Determination of Structure and Cytotoxicity of Ten Undescribed Steroidal Glycosides from Allium cristophii × A. macleanii ‘Globemaster’
Abstract
:1. Introduction
2. Results
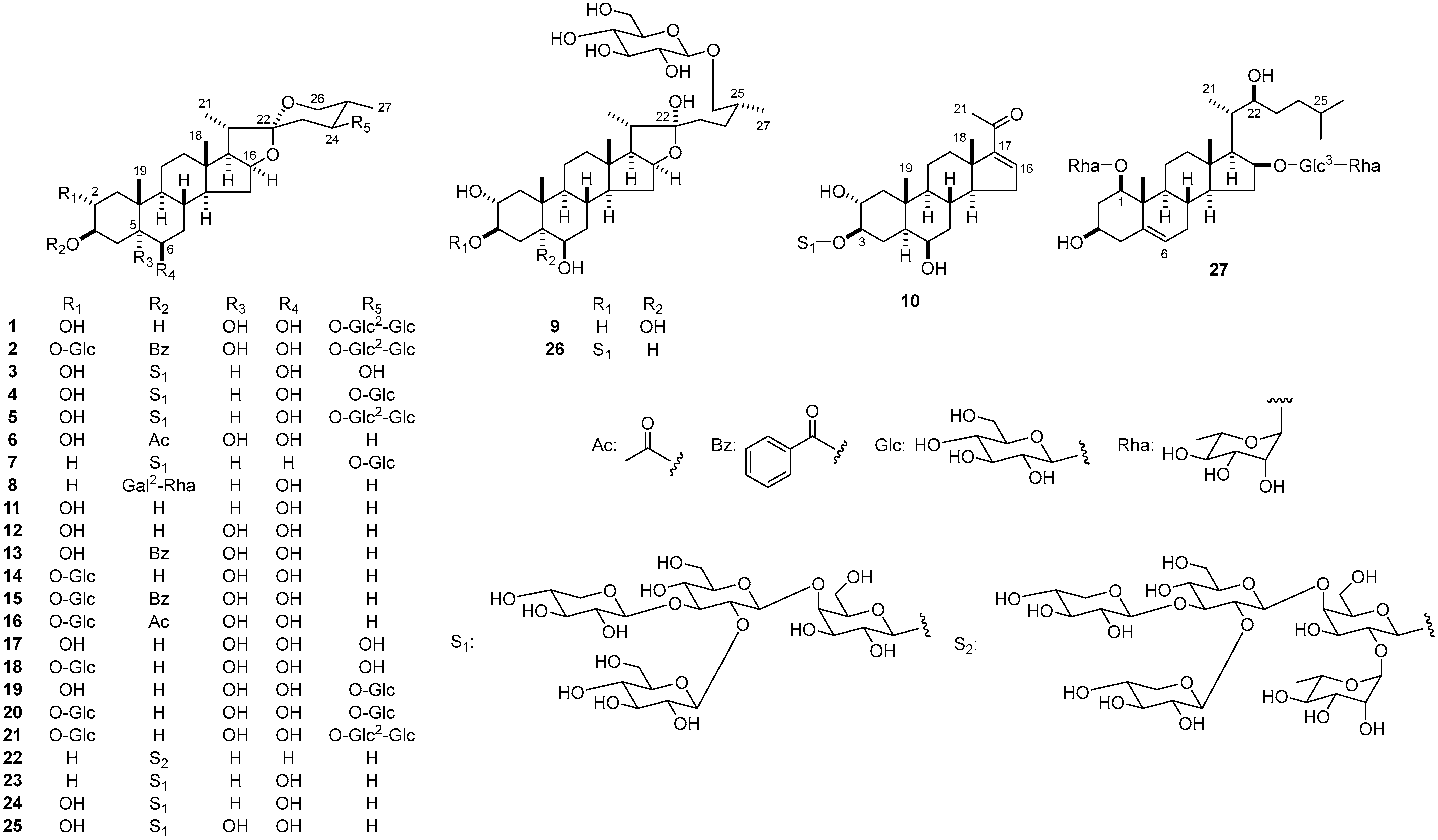
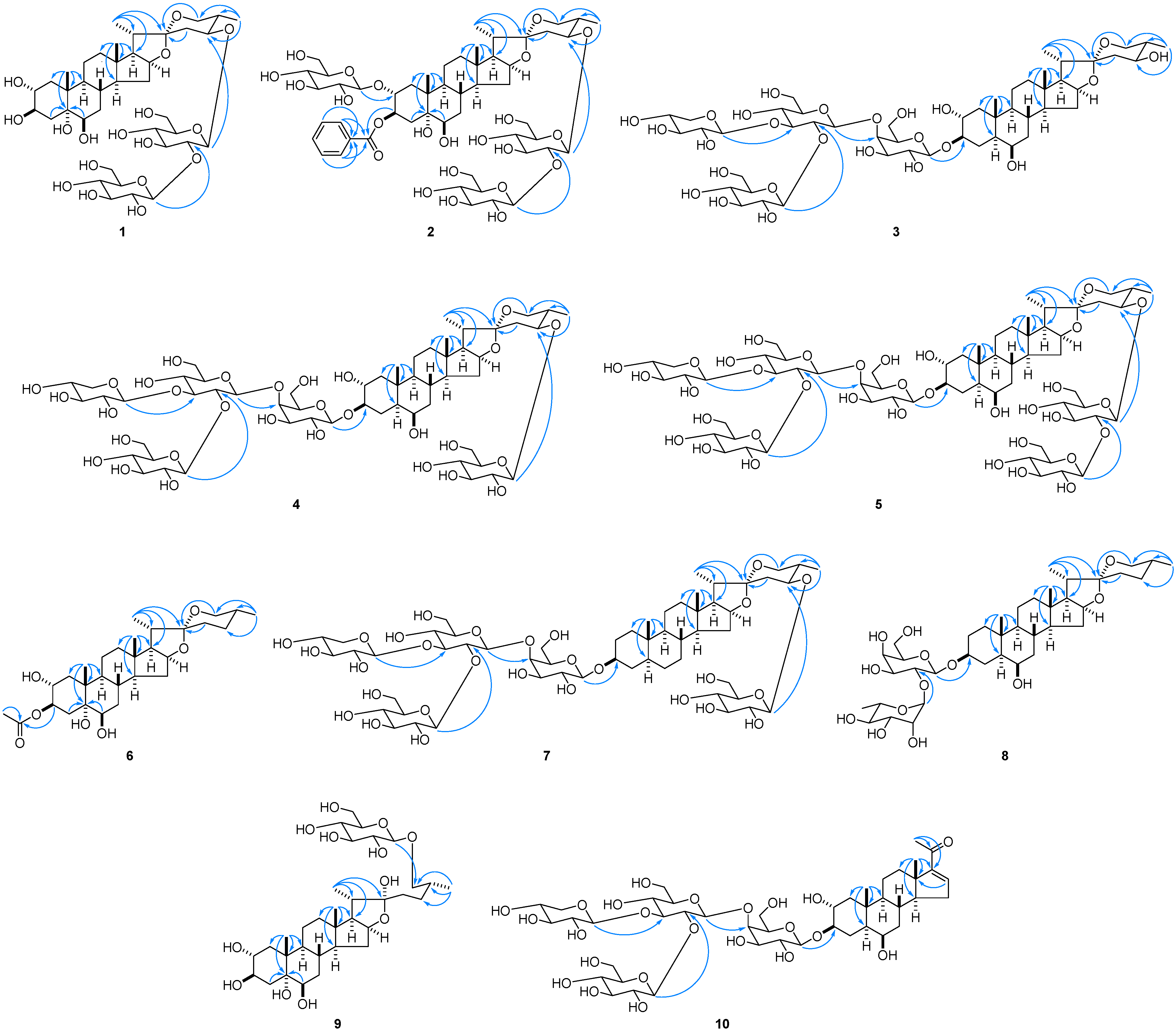
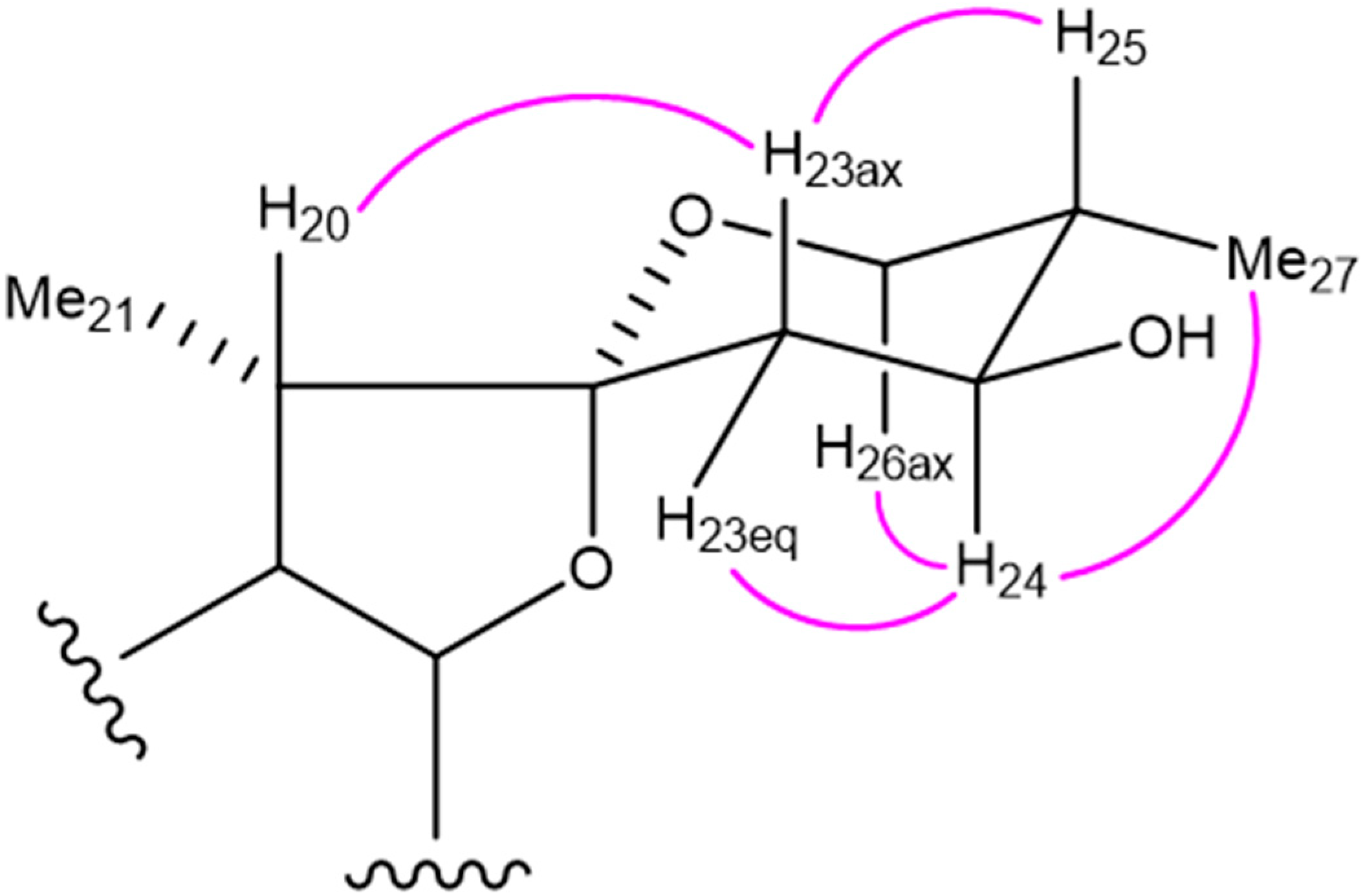
2.1. Structure Determination
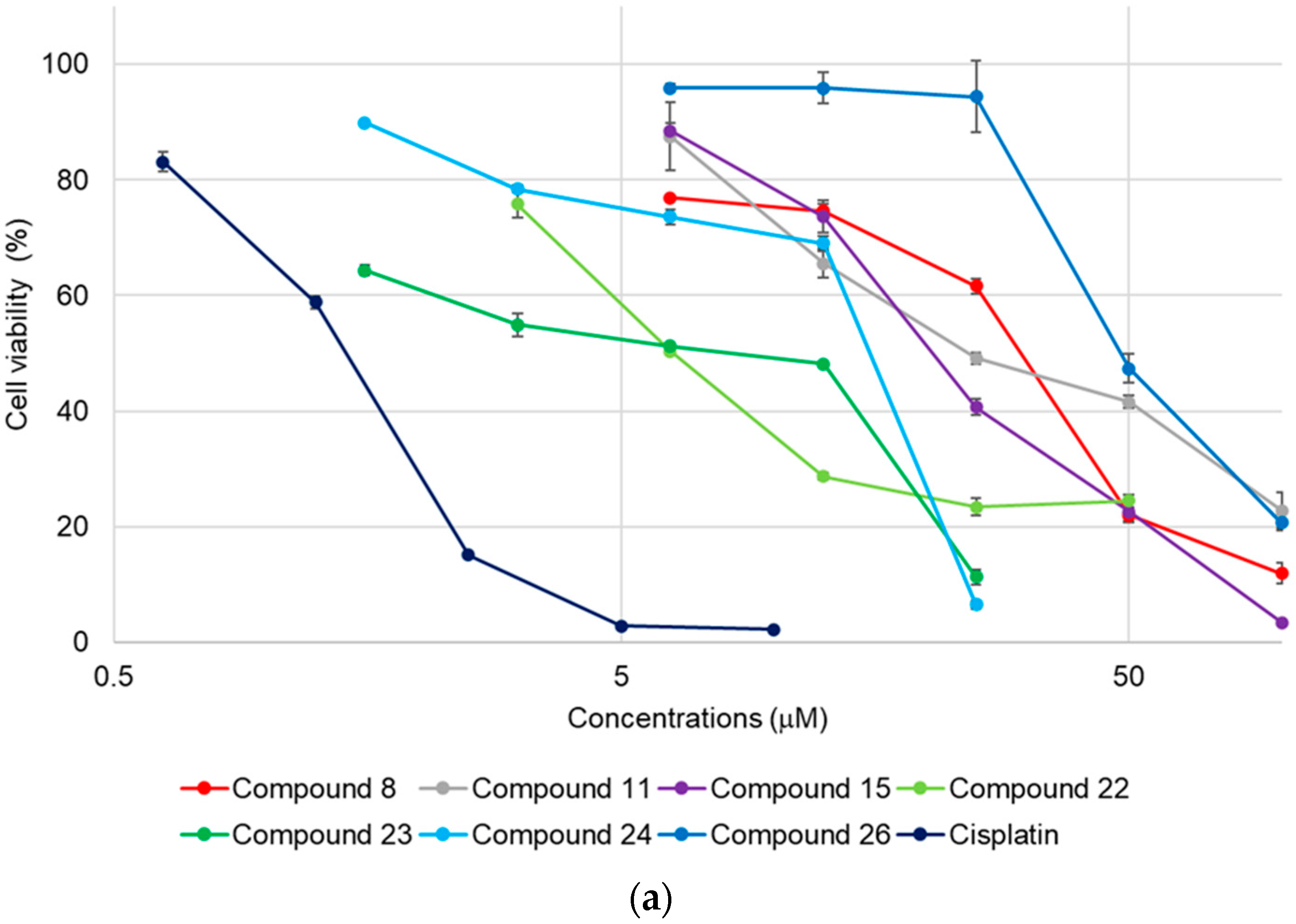
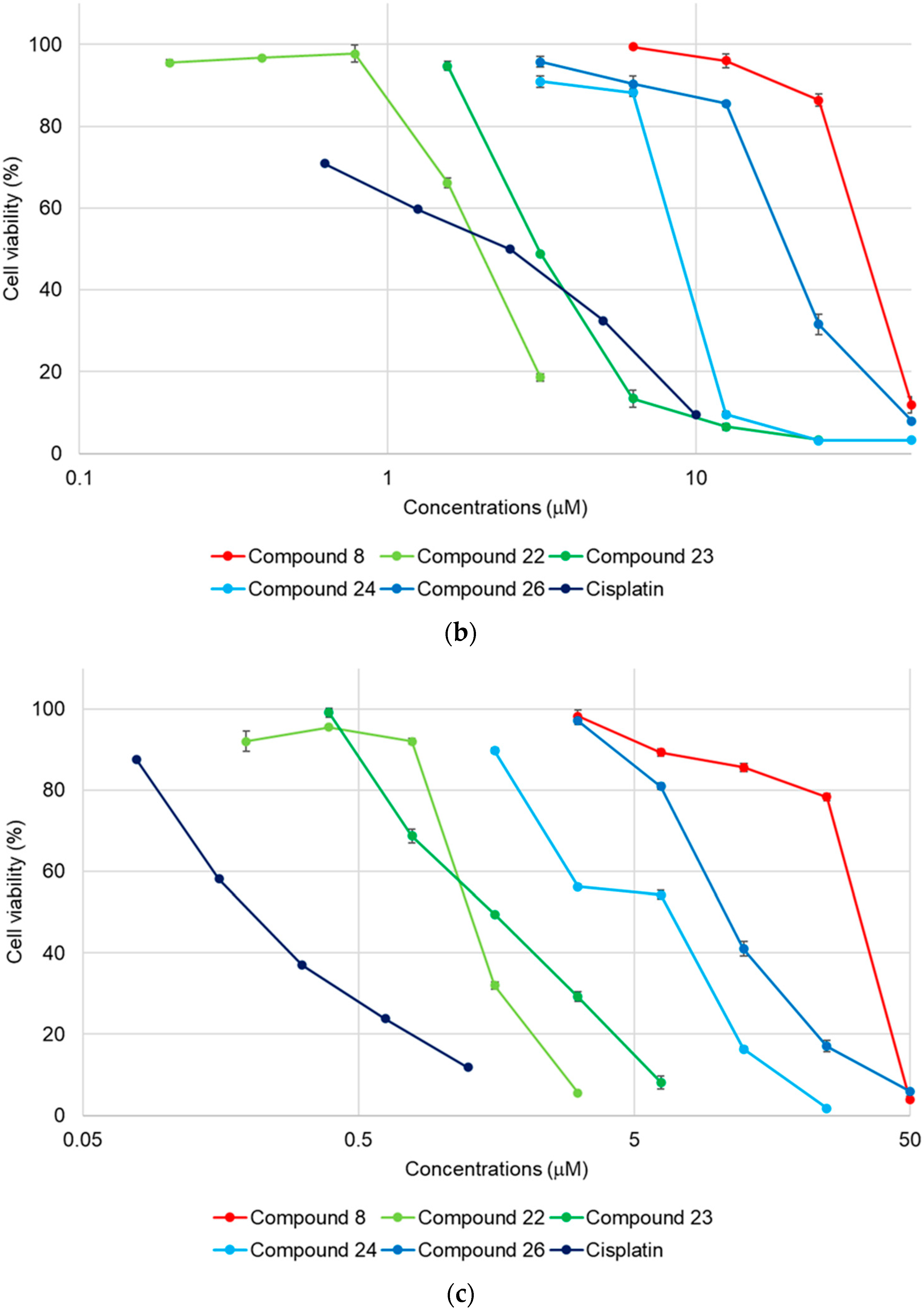
2.2. Cytotoxicity of 1–27
3. Experimental Section
3.1. General
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Structure Characterization
3.4.1. Compound 1
3.4.2. Compound 2
3.4.3. Compound 3
3.4.4. Compound 4
3.4.5. Compound 5
3.4.6. Compound 6
3.4.7. Compound 7
3.4.8. Compound 8
3.4.9. Compound 9
3.4.10. Compound 10
3.4.11. Enzymatic Hydrolysis of 1 and 9
3.5. Cell Culture and Cytotoxic Activity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Yan, J.K.; Zhu, J.; Liu, Y.; Chen, X.; Wang, W.; Zhang, H.; Li, L. Recent advances in research on Allium plants: Functional ingredients, physiological activities, and applications in agricultural and food sciences. Crit. Rev. Food Sci. Nutr. 2022, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, K.; Mimaki, Y.; Sashida, Y. Steroidal saponins from Allium giganteum and A. aflatunense. Phytochemistry 1991, 30, 3063–3067. [Google Scholar] [CrossRef]
- Mimaki, Y.; Kuroda, M.; Sashida, Y. Steroidal saponins from the bulbs of Allium aflatunense. Nat. Med. 1999, 53, 88–93. [Google Scholar]
- Mimaki, Y.; Kawashima, K.; Kanmoto, T.; Sashida, Y. Steroidal glycosides from Allium albopilosum and A. ostrowskianum. Phytochemistry 1993, 34, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Mimaki, Y.; Kuroda, M.; Sashida, Y. Steroidal saponins from the bulbs of Allium ampeloprasum. Nat. Med. 1999, 53, 134–137. [Google Scholar]
- Kuroda, M.; Mimaki, Y.; Kameyama, A.; Sashida, Y.; Nikaido, T. Steroidal saponins from Allium chinense and their inhibitory activities on cyclic AMP phosphodiesterase and Na+/K+ ATPase. Phytochemistry 1995, 40, 1071–1076. [Google Scholar] [CrossRef]
- Sashida, Y.; Kawashima, K.; Mimaki, Y. Novel polyhydroxylated steroidal saponins from Allium giganteum. Chem. Pharm. Bull. 1991, 39, 698–703. [Google Scholar] [CrossRef]
- Mimaki, Y.; Nikaido, T.; Matsumoto, K.; Sashida, Y.; Ohmoto, T. New steroidal saponins from the bulbs of Allium giganteum exhibiting potent inhibition of cAMP phosphodiesterase activity. Chem. Pharm. Bull. 1994, 42, 710–714. [Google Scholar] [CrossRef]
- Mimaki, Y.; Kuroda, M.; Fukasawa, T.; Sashida, Y. Steroidal glycosides from the bulbs of Allium jesdianum. J. Nat. Prod. 1999, 62, 194–197. [Google Scholar] [CrossRef]
- Mimaki, Y.; Kuroda, M.; Fukasawa, T.; Sashida, Y. Steroidal saponins from the bulbs of Allium karataviense. Chem. Pharm. Bull. 1999, 47, 738–743. [Google Scholar] [CrossRef]
- Kuroda, M.; Ori, K.; Takayama, H.; Sakagami, H.; Mimaki, Y. Karataviosides G–K, five new bisdesmosidic steroidal glycosides from the bulbs of Allium karataviense. Steroids 2015, 93, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Mimaki, Y.; Sashida, Y.; Nishino, A.; Satomi, Y.; Nishino, H. Steroidal glycosides from Allium macleanii and A. senescens, and their inhibitory activity on tumour promoter-induced phospholipid metabolism of HeLa cells. Phytochemistry 1995, 40, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Mimaki, Y.; Satou, T.; Ohmura, M.; Sashida, Y. Steroidal saponins from the bulbs of Allium narcissiflorum. Nat. Med. 1996, 50, 308. [Google Scholar]
- Kawashima, K.; Mimaki, Y.; Sashida, Y. Schubertosides A-D, new (22S)-hydroxycholestane glycosides from Allium schubertii. Chem. Pharm. Bull. 1991, 39, 2761–2763. [Google Scholar] [CrossRef]
- Kawashima, K.; Mimaki, Y.; Sashida, Y. Steroidal saponins from the bulbs of Allium schubertii. Phytochemistry 1993, 32, 1267–1272. [Google Scholar] [CrossRef]
- Mimaki, Y.; Satou, T.; Kuroda, M.; Kameyama, A.; Sashida, Y.; Li, H.U.; Harada, N. A new furostanol saponin with six sugars from the bulbs of Allium sphaerosephalon structural elucidation by modern NMR techniques. Chem. Lett. 1996, 25, 431–432. [Google Scholar] [CrossRef]
- Friesen, N.; Fritsch, R.; Bachmann, K. Hybrid origin of some ornamentals of Allium subgenus Melanocrommyum verified with GISH and RAPD. Theor. Appl. Genet. 1997, 95, 1229–1238. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, L.R.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rudin, M.C.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Primers. 2021, 7, 3. [Google Scholar] [CrossRef]
- Morita, T.; Ushiroguchi, T.; Hayashi, N.; Matsuura, H.; Itakura, Y.; Fuwa, T. Steroidal saponins from elephant garlic, bulbs of Allium ampeloprasum L. Chem. Pharm. Bull. 1988, 36, 3480–3486. [Google Scholar] [CrossRef]
- Gorovits, M.B.; Khristulas, F.S.; Abubakirov, N.K. Steroid saponins and sapogenins of Allium IV. Karatavigenin-A new sapogenin from Allium karataviense. Khimiya Prirodnykh Soedinenii 1973, 6, 747–749. [Google Scholar] [CrossRef]
- Mimaki, Y.; Kanmoto, T.; Sashida, Y.; Nishino, A.; Satomi, Y.; Nishino, H. Steroidal saponins from the underground parts of Chlorophytum comosum and their inhibitory activity on tumour promoter-induced phospholipids metabolism of HeLa cells. Phytochemistry 1996, 41, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Janeczko, Z.; Sobolewska, D.; Podolak, I. Pregnadienolone glycoside from wild garlic Allium ursinum L. Acta Pol. Pharm. 2000, 57, 131–134. [Google Scholar] [PubMed]
| Compounds | HL-60 Cells | A549 Cells | SBC-3 Cells |
|---|---|---|---|
| IC50 (μM) | IC50 (μM) | IC50 (μM) | |
| 1 | >50 | >50 | >50 |
| 2 | >50 | >50 | >50 |
| 3 | >50 | >50 | >50 |
| 4 | >50 | >50 | >50 |
| 5 | >50 | >50 | >50 |
| 6 | >50 | >50 | >50 |
| 7 | >50 | >50 | >50 |
| 8 | 30 ± 0.52 | 35 ± 0.36 | 33 ± 0.17 |
| 9 | >50 | >50 | >50 |
| 10 | >50 | >50 | >50 |
| 11 | 25 ± 0.98 | >50 | >50 |
| 12 | >50 | >50 | >50 |
| 13 | >50 | >50 | >50 |
| 14 | >50 | >50 | >50 |
| 15 | 21 ± 0.49 | >50 | >50 |
| 16 | >50 | >50 | >50 |
| 17 | >50 | >50 | >50 |
| 18 | >50 | >50 | >50 |
| 19 | >50 | >50 | >50 |
| 20 | >50 | >50 | >50 |
| 21 | >50 | >50 | >50 |
| 22 | 6.3 ± 0.035 | 2.0 ± 0.019 | 1.3 ± 0.010 |
| 23 | 8.4 ± 0.13 | 3.1 ± 0.0067 | 1.5 ± 0.020 |
| 24 | 15 ± 0.18 | 8.8 ± 0.045 | 6.6 ± 0.12 |
| 25 | >50 | >50 | >50 |
| 26 | 49 ± 1.6 | 20 ± 0.48 | 11 ± 0.29 |
| 27 | >50 | >50 | >50 |
| Cisplatin | 1.3 ± 0.024 | 2.5 ± 0.058 | 0.20 ± 0.0033 |
| 1 | 2 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| 1 | a | 2.45 | dd | 11.9, 11.9 | 1 | a | 2.46 | dd | 12.2, 12.2 |
| b | 2.15 | dd | 11.9, 5.0 | b | 2.36 | dd | 12.2, 5.5 | ||
| 2 | 4.45 | m | 2 | 4.88 | m | ||||
| 3 | 4.83 | m | 3 | 6.29 | ddd | 11.1, 9.5, 6.2 | |||
| 4 | a | 3.05 | dd | 12.8, 11.9 | 4 | a | 2.90 | dd | 12.9, 11.1 |
| b | 2.41 | dd | 12.8, 5.6 | b | 2.57 | dd | 12.9, 6.2 | ||
| 5 | – | 5 | – | ||||||
| 6 | 4.22 | m | 6 | 4.18 | m | ||||
| 7 | a | 2.22 | m | 7 | a | 2.20 | m | ||
| b | 1.88 | m | b | 1.87 | m | ||||
| 8 | 2.26 | m | 8 | 2.21 | m | ||||
| 9 | 2.03 | m | 9 | 1.95 | m | ||||
| 10 | – | 10 | – | ||||||
| 11 | a | 1.58 | m | 11 | a | 1.41 | m | ||
| b | 1.46 | m | b | 1.33 | m | ||||
| 12 | a | 1.67 | m | 12 | a | 1.64 | m | ||
| b | 1.12 | ddd | 12.7, 12.7, 3.8 | b | 1.07 | m | |||
| 13 | – | 13 | – | ||||||
| 14 | 1.27 | m | 14 | 1.22 | m | ||||
| 15 | a | 2.07 | m | 15 | a | 2.07 | m | ||
| b | 1.40 | m | b | 1.39 | m | ||||
| 16 | 4.50 | q-like | 7.6 | 16 | 4.51 | q-like | 7.8 | ||
| 17 | 1.75 | dd | 7.6, 7.0 | 17 | 1.76 | dd | 7.8, 6.5 | ||
| 18 | 0.81 | s | 18 | 0.81 | s | ||||
| 19 | 1.65 | s | 19 | 1.56 | s | ||||
| 20 | 1.91 | m | 20 | 1.91 | m | ||||
| 21 | 1.00 | d | 7.0 | 21 | 1.01 | d | 7.0 | ||
| 22 | – | 22 | – | ||||||
| 23 | ax | 1.95 | dd | 12.9, 10.7 | 23 | ax | 1.97 | m | |
| eq | 2.61 | dd | 12.9, 4.9 | eq | 2.63 | dd | 12.9, 4.7 | ||
| 24 | 3.94 | ddd | 10.7, 10.7, 4.9 | 24 | 3.95 | m | |||
| 25 | 1.98 | m | 25 | 1.99 | m | ||||
| 26 | ax | 3.49 | dd | 11.4, 11.4 | 26 | ax | 3.50 | dd | 11.4, 11.4 |
| eq | 3.59 | dd | 11.4, 4.9 | eq | 3.61 | dd | 11.4, 5.0 | ||
| 27 | 1.22 | d | 6.4 | 27 | 1.23 | d | 6.5 | ||
| 3 | 4 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| 1 | a | 2.20 | dd | 12.0, 4.2 | 1 | a | 2.18 | dd | 12.7, 4.3 |
| b | 1.24 | m | b | 1.22 | m | ||||
| 2 | 4.10 | m | 2 | 4.08 | m | ||||
| 3 | 4.01 | m | 3 | 4.11 | m | ||||
| 4 | a | 2.38 | br dd | 12.6, 12.6 | 4 | a | 2.37 | br dd | 12.5, 12.5 |
| b | 2.12 | m | b | 2.13 | m | ||||
| 5 | 1.16 | m | 5 | 1.14 | m | ||||
| 6 | 3.96 | m | 6 | 3.94 | m | ||||
| 7 | a | 2.00 | m | 7 | a | 2.00 | m | ||
| b | 1.15 | m | b | 1.09 | m | ||||
| 8 | 2.14 | m | 8 | 2.08 | m | ||||
| 9 | 0.73 | ddd | 12.0, 12.0, 3.6 | 9 | 0.70 | m | |||
| 10 | – | 10 | – | ||||||
| 11 | a | 1.53 | m | 11 | a | 1.48 | m | ||
| b | 1.37 | m | b | 1.34 | m | ||||
| 12 | a | 1.66 | m | 12 | a | 1.59 | m | ||
| b | 1.07 | m | b | 1.02 | m | ||||
| 13 | – | 13 | – | ||||||
| 14 | 1.12 | m | 14 | 1.07 | m | ||||
| 15 | a | 2.06 | m | 15 | a | 2.11 | m | ||
| b | 1.42 | m | b | 1.36 | m | ||||
| 16 | 4.55 | m | 16 | 4.50 | m | ||||
| 17 | 1.82 | dd | 8.4, 6.6 | 17 | 1.75 | dd | 8.0, 6.8 | ||
| 18 | 0.82 | s | 18 | 0.74 | s | ||||
| 19 | 1.26 | s | 19 | 1.24 | s | ||||
| 20 | 2.03 | m | 20 | 1.89 | m | ||||
| 21 | 1.15 | d | 6.6 | 21 | 1.03 | d | 6.8 | ||
| 22 | – | 22 | – | ||||||
| 23 | ax | 1.98 | dd | 12.6, 12.6 | 23 | ax | 1.94 | dd | 13.0, 13.0 |
| eq | 2.29 | dd | 12.6, 4.8 | eq | 2.61 | dd | 13.0, 4.8 | ||
| 24 | 3.99 | m | 24 | 3.99 | m | ||||
| 25 | 1.81 | m | 25 | 1.87 | m | ||||
| 26 | ax | 3.58 | dd | 10.8, 10.8 | 26 | ax | 3.54 | dd | 11.5, 11.5 |
| eq | 3.70 | dd | 10.8, 4.8 | eq | 3.61 | dd | 11.5, 5.0 | ||
| 27 | 1.07 | d | 6.6 | 27 | 1.12 | d | 6.4 | ||
| 5 | 6 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| 1 | a | 2.19 | dd | 12.3, 4.2 | 1 | a | 2.50 | dd | 11.5, 11.5 |
| b | 1.21 | m | b | 2.18 | dd | 11.5, 5.5 | |||
| 2 | 4.09 | m | 2 | 4.53 | ddd | 11.5, 9.4, 5.5 | |||
| 3 | 4.01 | m | 3 | 6.08 | ddd | 11.5, 9.4, 5.9 | |||
| 4 | a | 2.38 | br dd | 12.7, 12.7 | 4 | a | 2.83 | dd | 13.2, 11.5 |
| b | 2.13 | m | b | 2.38 | dd | 13.2, 5.9 | |||
| 5 | 1.14 | m | 5 | – | |||||
| 6 | 3.94 | m | 6 | 4.21 | dd | 2.5, 2.5 | |||
| 7 | a | 1.94 | m | 7 | a | 2.26 | m | ||
| b | 1.10 | m | b | 1.93 | m | ||||
| 8 | 2.10 | m | 8 | 2.30 | m | ||||
| 9 | 0.71 | m | 9 | 2.04 | m | ||||
| 10 | – | 10 | – | ||||||
| 11 | a | 1.49 | m | 11 | a | 1.58 | m | ||
| b | 1.34 | m | b | 1.49 | m | ||||
| 12 | a | 1.60 | m | 12 | a | 1.73 | m | ||
| b | 1.02 | m | b | 1.16 | m | ||||
| 13 | – | 13 | – | ||||||
| 14 | 1.07 | m | 14 | 1.31 | m | ||||
| 15 | a | 2.01 | m | 15 | a | 2.14 | m | ||
| b | 1.36 | m | b | 1.46 | m | ||||
| 16 | 4.49 | m | 16 | 4.57 | q-like | 6.5 | |||
| 17 | 1.74 | dd | 8.0, 6.7 | 17 | 1.84 | dd | 8.5, 6.5 | ||
| 18 | 0.76 | s | 18 | 0.90 | s | ||||
| 19 | 1.25 | s | 19 | 1.64 | s | ||||
| 20 | 1.89 | m | 20 | 1.95 | m | ||||
| 21 | 1.01 | d | 6.9 | 21 | 1.12 | d | 7.0 | ||
| 22 | – | 22 | – | ||||||
| 23 | ax | 1.93 | m | 23 | a | 1.68 | m | ||
| eq | 2.61 | dd | 12.9, 4.6 | b | 1.62 | m | |||
| 24 | 3.93 | m | 24 | a | 1.53 | m | |||
| 25 | 1.98 | m | b | 1.17 | m | ||||
| 26 | ax | 3.51 | dd | 11.4, 11.4 | 25 | 1.56 | m | ||
| eq | 3.61 | dd | 11.4, 5.6 | 26 | ax | 3.47 | dd | 10.7, 10.7 | |
| 27 | 1.23 | d | 6.4 | eq | 3.56 | dd | 10.7, 3.9 | ||
| 27 | 0.67 | d | 6.0 | ||||||
| 7 | 8 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| 1 | a | 1.50 | m | 1 | a | 1.59 | m | ||
| b | 0.79 | m | b | 0.88 | m | ||||
| 2 | a | 1.58 | br d | 12.0 | 2 | a | 2.14 | m | |
| b | 1.31 | m | b | 1.94 | m | ||||
| 3 | 3.89 | m | 3 | 4.10 | m | ||||
| 4 | a | 1.77 | br d | 13.2 | 4 | a | 2.40 | br dd | 12.6, 12.6 |
| b | 1.35 | m | b | 2.16 | m | ||||
| 5 | 0.87 | m | 5 | 1.05 | m | ||||
| 6 | (2H) | 1.09 | m | 6 | 3.97 | m | |||
| 7 | a | 1.48 | m | 7 | a | 2.02 | m | ||
| b | 0.77 | m | b | 1.15 | m | ||||
| 8 | 1.33 | m | 8 | 2.19 | m | ||||
| 9 | 0.47 | ddd | 12.0, 12.0, 3.6 | 9 | 0.63 | m | |||
| 10 | – | 10 | – | ||||||
| 11 | a | 1.37 | m | 11 | a | 1.47 | m | ||
| b | 1.14 | m | b | 1.40 | m | ||||
| 12 | a | 1.60 | m | 12 | a | 1.70 | m | ||
| b | 1.01 | m | b | 1.11 | m | ||||
| 13 | – | 13 | – | ||||||
| 14 | 1.00 | m | 14 | 1.13 | m | ||||
| 15 | a | 1.98 | m | 15 | a | 2.07 | m | ||
| b | 1.34 | m | b | 1.42 | m | ||||
| 16 | 4.51 | m | 16 | 4.54 | m | ||||
| 17 | 1.73 | dd | 8.4, 6.6 | 17 | 1.82 | dd | 8.4, 6.6 | ||
| 18 | 0.73 | s | 18 | 0.83 | s | ||||
| 19 | 0.62 | s | 19 | 1.36 | s | ||||
| 20 | 1.94 | m | 20 | 1.94 | m | ||||
| 21 | 1.05 | d | 7.2 | 21 | 1.13 | d | 6.9 | ||
| 22 | – | 22 | – | ||||||
| 23 | ax | 1.96 | dd | 13.2, 13.2 | 23 | a | 1.66 | m | |
| eq | 2.67 | dd | 13.2, 4.8 | b | 1.58 | m | |||
| 24 | 4.03 | m | 24 | a | 1.53 | m | |||
| 25 | 1.90 | m | b | 1.50 | m | ||||
| 26 | ax | 3.57 | dd | 11.4, 11.4 | 25 | 1.54 | m | ||
| eq | 3.63 | dd | 11.4, 5.4 | 26 | ax | 3.48 | dd | 10.7, 10.7 | |
| 27 | 1.14 | d | 6.6 | eq | 3.57 | dd | 10.7, 3.8 | ||
| 27 | 0.67 | d | 6.0 | ||||||
| 9 | 10 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| 1 | a | 2.45 | dd | 12.1, 12.1 | 1 | a | 2.13 | m | |
| b | 2.16 | dd | 12.1, 5.3 | b | 1.20 | m | |||
| 2 | 4.44 | m | 2 | 3.79 | t-like | 8.8 | |||
| 3 | 4.85 | m | 3 | 4.01 | m | ||||
| 4 | a | 3.06 | dd | 13.3, 11.8 | 4 | a | 2.38 | m | |
| b | 2.41 | dd | 13.3, 5.6 | b | 2.15 | m | |||
| 5 | – | 5 | 1.14 | m | |||||
| 6 | 4.22 | dd | 2.7, 2.2 | 6 | 3.99 | m | |||
| 7 | a | 2.21 | m | 7 | a | 2.00 | m | ||
| b | 1.91 | m | b | 1.22 | m | ||||
| 8 | 2.31 | m | 8 | 2.13 | m | ||||
| 9 | 2.03 | m | 9 | 0.78 | m | ||||
| 10 | – | 10 | – | ||||||
| 11 | a | 1.59 | m | 11 | a | 1.57 | m | ||
| b | 1.51 | m | b | 1.48 | m | ||||
| 12 | a | 1.78 | br d | 12.6 | 12 | a | 2.56 | m | |
| b | 1.19 | ddd | 12.6, 12.6, 3.9 | b | 1.37 | m | |||
| 13 | – | 13 | – | ||||||
| 14 | 1.31 | m | 14 | 1.38 | m | ||||
| 15 | a | 2.09 | m | 15 | a | 2.14 | m | ||
| b | 1.45 | m | b | 1.86 | br dd | 16.4, 12.5 | |||
| 16 | 4.93 | q-like | 7.3 | 16 | 6.57 | br s | |||
| 17 | 1.95 | dd | 8.7, 7.3 | 17 | – | ||||
| 18 | 0.93 | s | 18 | 0.90 | s | ||||
| 19 | 1.66 | s | 19 | 1.26 | s | ||||
| 20 | 2.23 | m | 20 | – | |||||
| 21 | 1.30 | d | 7.0 | 21 | 2.22 | s | |||
| 22 | – | ||||||||
| 23 | a | 2.04 | m | ||||||
| b | 1.98 | m | |||||||
| 24 | a | 2.01 | m | ||||||
| b | 1.66 | m | |||||||
| 25 | 1.90 | m | |||||||
| 26 | a | 3.92 | dd | 9.6, 7.2 | |||||
| b | 3.62 | dd | 9.6, 5.9 | ||||||
| 27 | 0.97 | d | 6.7 | ||||||
| 1 | 2 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| Glc (I) 1′ | 4.90 | d | 7.7 | Glc (I) 1′ | 5.22 | d | 7.8 | ||
| 2′ | 4.22 | dd | 9.0, 7.7 | 2′ | 3.96 | dd | 9.0, 7.8 | ||
| 3′ | 4.24 | dd | 9.0, 9.0 | 3′ | 4.30 | dd | 9.0, 9.0 | ||
| 4′ | 4.29 | dd | 9.0, 9.0 | 4′ | 4.20 | dd | 9.0, 9.0 | ||
| 5′ | 3.90 | m | 5′ | 3.89 | m | ||||
| 6′ | a | 4.44 | m | 6′ | a | 4.40 | dd | 11.6, 2.7 | |
| b | 4.39 | dd | 11.6, 4.5 | b | 4.31 | dd | 11.6, 4.6 | ||
| Glc (II) 1″ | 5.37 | d | 7.7 | Glc (II) 1″ | 4.91 | d | 7.7 | ||
| 2″ | 4.11 | dd | 8.8, 7.7 | 2″ | 4.22 | dd | 8.9, 7.7 | ||
| 3″ | 4.30 | dd | 9.1, 8.8 | 3″ | 4.25 | dd | 8.9, 8.9 | ||
| 4″ | 4.25 | dd | 9.1, 9.1 | 4″ | 4.28 | m | |||
| 5″ | 3.77 | m | 5″ | 3.91 | m | ||||
| 6″ | a | 4.46 | m | 6″ | a | 4.45 | dd | 11.7, 2.9 | |
| b | 4.34 | dd | 11.7, 5.1 | b | 4.38 | dd | 11.7, 5.0 | ||
| Glc (III) 1″′ | 5.37 | d | 7.7 | ||||||
| 2″′ | 4.11 | dd | 8.9, 7.7 | ||||||
| 3″′ | 4.29 | dd | 8.9, 8.9 | ||||||
| 4″′ | 4.23 | dd | 8.9, 8.9 | ||||||
| 5″′ | 3.78 | m | |||||||
| 6″′ | a | 4.46 | dd | 11.8, 2.5 | |||||
| b | 4.34 | dd | 11.8, 5.3 | ||||||
| Bz 1″″ | – | ||||||||
| 2″″ | 8.43 | dd | 7.8, 1.9 | ||||||
| 3″″ | 7.44 | m | |||||||
| 4″″ | 7.45 | m | |||||||
| 5″″ | 7.44 | m | |||||||
| 6″″ | 8.43 | dd | 7.8, 1.9 | ||||||
| 7″″ | – | ||||||||
| 3 | 4 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| Gal 1′ | 4.96 | d | 7.8 | Gal 1′ | 4.97 | d | 7.7 | ||
| 2′ | 4.53 | m | 2′ | 4.52 | dd | 9.1, 7.7 | |||
| 3′ | 4.12 | m | 3′ | 4.13 | dd | 9.1, 3.4 | |||
| 4′ | 4.58 | br s | 4′ | 4.58 | br d | 3.4 | |||
| 5′ | 4.03 | m | 5′ | 4.04 | m | ||||
| 6′ | a | 4.60 | dd | 10.8, 8.4 | 6′ | a | 4.59 | dd | 10.7, 8.8 |
| b | 4.20 | dd | 10.8, 5.4 | b | 4.20 | m | |||
| Glc (I) 1″ | 5.18 | d | 7.8 | Glc (I) 1″ | 5.18 | d | 7.9 | ||
| 2″ | 4.32 | dd | 8.4, 7.8 | 2″ | 4.31 | dd | 8.7, 7.9 | ||
| 3″ | 4.13 | m | 3″ | 4.12 | dd | 8.7, 8.7 | |||
| 4″ | 3.79 | dd | 8.4, 8.4 | 4″ | 3.79 | dd | 8.7, 8.7 | ||
| 5″ | 3.84 | m | 5″ | 3.84 | m | ||||
| 6″ | a | 4.48 | dd | 11.4, 1.8 | 6″ | a | 4.37 | dd | 11.8, 5.1 |
| b | 4.05 | m | b | 4.03 | dd | 11.8, 3.5 | |||
| Glc (II) 1″′ | 5.57 | d | 7.8 | Glc (II) 1″′ | 5.58 | d | 7.9 | ||
| 2″′ | 4.03 | dd | 9.0, 7.8 | 2″′ | 4.02 | dd | 8.9, 7.9 | ||
| 3″′ | 4.16 | dd | 9.0, 9.0 | 3″′ | 4.17 | dd | 8.9, 8.9 | ||
| 4″′ | 4.07 | dd | 9.0, 9.0 | 4″′ | 4.09 | dd | 8.9, 8.9 | ||
| 5″′ | 3.90 | m | 5″′ | 3.89 | m | ||||
| 6″′ | a | 4.52 | dd | 12.0, 1.8 | 6″′ | a | 4.48 | dd | 12.1, 2.0 |
| b | 4.41 | dd | 12.0, 5.4 | b | 4.41 | dd | 12.1, 5.4 | ||
| Xyl 1″″ | 5.24 | d | 7.8 | Xyl 1″″ | 5.24 | d | 7.8 | ||
| 2″″ | 3.95 | dd | 8.4, 7.8 | 2″″ | 3.95 | dd | 8.7, 7.8 | ||
| 3″″ | 4.10 | m | 3″″ | 4.11 | dd | 8.7, 8.7 | |||
| 4″″ | 4.11 | m | 4″″ | 4.14 | m | ||||
| 5″″ | a | 4.22 | m | 5″″ | a | 4.21 | dd | 10.8, 4.3 | |
| b | 3.66 | dd | 10.2, 10.2 | b | 3.66 | dd | 10.8, 10.8 | ||
| Glc (III) 1″″′ | 4.90 | d | 7.7 | ||||||
| 2″″′ | 4.06 | dd | 8.8, 7.7 | ||||||
| 3″″′ | 4.23 | dd | 8.8, 8.8 | ||||||
| 4″″′ | 4.26 | dd | 8.8, 8.8 | ||||||
| 5″″′ | 3.86 | m | |||||||
| 6″″′ | a | 4.52 | dd | 12.1, 2.0 | |||||
| b | 4.39 | dd | 12.1, 5.1 | ||||||
| 5 | 6 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| Gal 1′ | 4.97 | d | 7.8 | Ac | 1.99 | s | |||
| 2′ | 4.55 | dd | 9.0, 7.8 | ||||||
| 3′ | 4.03 | m | |||||||
| 4′ | 4.59 | br d | 2.9 | ||||||
| 5′ | 3.85 | m | |||||||
| 6′ | a | 4.61 | dd | 10.6, 9.0 | |||||
| b | 4.20 | br d | 10.6 | ||||||
| Glc (I) 1″ | 5.20 | d | 7.9 | ||||||
| 2″ | 4.34 | dd | 8.8, 7.9 | ||||||
| 3″ | 4.13 | dd | 8.8, 8.8 | ||||||
| 4″ | 3.80 | dd | 8.8, 8.8 | ||||||
| 5″ | 3.84 | m | |||||||
| 6″ | a | 4.49 | dd | 11.3, 2.2 | |||||
| b | 4.03 | m | |||||||
| Glc (II) 1″′ | 5.59 | d | 7.5 | ||||||
| 2″′ | 4.04 | dd | 9.0, 7.5 | ||||||
| 3″′ | 4.15 | dd | 9.0, 9.0 | ||||||
| 4″′ | 4.07 | dd | 9.0, 9.0 | ||||||
| 5″′ | 3.90 | m | |||||||
| 6″′ | a | 4.54 | dd | 12.3, 2.3 | |||||
| b | 4.41 | dd | 12.3, 5.7 | ||||||
| Xyl 1″″ | 5.25 | d | 7.8 | ||||||
| 2″″ | 3.95 | dd | 8.2, 7.8 | ||||||
| 3″″ | 4.10 | m | |||||||
| 4″″ | 4.11 | m | |||||||
| 5″″ | a | 4.21 | dd | 10.9, 4.5 | |||||
| b | 3.66 | dd | 10.9, 10.9 | ||||||
| Glc (III) 1″″′ | 4.91 | d | 7.5 | ||||||
| 2″″′ | 4.23 | dd | 9.2, 7.5 | ||||||
| 3″″′ | 4.28 | dd | 9.2, 9.2 | ||||||
| 4″″′ | 4.27 | m | |||||||
| 5″″′ | 3.92 | m | |||||||
| 6″″′ | a | 4.46 | dd | 11.9, 2.5 | |||||
| b | 4.32 | dd | 11.9, 4.5 | ||||||
| Glc (IV) 1″″″ | 5.38 | d | 7.7 | ||||||
| 2″″″ | 4.10 | dd | 8.8, 7.7 | ||||||
| 3″″″ | 4.24 | dd | 8.8, 8.8 | ||||||
| 4″″″ | 4.22 | dd | 8.8, 8.8 | ||||||
| 5″″″ | 3.79 | m | |||||||
| 6″″″ | a | 4.45 | br d | 11.9 | |||||
| b | 4.37 | dd | 11.9, 4.9 | ||||||
| 7 | 8 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| Gal 1′ | 4.89 | d | 7.8 | Gal 1′ | 4.99 | d | 7.9 | ||
| 2′ | 4.40 | dd | 9.0, 7.8 | 2′ | 4.62 | dd | 9.5, 7.9 | ||
| 3′ | 4.11 | m | 3′ | 4.24 | dd | 9.5, 3.5 | |||
| 4′ | 4.59 | br d | 3.0 | 4′ | 4.53 | br d | 3.5 | ||
| 5′ | 4.04 | m | 5′ | 4.05 | m | ||||
| 6′ | a | 4.70 | dd | 10.2, 9.0 | 6′ | a | 4.55 | m | |
| b | 4.21 | m | b | 4.43 | dd | 11.4, 5.3 | |||
| Glc (I) 1″ | 5.16 | d | 7.8 | Rha 1″ | 6.26 | d | 1.3 | ||
| 2″ | 4.37 | dd | 9.0, 7.8 | 2″ | 4.80 | dd | 3.4, 1.3 | ||
| 3″ | 4.14 | dd | 9.0, 9.0 | 3″ | 4.66 | dd | 9.4, 3.4 | ||
| 4″ | 3.80 | dd | 9.0, 9.0 | 4″ | 4.28 | dd | 9.4, 9.4 | ||
| 5″ | 3.90 | m | 5″ | 4.98 | m | ||||
| 6″ | a | 4.50 | m | 6″ | 1.67 | d | 6.2 | ||
| b | 4.04 | m | |||||||
| Glc (II) 1″′ | 5.55 | d | 7.8 | ||||||
| 2″′ | 4.02 | m | |||||||
| 3″′ | 4.12 | m | |||||||
| 4″′ | 4.20 | dd | 9.0, 9.0 | ||||||
| 5″′ | 3.89 | m | |||||||
| 6″′ | a | 4.53 | dd | 12.0, 1.8 | |||||
| b | 4.38 | m | |||||||
| Xyl 1″″ | 5.22 | d | 7.8 | ||||||
| 2″″ | 3.96 | dd | 8.4, 7.8 | ||||||
| 3″″ | 4.10 | m | |||||||
| 4″″ | 4.12 | m | |||||||
| 5″″ | a | 4.22 | m | ||||||
| b | 3.67 | dd | 11.4, 11.4 | ||||||
| Glc (III) 1″″′ | 4.91 | d | 7.8 | ||||||
| 2″″′ | 4.04 | dd | 9.0, 7.8 | ||||||
| 3″″′ | 4.21 | m | |||||||
| 4″″′ | 4.25 | dd | 9.0, 9.0 | ||||||
| 5″″′ | 3.85 | m | |||||||
| 6″″′ | a | 4.48 | dd | 12.0, 2.4 | |||||
| b | 4.35 | dd | 12.0, 5.4 | ||||||
| 9 | 10 | ||||||||
| Positions | δH | J (Hz) | Positions | δH | J (Hz) | ||||
| Glc 1′ | 4.81 | d | 7.8 | Gal 1′ | 4.97 | d | 7.7 | ||
| 2′ | 4.02 | dd | 8.6, 7.8 | 2′ | 4.53 | dd | 8.8, 7.7 | ||
| 3′ | 4.26 | dd | 8.6, 8.6 | 3′ | 4.14 | m | |||
| 4′ | 4.23 | dd | 8.6, 8.6 | 4′ | 4.58 | br d | 3.4 | ||
| 5′ | 3.93 | m | 5′ | 4.00 | m | ||||
| 6′ | a | 4.54 | dd | 11.8, 2.4 | 6′ | a | 4.60 | dd | 10.7, 8.9 |
| b | 4.39 | dd | 11.8, 5.3 | b | 4.20 | m | |||
| Glc (I) 1″ | 5.18 | d | 7.8 | ||||||
| 2″ | 4.32 | dd | 8.5, 7.8 | ||||||
| 3″ | 4.13 | m | |||||||
| 4″ | 4.07 | dd | 9.1, 9.1 | ||||||
| 5″ | 3.84 | m | |||||||
| 6″ | a | 4.49 | br d | 11.3 | |||||
| b | 4.20 | m | |||||||
| Glc (II) 1″′ | 5.58 | d | 7.8 | ||||||
| 2″′ | 4.03 | dd | 9.0, 7.8 | ||||||
| 3″′ | 4.17 | dd | 9.0, 9.0 | ||||||
| 4″′ | 4.12 | m | |||||||
| 5″′ | 3.90 | m | |||||||
| 6″′ | a | 4.52 | dd | 12.1, 3.8 | |||||
| b | 4.41 | dd | 12.1, 5.3 | ||||||
| Xyl 1″″ | 5.24 | d | 7.8 | ||||||
| 2″″ | 3.95 | dd | 8.3, 7.8 | ||||||
| 3″″ | 4.11 | m | |||||||
| 4″″ | 4.08 | m | |||||||
| 5″″ | a | 4.21 | m | ||||||
| b | 3.66 | dd | 11.3, 11.3 | ||||||
| Positions | 1 | 2 | 3 | 4 | 5 |
| δC | δC | δC | δC | δC | |
| 1 | 42.2 | 38.9 | 47.1 | 47.1 | 47.1 |
| 2 | 73.7 | 77.7 | 70.5 | 70.4 | 70.5 |
| 3 | 73.6 | 76.4 | 84.6 | 84.5 | 84.5 |
| 4 | 41.1 | 37.8 | 31.9 | 31.8 | 31.9 |
| 5 | 75.6 | 75.0 | 47.8 | 47.7 | 47.8 |
| 6 | 75.5 | 74.9 | 69.9 | 69.9 | 69.9 |
| 7 | 35.7 | 35.6 | 40.7 | 40.5 | 40.6 |
| 8 | 30.1 | 30.0 | 29.9 | 29.8 | 29.9 |
| 9 | 45.8 | 45.5 | 54.5 | 54.4 | 54.4 |
| 10 | 40.9 | 40.4 | 37.0 | 36.9 | 37.0 |
| 11 | 21.6 | 21.4 | 21.3 | 21.2 | 21.3 |
| 12 | 40.4 | 40.2 | 40.0 | 39.9 | 40.0 |
| 13 | 40.9 | 40.9 | 40.8 | 40.7 | 40.7 |
| 14 | 56.3 | 56.1 | 56.2 | 56.1 | 56.1 |
| 15 | 32.1 | 32.1 | 32.1 | 32.0 | 32.0 |
| 16 | 81.5 | 81.5 | 81.4 | 81.4 | 81.4 |
| 17 | 62.6 | 62.6 | 62.6 | 62.4 | 62.5 |
| 18 | 16.7 | 16.6 | 16.5 | 16.5 | 16.5 |
| 19 | 18.5 | 17.9 | 17.1 | 17.1 | 17.1 |
| 20 | 42.1 | 42.1 | 42.2 | 42.0 | 42.1 |
| 21 | 14.8 | 14.8 | 14.9 | 14.8 | 148.0 |
| 22 | 111.5 | 111.5 | 111.8 | 111.5 | 111.5 |
| 23 | 40.6 | 40.6 | 41.8 | 40.7 | 40.6 |
| 24 | 81.7 | 81.7 | 70.5 | 81.3 | 81.7 |
| 25 | 38.0 | 38.0 | 39.9 | 38.1 | 38.0 |
| 26 | 65.2 | 65.2 | 65.3 | 65.0 | 65.2 |
| 27 | 13.6 | 13.6 | 13.6 | 13.4 | 13.6 |
| Glc (I) | Glc (I) | Gal | Gal | Gal | |
| 1′ | 104.2 | 103.3 | 103.1 | 103.0 | 103.0 |
| 2′ | 83.7 | 75.3 | 72.5 | 72.5 | 72.5 |
| 3′ | 78.0 | 78.5 | 75.5 | 75.4 | 75.5 |
| 4′ | 71.6 | 71.6 | 79.5 | 79.6 | 79.4 |
| 5′ | 78.4 | 78.4 | 75.7 | 75.6 | 75.7 |
| 6′ | 62.7 | 62.8 | 60.6 | 60.6 | 60.6 |
| Glc (II) | Glc (II) | Glc (I) | Glc (I) | Glc (I) | |
| 1″ | 106.1 | 104.2 | 104.6 | 104.6 | 104.6 |
| 2″ | 76.9 | 83.7 | 81.2 | 81.1 | 81.2 |
| 3″ | 78.2 | 78.0 | 87.1 | 87.0 | 87.0 |
| 4″ | 71.3 | 71.6 | 70.3 | 70.2 | 70.3 |
| 5″ | 77.8 | 78.4 | 77.5 | 77.5 | 77.5 |
| 6″ | 62.5 | 62.7 | 62.8 | 62.8 | 62.8 |
| Glc (III) | Glc (II) | Glc (II) | Glc (II) | ||
| 1″′ | 106.1 | 104.7 | 104.6 | 104.7 | |
| 2″′ | 76.9 | 76.0 | 75.9 | 76.0 | |
| 3″′ | 78.2 | 78.1 | 78.0 | 78.0 | |
| 4″′ | 71.3 | 71.3 | 71.2 | 71.3 | |
| 5″′ | 77.8 | 78.4 | 78.3 | 78.3 | |
| 6″′ | 62.5 | 62.6 | 62.5 | 62.6 | |
| Bz | Xyl | Xyl | Xyl | ||
| 1″″ | 131.9 | 104.9 | 104.8 | 104.9 | |
| 2″″ | 130.3 | 75.1 | 75.0 | 75.1 | |
| 3″″ | 128.6 | 78.6 | 78.6 | 78.6 | |
| 4″″ | 132.9 | 70.8 | 70.8 | 70.7 | |
| 5″″ | 128.6 | 67.2 | 67.2 | 67.2 | |
| 6″″ | 130.3 | ||||
| 7″″ | 166.8 | ||||
| Glc (III) | Glc (III) | ||||
| 1″″′ | 106.3 | 104.2 | |||
| 2″″′ | 75.6 | 83.6 | |||
| 3″″′ | 78.5 | 78.1 | |||
| 4″″′ | 71.6 | 71.6 | |||
| 5″″′ | 77.9 | 78.4 | |||
| 6″″′ | 62.7 | 62.5 | |||
| Glc (IV) | |||||
| 1″″″ | 106.0 | ||||
| 2″″″ | 76.9 | ||||
| 3″″″ | 78.3 | ||||
| 4″″″ | 71.4 | ||||
| 5″″″ | 77.8 | ||||
| 6″″″ | 62.7 | ||||
| Positions | 6 | 7 | 8 | 9 | 10 |
| δC | δC | δC | δC | δC | |
| 1 | 42.4 | 37.1 | 38.8 | 42.2 | 46.9 |
| 2 | 69.7 | 29.9 | 30.1 | 73.7 | 70.3 |
| 3 | 78.2 | 77.4 | 77.9 | 73.6 | 84.4 |
| 4 | 37.7 | 34.8 | 32.5 | 41.1 | 31.9 |
| 5 | 75.3 | 44.6 | 47.8 | 75.6 | 48.0 |
| 6 | 75.1 | 28.9 | 70.8 | 75.5 | 69.9 |
| 7 | 35.8 | 32.3 | 40.7 | 35.8 | 40.2 |
| 8 | 30.1 | 35.2 | 30.6 | 30.1 | 28.6 |
| 9 | 45.7 | 54.3 | 54.6 | 45.9 | 55.0 |
| 10 | 40.6 | 35.8 | 36.1 | 41.3 | 37.1 |
| 11 | 21.5 | 21.2 | 21.2 | 21.7 | 21.3 |
| 12 | 40.3 | 40.0 | 40.2 | 40.5 | 35.2 |
| 13 | 41.0 | 40.6 | 40.8 | 40.9 | 46.6 |
| 14 | 56.2 | 56.4 | 56.3 | 56.2 | 56.1 |
| 15 | 32.3 | 31.9 | 32.2 | 32.5 | 32.3 |
| 16 | 81.2 | 81.5 | 81.1 | 81.1 | 144.7 |
| 17 | 63.1 | 62.5 | 63.0 | 63.9 | 155.3 |
| 18 | 16.7 | 16.5 | 16.5 | 16.8 | 16.1 |
| 19 | 18.2 | 12.2 | 16.0 | 18.5 | 16.9 |
| 20 | 42.0 | 42.0 | 41.9 | 40.6 | 196.3 |
| 21 | 15.0 | 14.8 | 15.0 | 16.3 | 27.1 |
| 22 | 109.2 | 111.6 | 109.2 | 110.6 | |
| 23 | 31.8 | 40.8 | 31.7 | 37.1 | |
| 24 | 29.2 | 81.4 | 29.2 | 28.3 | |
| 25 | 30.5 | 38.1 | 30.5 | 34.2 | |
| 26 | 66.8 | 65.1 | 66.8 | 75.2 | |
| 27 | 17.3 | 13.4 | 17.2 | 17.4 | |
| Ac | Gal | Gal | Glc | Gal | |
| 1′ | 21.4 | 102.4 | 100.5 | 104.8 | 103.0 |
| 2′ | 171.0 | 73.1 | 76.3 | 75.1 | 72.4 |
| 3′ | 75.5 | 76.5 | 78.5 | 75.4 | |
| 4′ | 79.9 | 70.7 | 71.6 | 79.6 | |
| 5′ | 76.1 | 76.6 | 78.4 | 75.7 | |
| 6′ | 60.6 | 62.2 | 62.7 | 60.6 | |
| Glc (I) | Rha | Glc (I) | |||
| 1″ | 105.0 | 102.0 | 104.6 | ||
| 2″ | 81.3 | 72.4 | 81.2 | ||
| 3″ | 86.9 | 72.7 | 87.1 | ||
| 4″ | 70.4 | 74.3 | 70.4 | ||
| 5″ | 77.5 | 69.3 | 77.5 | ||
| 6″ | 62.9 | 18.6 | 62.8 | ||
| Glc (II) | Glc (II) | ||||
| 1″′ | 104.7 | 104.6 | |||
| 2″′ | 75.3 | 76.0 | |||
| 3″′ | 77.7 | 78.0 | |||
| 4″′ | 71.0 | 71.2 | |||
| 5″′ | 78.6 | 78.4 | |||
| 6″′ | 62.4 | 62.5 | |||
| Xyl | Xyl | ||||
| 1″″ | 104.9 | 104.8 | |||
| 2″″ | 75.0 | 75.0 | |||
| 3″″ | 78.6 | 78.6 | |||
| 4″″ | 70.7 | 70.8 | |||
| 5″″ | 67.2 | 67.2 | |||
| Glc (III) | |||||
| 1″″′ | 106.3 | ||||
| 2″″′ | 75.6 | ||||
| 3″″′ | 78.5 | ||||
| 4″″′ | 71.7 | ||||
| 5″″′ | 78.0 | ||||
| 6″″′ | 62.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimazaki, T.; Iguchi, T.; Takahashi, Y.; Yamamoto, K.; Takahashi, N.; Mimaki, Y. Determination of Structure and Cytotoxicity of Ten Undescribed Steroidal Glycosides from Allium cristophii × A. macleanii ‘Globemaster’. Molecules 2023, 28, 6248. https://doi.org/10.3390/molecules28176248
Shimazaki T, Iguchi T, Takahashi Y, Yamamoto K, Takahashi N, Mimaki Y. Determination of Structure and Cytotoxicity of Ten Undescribed Steroidal Glycosides from Allium cristophii × A. macleanii ‘Globemaster’. Molecules. 2023; 28(17):6248. https://doi.org/10.3390/molecules28176248
Chicago/Turabian StyleShimazaki, Tamami, Tomoki Iguchi, Yuna Takahashi, Kie Yamamoto, Naoki Takahashi, and Yoshihiro Mimaki. 2023. "Determination of Structure and Cytotoxicity of Ten Undescribed Steroidal Glycosides from Allium cristophii × A. macleanii ‘Globemaster’" Molecules 28, no. 17: 6248. https://doi.org/10.3390/molecules28176248
APA StyleShimazaki, T., Iguchi, T., Takahashi, Y., Yamamoto, K., Takahashi, N., & Mimaki, Y. (2023). Determination of Structure and Cytotoxicity of Ten Undescribed Steroidal Glycosides from Allium cristophii × A. macleanii ‘Globemaster’. Molecules, 28(17), 6248. https://doi.org/10.3390/molecules28176248






