T.K.; Brew, B.J.; Tourtellotte, W.; Morgello, S.; Gelman, B.B.; Saksena, N.K. Independent Evolution of Human Immunodeficiency Virus (HIV) Drug Resistance Mutations in Diverse Areas of the Brain in HIV-Infected Patients, with and without Dementia, on Antiretroviral Treatment. J. Virol. 2004, 78, 10133–10148. [Google Scholar] [CrossRef]
- Asahchop, E.L.; Meziane, O.; Mamik, M.K.; Chan, W.F.; Branton, W.G.; Resch, L.; Gill, M.J.; Haddad, E.; Guimond, J.V.; Wainberg, M.A.; et al. Reduced antiretroviral drug efficacy and concentration in HIV-infected microglia contributes to viral persistence in brain. Retrovirology 2017, 14, 47. [Google Scholar] [CrossRef]
- Taylor, S.; Back, D.J.; Workman, J.; Drake, S.M.; White, D.J.; Choudhury, B.; Cane, P.A.; Beards, G.M.; Halifax, K.; Pillay, D. Poor penetration of the male genital tract by HIV-1 protease inhibitors. AIDS 1999, 13, 859–860. [Google Scholar] [CrossRef]
- Eyre, R.C.; Zheng, G.; Kiessling, A.A. Multiple drug resistance mutations in human immunodeficiency virus in semen but not blood of a man on antiretroviral therapy. Urology 2000, 55, 591. [Google Scholar] [CrossRef]
- Glynn, S.L.; Yazdanian, M. In Vitro Blood−Brain Barrier Permeability of Nevirapine Compared to Other HIV Antiretroviral Agents. J. Pharm. Sci. 1998, 87, 306–310. [Google Scholar] [CrossRef]
- Plagemann, P.G.; Wohlhueter, R.M.; Woffendin, C. Nucleoside and nucleobase transport in animal cells. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 1988, 947, 405–443. [Google Scholar] [CrossRef]
- Nightingale, S.L. Zalcitabine Approved for Use in Combination with Zidovudine for HIV Infection. JAMA 1992, 268, 705. [Google Scholar] [CrossRef]
- Whittington, R.; Brogden, R.N. Zalcitabine. Drugs 1992, 44, 656–683. [Google Scholar] [CrossRef]
- Mansuri, M.M.; Starrett, J.E.; Ghazzouli, I.; Hitchcock, M.J.M.; Sterzycki, R.Z.; Brankovan, V.; Lin, T.S.; August, E.M.; Prusoff, W.H. 1-(2,3-Dideoxy-.beta.-D-glycero-pent-2-enofuranosyl)thymine. A highly potent and selective anti-HIV agent. J. Med. Chem. 1989, 32, 461–466. [Google Scholar] [CrossRef]
- Mitsuya, H.; Broder, S. Inhibition of the in vitro infectivity and cytopathic effect of human T-lymphotrophic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV) by 2′,3′-dideoxynucleosides (acquired immunodeficiency syndrome). Proc. Natl. Acad. Sci. USA 1986, 83, 1911–1915. [Google Scholar] [CrossRef]
- Gaffney, M.M.; Belliveau, P.P.; Spooner, L.M. Apricitabine: A Nucleoside Reverse Transcriptase Inhibitor for HIV Infection. Ann. Pharmacother. 2009, 43, 1676–1683. [Google Scholar] [CrossRef]
- Martin, J.C.; Hitchcock, M.J.; De Clercq, E.; Prusoff, W.H. Early nucleoside reverse transcriptase inhibitors for the treatment of HIV: A brief history of stavudine (D4T) and its comparison with other dideoxynucleosides. Antivir. Res. 2010, 85, 34–38. [Google Scholar] [CrossRef]
- Maagaard, A.; Kvale, D. Long term adverse effects related to nucleoside reverse transcriptase inhibitors: Clinical impact of mitochondrial toxicity. Scand. J. Infect. Dis. 2009, 41, 808–817. [Google Scholar] [CrossRef]
- Gu, Z.; Allard, B.; de Muys, J.M.; Lippens, J.; Rando, R.F.; Nguyen-Ba, N.; Ren, C.; McKenna, P.; Taylor, D.L.; Bethell, R.C. In Vitro Antiretroviral Activity and In Vitro Toxicity Profile of SPD754, a New Deoxycytidine Nucleoside Reverse Transcriptase Inhibitor for Treatment of Human Immunodeficiency Virus Infection. Antimicrob. Agents Chemother. 2006, 50, 625–631. [Google Scholar] [CrossRef][Green Version]
- Cahn, P.; Wainberg, M.A. Resistance profile of the new nucleoside reverse transcriptase inhibitor apricitabine. J. Antimicrob. Chemother. 2010, 65, 213–217. [Google Scholar] [CrossRef]
- Kwiecińska, K.; Stachowicz-Kuśnierz, A.; Korchowiec, B.; Roman, M.; Kwiatek, W.M.; Jagusiak, A.; Roterman, I.; Korchowiec, J. Congo Red as a Supramolecular Carrier System for Doxorubicin: An Approach to Understanding the Mechanism of Action. Int. J. Mol. Sci. 2022, 23, 8935. [Google Scholar] [CrossRef]
- Korchowiec, B.; Korchowiec, J.; Kwiecińska, K.; Gevrenova, R.; Bouguet-Bonnet, S.; Deng, C.; Henry, M.; Rogalska, E. The Molecular Bases of the Interaction between a Saponin from the Roots of Gypsophila paniculata L. and Model Lipid Membranes. Int. J. Mol. Sci. 2022, 23, 3397. [Google Scholar] [CrossRef]
- Singh, V.K.; Srivastava, R.; Gupta, P.S.S.; Naaz, F.; Chaurasia, H.; Mishra, R.; Rana, M.K.; Singh, R.K. Anti-HIV potential of diarylpyrimidine derivatives as non-nucleoside reverse transcriptase inhibitors: Design, synthesis, docking, TOPKAT analysis and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2021, 39, 2430–2446. [Google Scholar] [CrossRef] [PubMed]
- Frey, K.M.; Bertoletti, N.; Chan, A.H.; Ippolito, J.A.; Bollini, M.; Spasov, K.A.; Jorgensen, W.L.; Anderson, K.S. Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions with HIV-1 Reverse Transcriptase. Front. Mol. Biosci. 2022, 9, 805187. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Tian, Y.; Wang, W.; Gu, S.; Ju, X.; Liu, G. In silico studies of diarylpyridine derivatives as novel HIV-1 NNRTIs using docking-based 3D-QSAR, molecular dynamics, and pharmacophore modeling approaches. RSC Adv. 2018, 8, 40529–40543. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.W.; Hall, B.A.; Kellam, P.; Coveney, P.V. Global Conformational Dynamics of HIV-1 Reverse Transcriptase Bound to Non-Nucleoside Inhibitors. Biology 2012, 1, 222–244. [Google Scholar] [CrossRef]
- Vijayan, R.S.K.; Arnold, E.; Das, K. Molecular dynamics study of HIV-1 RT-DNA-nevirapine complexes explains NNRTI inhibition and resistance by connection mutations. Proteins Struct. Funct. Bioinform. 2014, 82, 815–829. [Google Scholar] [CrossRef]
- Leach, A.R. Molecular Modeling: Principles and Applications; Pearson: London, UK, 2001. [Google Scholar]
- Arunan, E.; Desiraju, G.R.; Klein, R.A.; Sadlej, J.; Scheiner, S.; Alkorta, I.; Clary, D.C.; Crabtree, R.H.; Dannenberg, J.J.; Hobza, P.; et al. Definition of the hydrogen bond (IUPAC Recommendations 2011). Pure Appl. Chem. 2011, 83, 1637–1641. [Google Scholar] [CrossRef]
- Wiȩcław, K.; Korchowiec, B.; Corvis, Y.; Korchowiec, J.; Guermouche, H.; Rogalska, E. Meloxicam and Meloxicam-β-Cyclodextrin Complex in Model Membranes: Effects on the Properties and Enzymatic Lipolysis of Phospholipid Monolayers in Relation to Anti-inflammatory Activity. Langmuir 2009, 25, 1417–1426. [Google Scholar] [CrossRef]
- Kwiecińska, K.; Stachowicz-Kuśnierz, A.; Jagusiak, A.; Roterman, I.; Korchowiec, J. Impact of Doxorubicin on Self-Organization of Congo Red: Quantum Chemical Calculations and Molecular Dynamics Simulations. ACS Omega 2020, 5, 19377–19384. [Google Scholar] [CrossRef]
- Geerlings, P.; Chamorro, E.; Chattaraj, P.K.; De Proft, F.; Gázquez, J.L.; Liu, S.; Morell, C.; Toro-Labbé, A.; Vela, A.; Ayers, P. Conceptual density functional theory: Status, prospects, issues. Theor. Chem. Acc. 2020, 139, 36. [Google Scholar] [CrossRef]
- Geerlings, P.; De Proft, F. Conceptual DFT: The chemical relevance of higher response functions. Phys. Chem. Chem. Phys. 2008, 10, 3028–3042. [Google Scholar] [CrossRef] [PubMed]
- Stachowicz-Kuśnierz, A.; Korchowiec, J. Nucleophilic properties of purine bases: Inherent reactivity versus reaction conditions. Struct. Chem. 2016, 27, 543–555. [Google Scholar] [CrossRef]
- Stachowicz-Kuśnierz, A.; Korchowiec, B.; Rogalska, E.; Korchowiec, J. The lung surfactant activity probed with molecular dynamics simulations. Adv. Colloid Interface Sci. 2022, 304, 102659. [Google Scholar] [CrossRef]
- Lee, J.; Patel, D.S.; Ståhle, J.; Park, S.-J.; Kern, N.R.; Kim, S.H.; Lee, J.; Cheng, X.; Valvano, M.A.; Holst, O.; et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J. Chem. Theory Comput. 2019, 15, 775–786. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Beglov, D.; Roux, B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. J. Chem. Phys. 1994, 100, 9050–9063. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Allinger, N.L. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 1977, 99, 8127–8134. [Google Scholar] [CrossRef]
- Schnur, D.M.; Grieshaber, M.V.; Bowen, J.P. Development of an internal searching algorithm for parameterization of the MM2/MM3 force fields. J. Comput. Chem. 1991, 12, 844–849. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2009, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Mayne, C.G.; Saam, J.; Schulten, K.; Tajkhorshid, E.; Gumbart, J.C. Rapid parameterization of small molecules using the force field toolkit. J. Comput. Chem. 2013, 34, 2757–2770. [Google Scholar] [CrossRef]
- Kučerka, N.; Tristram-Nagle, S.; Nagle, J.F. Structure of Fully Hydrated Fluid Phase Lipid Bilayers with Monounsaturated Chains. J. Membr. Biol. 2006, 208, 193–202. [Google Scholar] [CrossRef]
- Smaby, J.; Momsen, M.; Brockman, H.; Brown, R. Phosphatidylcholine acyl unsaturation modulates the decrease in interfacial elasticity induced by cholesterol. Biophys. J. 1997, 73, 1492–1505. [Google Scholar] [CrossRef]
- Hyslop, P.A.; Morel, B.; Sauerheber, R.D. Organization and interaction of cholesterol and phosphatidylcholine in model bilayer membranes. Biochemistry 1990, 29, 1025–1038. [Google Scholar] [CrossRef]
- Kučerka, N.; Holland, B.W.; Gray, C.G.; Tomberli, B.; Katsaras, J. Scattering Density Profile Model of POPG Bilayers as Determined by Molecular Dynamics Simulations and Small-Angle Neutron and X-ray Scattering Experiments. J. Phys. Chem. B 2012, 116, 232–239. [Google Scholar] [CrossRef]
- Kwon, B.; Waring, A.J.; Hong, M. A 2H Solid-State NMR Study of Lipid Clustering by Cationic Antimicrobial and Cell-Penetrating Peptides in Model Bacterial Membranes. Biophys. J. 2013, 105, 2333–2342. [Google Scholar] [CrossRef]
- Shahane, G.; Ding, W.; Palaiokostas, M.; Orsi, M. Physical properties of model biological lipid bilayers: Insights from all-atom molecular dynamics simulations. J. Mol. Model. 2019, 25, 76. [Google Scholar] [CrossRef]
- Janosi, L.; Gorfe, A.A. Simulating POPC and POPC/POPG Bilayers: Conserved Packing and Altered Surface Reactivity. J. Chem. Theory Comput. 2010, 6, 3267–3273. [Google Scholar] [CrossRef]
- Hong, C.; Tieleman, D.P.; Wang, Y. Microsecond Molecular Dynamics Simulations of Lipid Mixing. Langmuir 2014, 30, 11993–12001. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Du, Y.; Cang, X.; Wang, J.; Chen, Z.; Yang, H.; Jiang, H. Binding Competition to the POPG Lipid Bilayer of Ca2+, Mg2+, Na+, and K+ in Different Ion Mixtures and Biological Implication. J. Phys. Chem. B 2013, 117, 850–858. [Google Scholar] [CrossRef]
- Kang, H.; Klauda, J.B. Molecular dynamics simulations of palmitoyloleoylphosphatidylglycerol bilayers. Mol. Simul. 2015, 41, 948–954. [Google Scholar] [CrossRef]
- Yu, Y.; Krämer, A.; Venable, R.M.; Simmonett, A.C.; MacKerell, A.D., Jr.; Klauda, J.B.; Pastor, R.W.; Brooks, B.R. Semi-automated Optimization of the CHARMM36 Lipid Force Field to Include Explicit Treatment of Long-Range Dispersion. J. Chem. Theory Comput. 2021, 17, 1562–1580. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Breneman, C.M.; Wiberg, K.B. Determining atom-centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. J. Comput. Chem. 1990, 11, 361–373. [Google Scholar] [CrossRef]

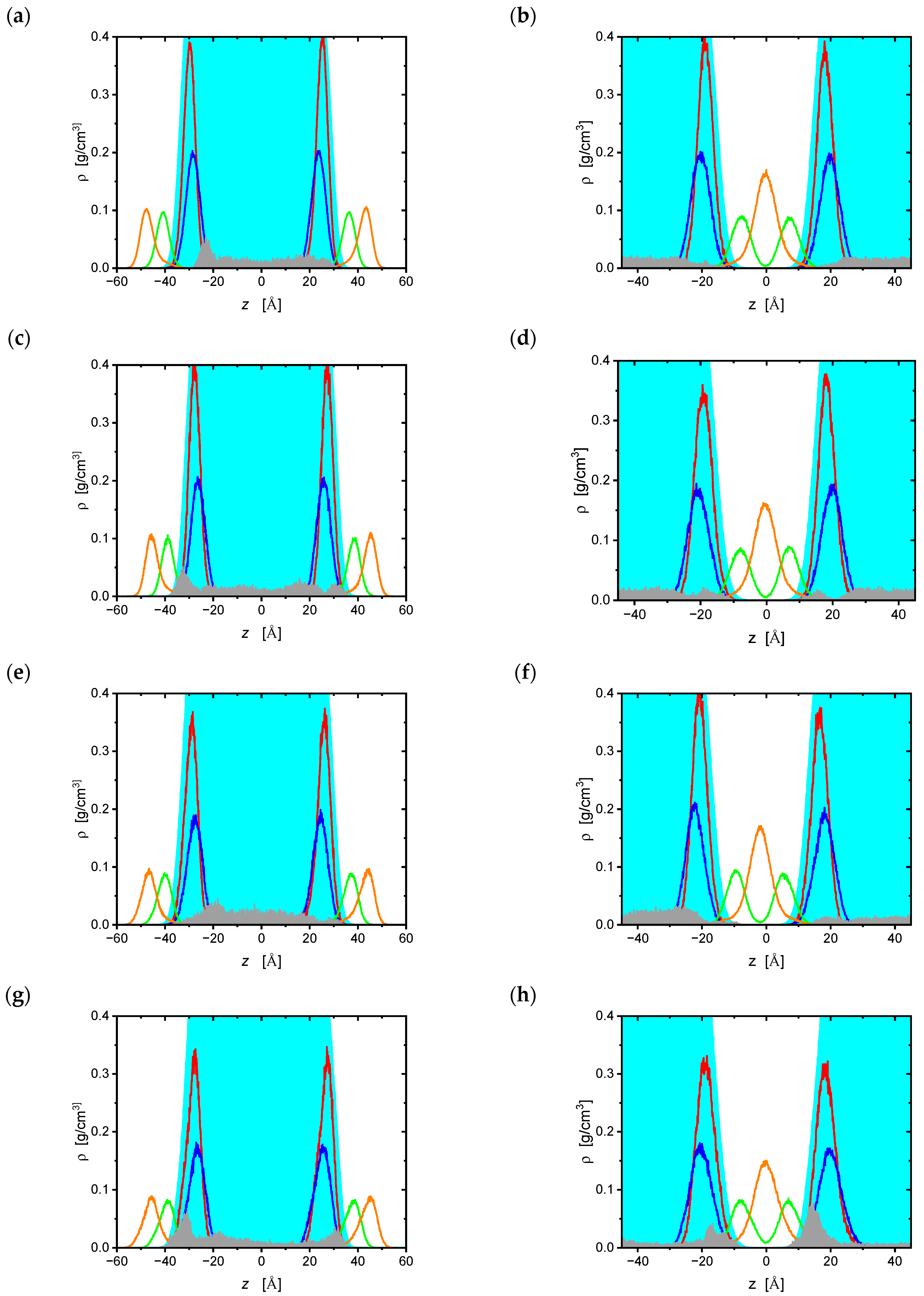
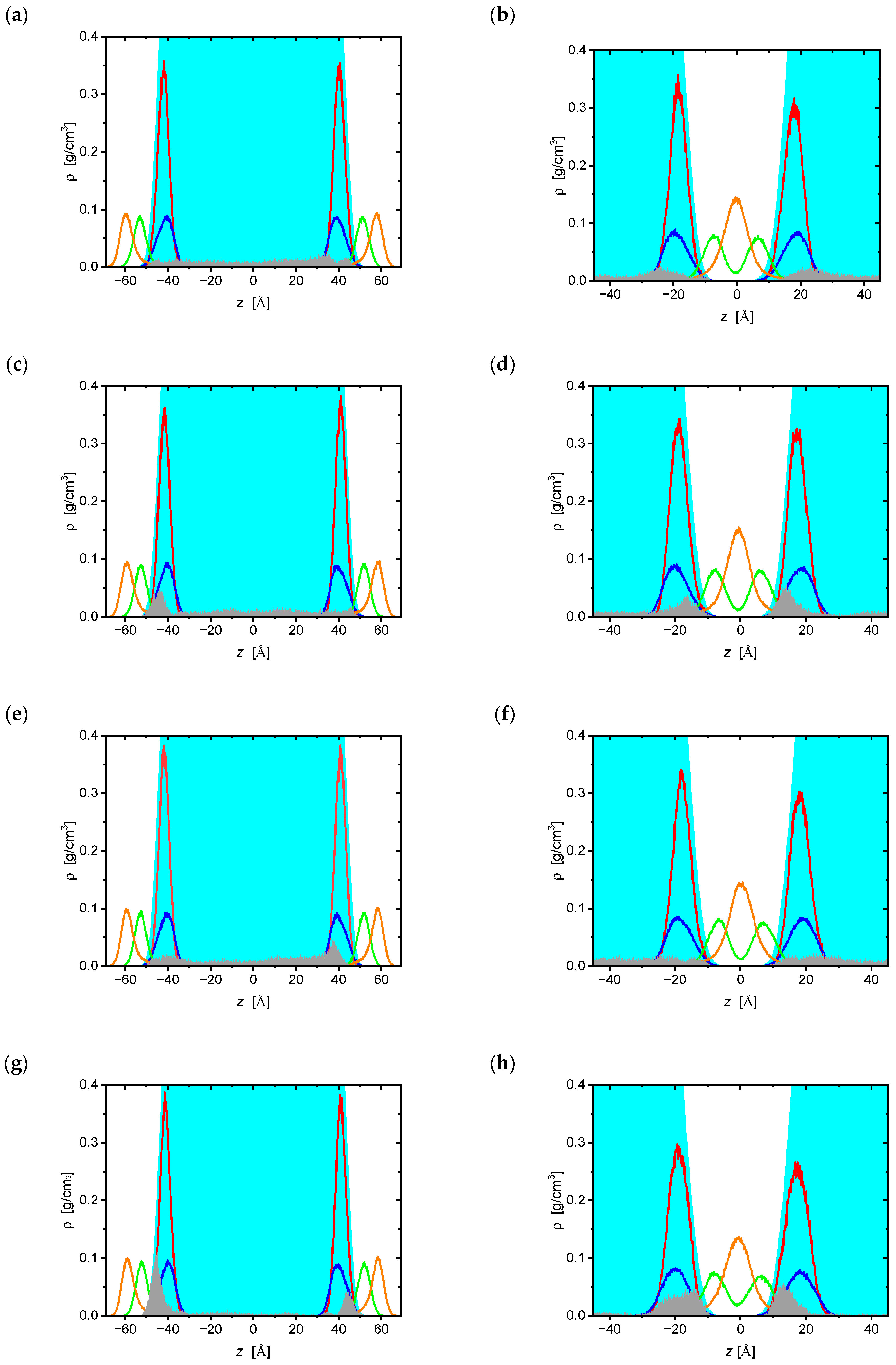
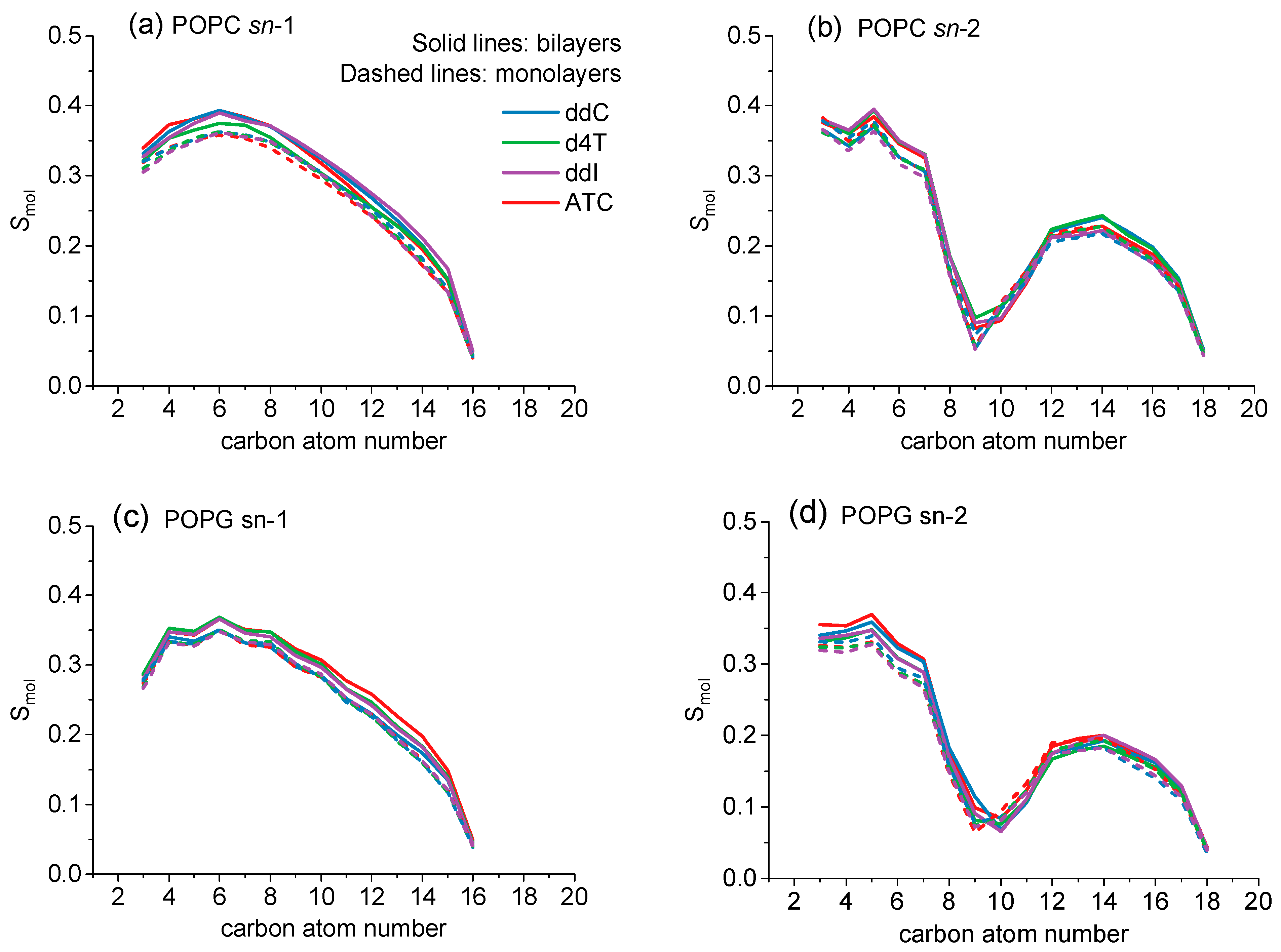
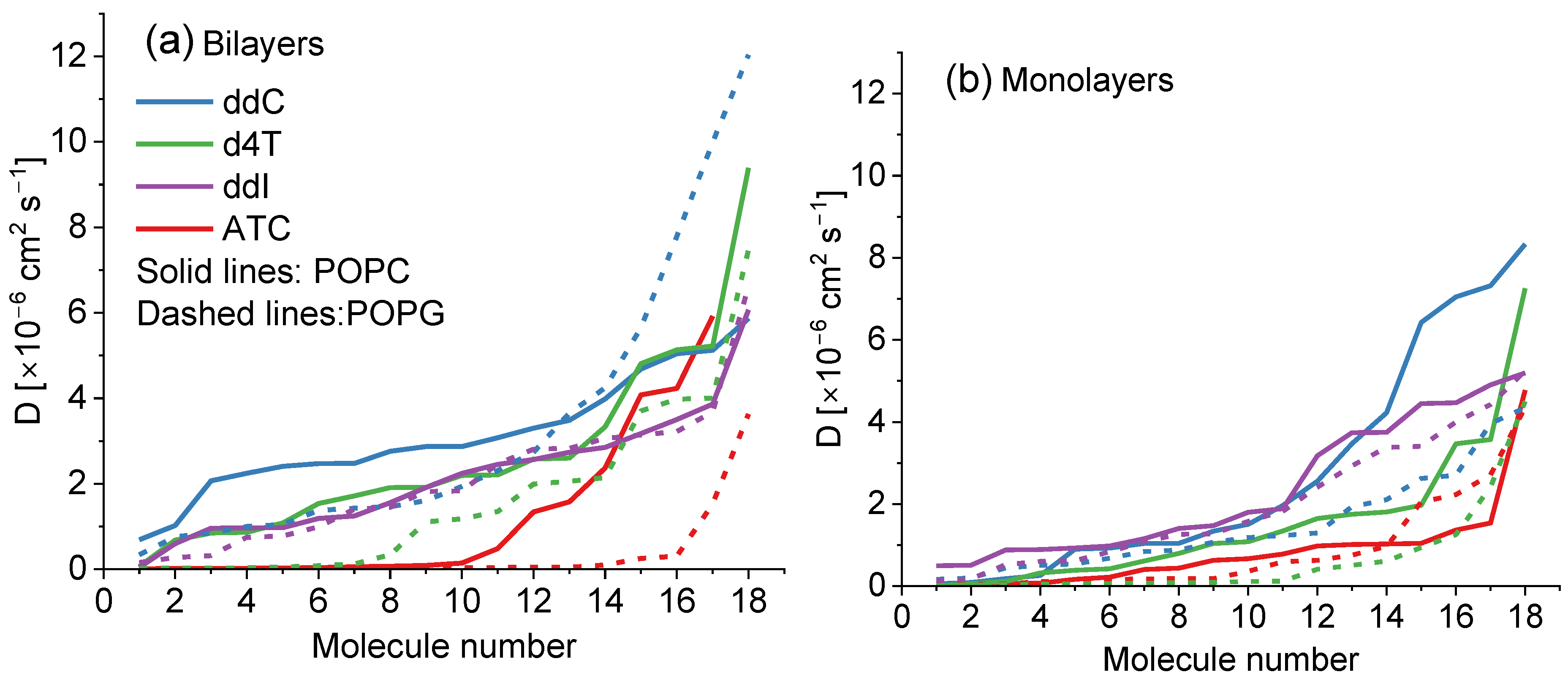
| Bilayer | ddC | d4T | ddl | ATC |
|---|---|---|---|---|
| POPC: | 37.2 ± 0.2 | 37.3 ± 0.2 | 37.3 ± 0.2 | 37.4 ± 0.2 |
| POPC: | 31.1 ± 0.1 | 31.2 ± 0.2 | 31.1 ± 0.2 | 31.3 ± 0.2 |
| POPG: | 36.0 ± 0.2 | 36.1 ± 0.2 | 35.9 ± 0.3 | 36.2 ± 0.2 |
| POPG: | 29.4 ± 0.5 | 30.0 ± 0.3 | 29.2 ± 0.5 | 30.4 ± 0.2 |
| Monolayer | ||||
| POPC: | 17.0 ± 0.3 | 17.0 ± 0.2 | 17.0 ± 0.2 | 17.2 ± 0.2 |
| POPC: | 13.9 ± 0.3 | 13.9 ± 0.2 | 13.9 ± 0.2 | 14.1 ± 0.2 |
| POPG: | 16.5 ± 0.2 | 16.4 ± 0.2 | 16.4 ± 0.2 | 16.6 ± 0.2 |
| POPG: | 13.6 ± 0.2 | 13.6 ± 0.2 | 13.6 ± 0.3 | 13.7 ± 0.2 |
| Monolayer | Bilayer | |||
|---|---|---|---|---|
| System | HB with Lipid | HB with Water | HB with Lipid | HB with Water |
| ddC/POPC | 1.57 ± 1.22 1.48 ± 1.19 0.09 ± 0.30 | 27.35 ± 4.93 7.41 ± 2.47 19.94 ± 3.94 | 0.21 ± 0.47 0.18 ± 0.43 0.03 ± 0.20 | 28.93 ± 5.10 7.29 ± 2.54 21.64 ± 4.28 |
| d4T/POPC | 2.65 ± 1.53 2.46 ± 1.29 0.19 ± 0.42 | 29.87 ± 5.11 8.56 ± 2.57 21.31 ± 4.30 | 0.87 ± 0.94 0.79 ± 0.88 0.09 ± 0.30 | 31.38 ± 4.87 8.71 ± 2.46 22.67 ± 4.07 |
| ddI/POPC | 1.13 ± 0.97 0.99 ± 0.88 0.14 ± 0.26 | 34.77 ± 5.25 8.59 ± 2.45 26.18 ± 4.62 | 0.64 ± 0.80 0.61 ± 0.78 0.03 ± 0.18 | 34.86 ± 5.44 8.63 ± 2.47 26.23 ± 4.64 |
| ATC/POPC | 4.79 ± 2.11 4.62 ± 2.24 0.17 ± 0.25 | 31.13 ± 4.84 12.11 ± 3.14 19.02 ± 3.85 | 6.07 ± 2.12 5.98 ± 2.09 0.09 ± 0.27 | 28.76 ± 4.76 11.42 ± 2.97 17.34 ± 3.72 |
| ddC/POPG | 1.30 ± 1.16 0.69 ± 0.77 0.61 ± 0.73 | 27.68 ± 4.78 6.23 ± 2.26 21.45 ± 4.14 | 1.44 ± 1.19 1.20 ± 1.07 0.24 ± 0.50 | 25.48 ± 4.55 5.93 ± 2.09 19.55 ± 3.95 |
| d4T/POPG | 3.39 ± 1.68 3.09 ± 1.90 0.30 ± 0.62 | 24.99 ± 4.67 6.86 ± 2.36 18.13 ± 4.31 | 5.89 ± 2.10 5.49 ± 1.96 0.41 ± 0.62 | 21.26 ± 4.73 5.95 ± 2.20 15.31 ± 3.58 |
| ddI/POPG | 1.87 ± 1.23 1.66 ± 1.21 0.21 ± 0.31 | 26.58 ± 4.94 6.52 ± 2.21 20.06 ± 3.97 | 1.20 ± 1.10 1.12 ± 1.04 0.08 ± 0.30 | 28.66 ± 5.17 7.30 ± 2.50 21.36 ± 4.63 |
| ATC/POPG | 7.80 ± 2.56 6.93 ± 2.25 0.87 ± 0.58 | 17.59 ± 4.02 7.46 ± 2.81 10.13 ± 2.97 | 7.07 ± 2.62 6.84 ± 2.56 0.23 ± 0.49 | 20.33 ± 4.15 10.22 ± 2.88 10.11 ± 2.96 |
| Vacuum | Water | |||
|---|---|---|---|---|
| Molecule | [eV] | [eV] | [eV] | [eV] |
| ddC | −3.6 | 9.0 | −3.8 | 5.4 |
| d4T | −3.9 | 9.1 | −3.9 | 5.4 |
| ddI | −3.5 | 9.0 | −3.8 | 5.3 |
| ATC | −3.9 | 8.7 | −3.9 | 5.1 |
| Bilayer | ddC | d4T | ddl | ATC |
|---|---|---|---|---|
| POPC | −2.0 ± 0.7 | −2.2 ± 0.6 | −1.5 ± 0.7 | −3.2 ± 1.2 |
| POPG | −2.6 ± 1.4 | −3.8 ± 1.9 | −2.5 ± 1.4 | −4.9 ± 1.7 |
| Monolayer | ||||
| POPC | −2.5 ± 1.0 | −2.7 ± 0.7 | −3.3 ± 0.7 | −2.9 ± 1.1 |
| POPG | −2.4 ± 1.4 | −3.4 ± 1.6 | −1.3 ± 0.5 | −4.5 ± 1.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stachowicz-Kuśnierz, A.; Korchowiec, B.; Korchowiec, J. Nucleoside Analog Reverse-Transcriptase Inhibitors in Membrane Environment: Molecular Dynamics Simulations. Molecules 2023, 28, 6273. https://doi.org/10.3390/molecules28176273
Stachowicz-Kuśnierz A, Korchowiec B, Korchowiec J. Nucleoside Analog Reverse-Transcriptase Inhibitors in Membrane Environment: Molecular Dynamics Simulations. Molecules. 2023; 28(17):6273. https://doi.org/10.3390/molecules28176273
Chicago/Turabian StyleStachowicz-Kuśnierz, Anna, Beata Korchowiec, and Jacek Korchowiec. 2023. "Nucleoside Analog Reverse-Transcriptase Inhibitors in Membrane Environment: Molecular Dynamics Simulations" Molecules 28, no. 17: 6273. https://doi.org/10.3390/molecules28176273
APA StyleStachowicz-Kuśnierz, A., Korchowiec, B., & Korchowiec, J. (2023). Nucleoside Analog Reverse-Transcriptase Inhibitors in Membrane Environment: Molecular Dynamics Simulations. Molecules, 28(17), 6273. https://doi.org/10.3390/molecules28176273







