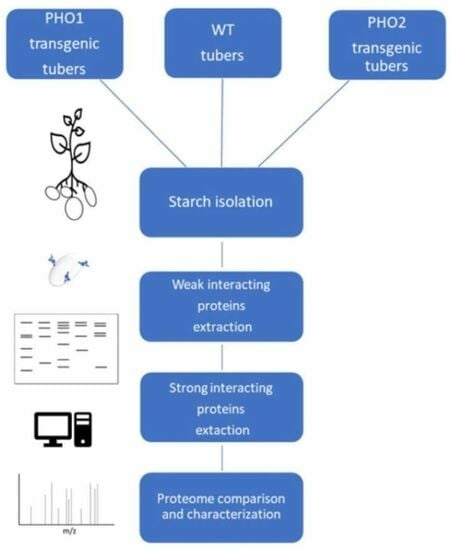
Gradual Analytics of Starch-Interacting Proteins Revealed the Involvement of Starch-Phosphorylating Enzymes during Synthesis of Storage Starch in Potato (Solanum tuberosum L.) Tubers
Abstract
:1. Introduction
2. Results
2.1. Proteome Profile of Starch-Interacting Proteins
2.2. Weak Interacting Proteins to Starch
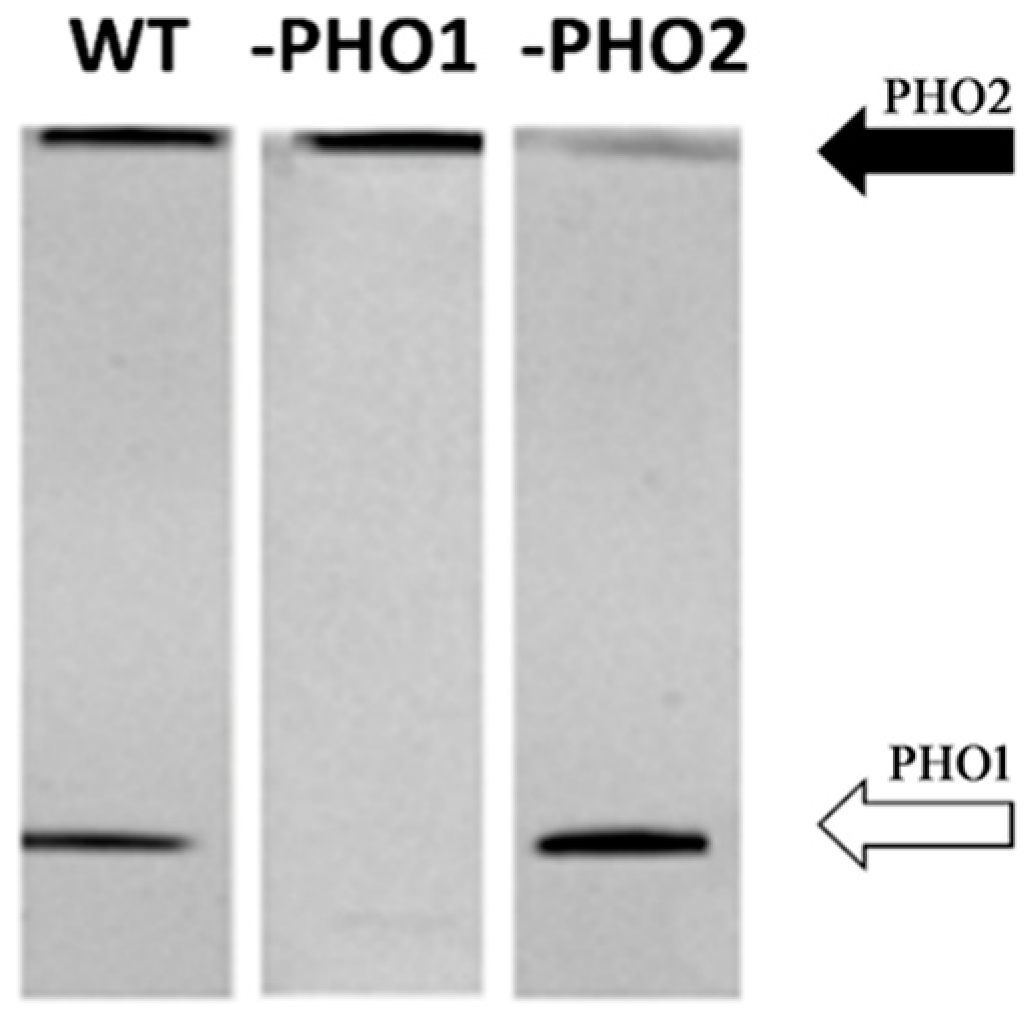
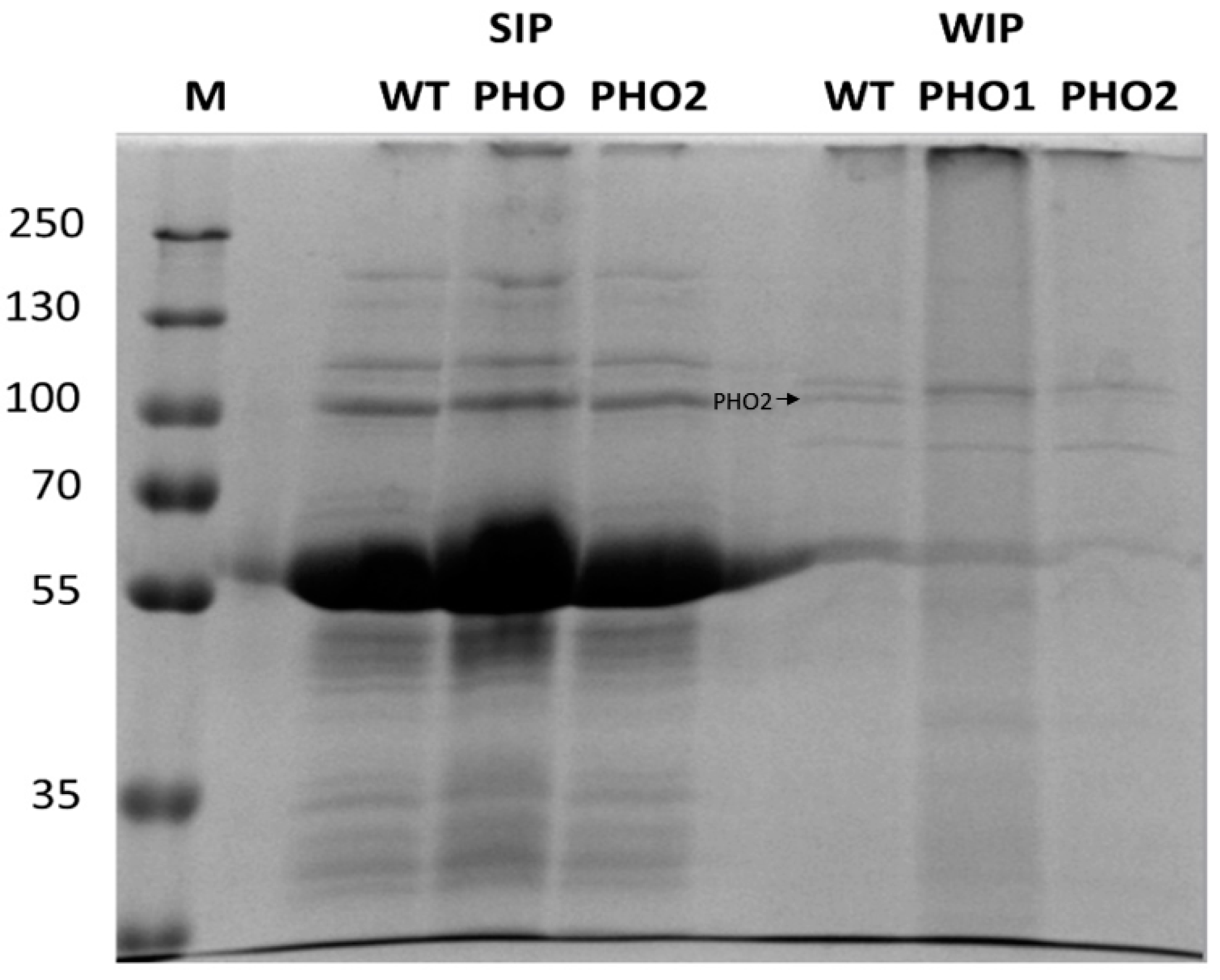
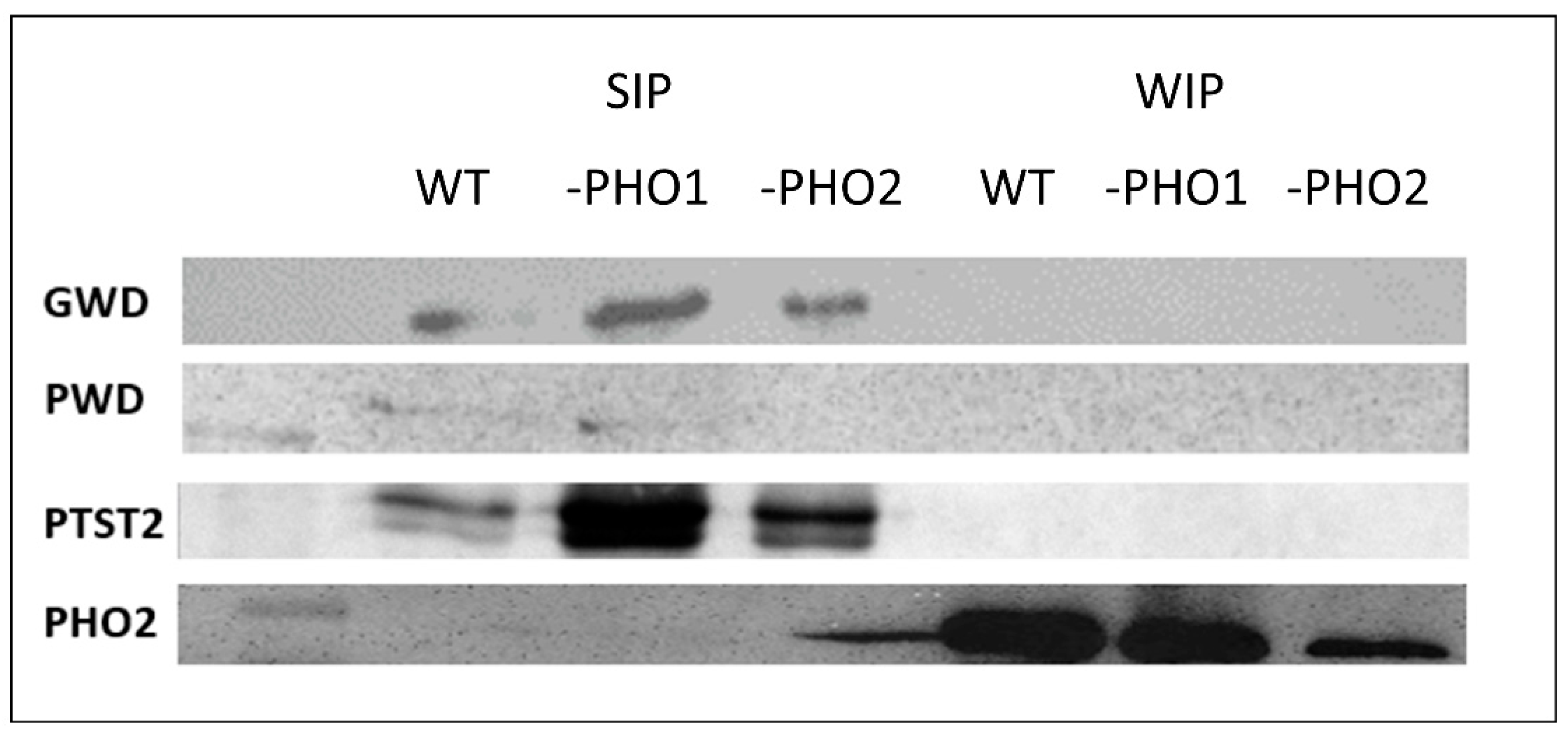
2.3. Starch-Interacting Proteins in Transgenic Lines
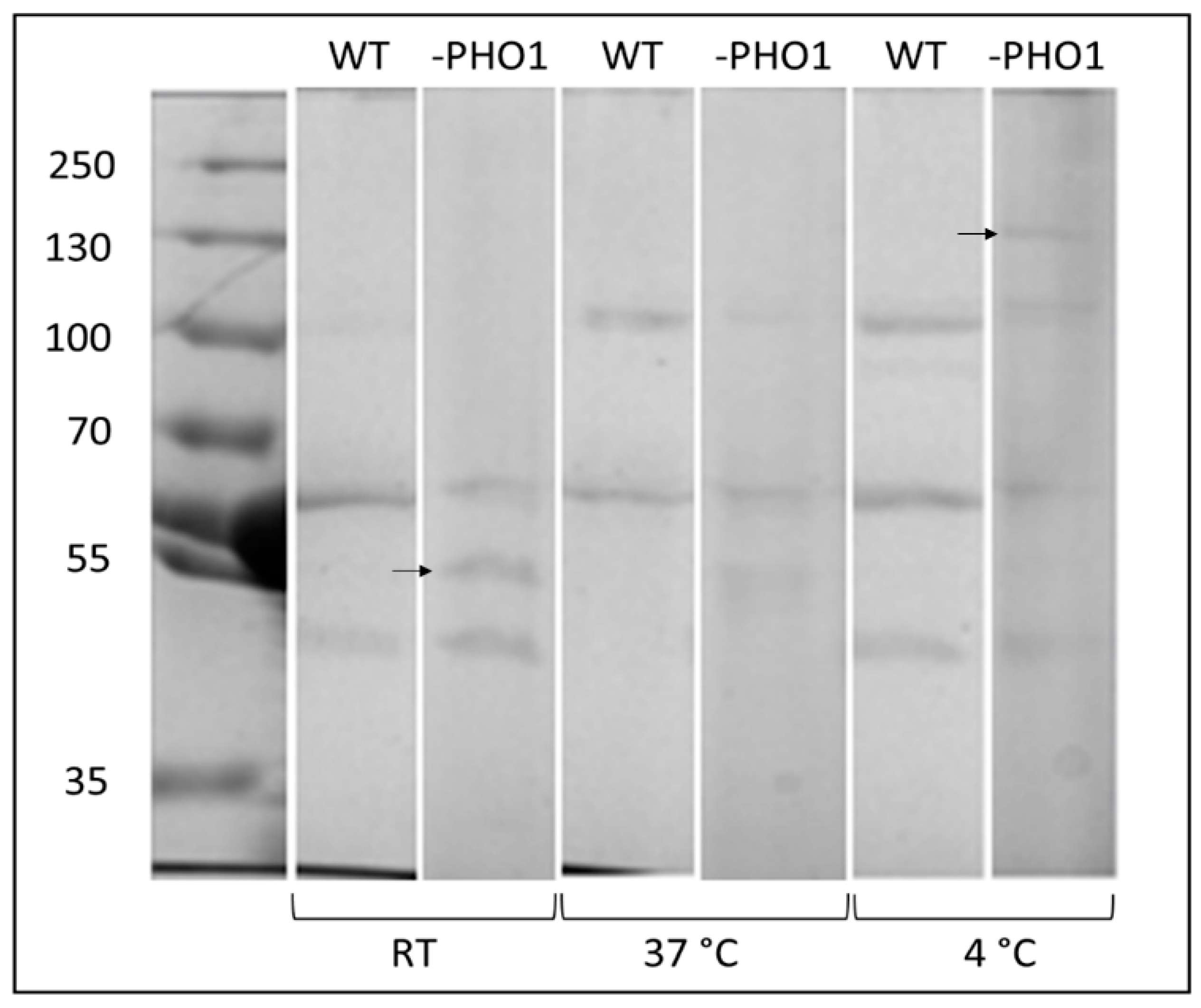
2.4. Changes in the Protein Band Pattern of WIP to Starch Was Distinguished When Tubers Were Stored under Different Conditions
3. Discussion
3.1. Proteins Were Mainly Contained in the SIP Fraction
3.2. PHO1 Is Not a Starch Granule-Bound Protein
3.3. Effects of Genotypes and Storing Conditions on Proteins Bound to Starch Granules
4. Conclusions
5. Materials and Methods
5.1. Biological Material
Potato Plants (Solanum tuberosum L.)
5.2. Antibodies
5.3. Methods
5.3.1. Starch Isolation
5.3.2. Detection of Phosphorylase Activity by Native PAGE
5.3.3. Extraction of Starch-Interacting Proteins
5.3.4. Concentration of Protein Samples
5.3.5. Determination of Protein Concentration
5.3.6. Electrophoretic Separation of Proteins
5.3.7. Quantitation of Protein Bands
5.3.8. Tryptic Digestion
5.3.9. Protein Identification
5.3.10. Western Blot
5.3.11. Immunodetection of Proteins
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ball, S.; Guan, H.P.; James, M.; Myers, A.; Keeling, P.; Mouille, G.; Buléon, A.; Colonna, P.; Preiss, J. From Glycogen to Amylopectin: A Model for the Biogenesis of the Plant Starch Granule. Cell 1996, 86, 349–352. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Bertoft, E. A Review of Starch Biosynthesis in Relation to the Building Block-Backbone Model. Int. J. Mol. Sci. 2020, 21, 7011. [Google Scholar] [CrossRef]
- Mérida, A.; Fettke, J. Starch Granule Initiation in Arabidopsis thaliana Chloroplasts. Plant J. 2021, 107, 688–697. [Google Scholar] [CrossRef]
- Shoaib, N.; Liu, L.; Ali, A.; Mughal, N.; Yu, G.; Huang, Y. Molecular Functions and Pathways of Plastidial Starch Phosphorylase (Pho1) in Starch Metabolism: Current and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 10450. [Google Scholar] [CrossRef] [PubMed]
- Helle, S.; Bray, F.; Verbeke, J.; Devassine, S.; Courseaux, A.; Facon, M.; Tokarski, C.; Rolando, C.; Szydlowski, N. Proteome Analysis of Potato Starch Reveals the Presence of New Starch Metabolic Proteins as Well as Multiple Protease Inhibitors. Front. Plant Sci. 2018, 9, 746. [Google Scholar] [CrossRef]
- Delvallé, D.; Dumez, S.; Wattebled, F.; Roldán, I.; Planchot, V.; Berbezy, P.; Colonna, P.; Vyas, D.; Chatterjee, M.; Ball, S.; et al. Soluble Starch Synthase I: A Major Determinant for the Synthesis of Amylopectin in Arabidopsis Thaliana Leaves. Plant J. 2005, 43, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Szydlowski, N.; Delvallé, D.; D’Hulst, C.; James, M.G.; Myers, A.M. Overlapping Functions of the Starch Synthases SSII and SSIII in Amylopectin Biosynthesis in Arabidopsis. BMC Plant Biol. 2008, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Seung, D.; Boudet, J.; Monroe, J.; Schreier, T.B.; David, L.C.; Abt, M.; Lu, K.J.; Zanella, M.; Zeeman, S.C. Homologs of PROTEIN TARGETING TO STARCH Control Starch Granule Initiation in Arabidopsis Leaves. Plant Cell 2017, 29, 1657–1677. [Google Scholar] [CrossRef] [PubMed]
- Sawada, T.; Itoh, M.; Nakamura, Y. Contributions of Three Starch Branching Enzyme Isozymes to the Fine Structure of Amylopectin in Rice Endosperm. Front. Plant Sci. 2018, 871, 1536. [Google Scholar] [CrossRef]
- Delatte, T.; Umhang, M.; Trevisan, M.; Eicke, S.; Thorneycroft, D.; Smith, S.M.; Zeeman, S.C. Evidence for Distinct Mechanisms of Starch Granule Breakdown in Plants. J. Biol. Chem. 2006, 281, 12050–12059. [Google Scholar] [CrossRef]
- Lin, Q.; Facon, M.; Putaux, J.L.; Dinges, J.R.; Wattebled, F.; D’Hulst, C.; Hennen-Bierwagen, T.A.; Myers, A.M. Function of Isoamylase-Type Starch Debranching Enzymes ISA1 and ISA2 in the Zea Mays Leaf. New Phytol. 2013, 200, 1009–1021. [Google Scholar] [CrossRef]
- Streb, S.; Zeeman, S.C. Replacement of the Endogenous Starch Debranching Enzymes ISA1 and ISA2 of Arabidopsis with the Rice Orthologs Reveals a Degree of Functional Conservation during Starch Synthesis. PLoS ONE 2014, 9, e92174. [Google Scholar] [CrossRef] [PubMed]
- Comparot-Moss, S.; Kötting, O.; Stettler, M.; Edner, C.; Graf, A.; Weise, S.E.; Streb, S.; Lue, W.L.; MacLean, D.; Mahlow, S.; et al. A Putative Phosphatase, LSF1, Is Required for Normal Starch Turnover in Arabidopsis Leaves. Plant Physiol. 2010, 152, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Santelia, D.; Kötting, O.; Seung, D.; Schubert, M.; Thalmann, M.; Bischof, S.; Meekins, D.A.; Lutz, A.; Patron, N.; Gentry, M.S.; et al. The Phosphoglucan Phosphatase like Sex Four2 Dephosphorylates Starch at the C3-Position in Arabidopsis. Plant Cell 2011, 23, 4096–4111. [Google Scholar] [CrossRef]
- Zhou, W.; He, S.; Naconsie, M.; Ma, Q.; Zeeman, S.C.; Gruissem, W.; Zhang, P. Alpha-Glucan, Water Dikinase 1 Affects Starch Metabolism and Storage Root Growth in Cassava (Manihot Esculenta Crantz). Sci. Rep. 2017, 7, 9863. [Google Scholar] [CrossRef] [PubMed]
- Mak, C.A.; Weis, K.; Henao, T.; Kuchtova, A.; Chen, T.; Sharma, S.; Meekins, D.A.; Thalmann, M.; Vander Kooi, C.W.; Raththagala, M. Cooperative Kinetics of the Glucan Phosphatase Starch Excess4. Biochemistry 2021, 60, 2425–2435. [Google Scholar] [CrossRef]
- Lorberth, R.; Ritte, G.; Willmitzer, L.; Kossmann, J. Inhibition of a Starch-Granule-Bound Protein Leads to Modified Starch and Repression of Cold Sweetening. Nat. Biotechnol. 1998, 16, 473–477. [Google Scholar] [CrossRef]
- Nielsen, T.H.; Wischmann, B.; Enevoldsen, K.; Møller, B.L. Starch Phosphorylation in Potato Tubers Proceeds Concurrently with de Novo Biosynthesis of Starch. Plant Physiol. 1994, 105, 111–117. [Google Scholar] [CrossRef]
- Ritte, G.; Heydenreich, M.; Mahlow, S.; Haebel, S.; Kötting, O.; Steup, M. Phosphorylation of C6- and C3-Positions of Glucosyl Residues in Starch Is Catalysed by Distinct Dikinases. FEBS Lett. 2006, 580, 4872–4876. [Google Scholar] [CrossRef]
- Samodien, E.; Jewell, J.F.; Loedolff, B.; Oberlander, K.; George, G.M.; Zeeman, S.C.; Damberger, F.F.; van der Vyver, C.; Kossmann, J.; Lloyd, J.R. Repression of SEX4 and LIKE SEX FOUR2 Orthologs in Potato Increases Tuber Starch Bound Phosphate with Concomitant Alterations in Starch Physical Properties. Front. Plant Sci. 2018, 9, 1044. [Google Scholar] [CrossRef]
- Zeeman, S.C.; Northrop, F.; Smith, A.M.; Ap Rees, T. A Starch-Accumulating Mutant of Arabidopsis thaliana Deficient in a Chloroplastic Starch-Hydrolysing Enzyme. Plant J. 1998, 15, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Zeeman, S.C.; Rees, T.A. Changes in Carbohydrate Metabolism and Assimilate Export in Starch-Excess Mutants of Arabidopsis. Plant Cell Environ. 1999, 22, 1445–1453. [Google Scholar] [CrossRef]
- Feike, D.; Seung, D.; Graf, A.; Bischof, S.; Ellick, T.; Coiro, M.; Soyk, S.; Eicke, S.; Mettler-Altmann, T.; Lu, K.J.; et al. The Starch Granule-Associated Protein EARLY STARVATION1 Is Required for the Control of Starch Degradation in Arabidopsis thaliana Leaves. Plant Cell 2016, 28, 1472–1489. [Google Scholar] [CrossRef] [PubMed]
- Helle, S.; Bray, F.; Putaux, J.L.; Verbeke, J.; Flament, S.; Rolando, C.; D’Hulst, C.; Szydlowski, N. Intra-Sample Heterogeneity of Potato Starch Reveals Fluctuation of Starch-Binding Proteins According to Granule Morphology. Plants 2019, 8, 324. [Google Scholar] [CrossRef]
- Abt, M.R.; Pfister, B.; Sharma, M.; Eicke, S.; Bürgy, L.; Neale, I.; Seung, D.; Zeeman, S.C. STARCH SYNTHASE5, a Noncanonical Starch Synthase-like Protein, Promotes Starch Granule Initiation in Arabidopsis. Plant Cell 2020, 32, 2543–2565. [Google Scholar] [CrossRef]
- Flores-Castellanos, J.; Fettke, J. The Plastidial Glucan Phosphorylase Affects the Maltooligosaccharide Metabolism in Parenchyma Cells of Potato (Solanum Tuberosum, L.) Tuber Discs. Plant Cell Physiol. 2023, 64, 422–432. [Google Scholar] [CrossRef]
- Orawetz, T.; Malinova, I.; Orzechowski, S.; Fettke, J. Reduction of the Plastidial Phosphorylase in Potato (Solanum Tuberosum, L.) Reveals Impact on Storage Starch Structure during Growth at Low Temperature. Plant Physiol. Biochem. PPB 2016, 100, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Green, A.R.; Nissen, M.S.; Mohan Kumar, G.N.; Knowles, N.R.; Kang, C.H. Characterization of Solanum Tuberosum Multicystatin and the Significance of Core Domains. Plant Cell 2013, 25, 5043–5052. [Google Scholar] [CrossRef]
- Rodis, P.; Hoff, J.E. Naturally Occurring Protein Crystals in the Potato. Plant Physiol. 1984, 74, 907–911. [Google Scholar] [CrossRef]
- Nissen, M.S.; Kumar, G.N.M.; Youn, B.; Knowles, D.B.; Lam, K.S.; Ballinger, W.J.; Knowles, N.R.; Kang, C. Characterization of Solanum Tuberosum Multicystatin and Its Structural Comparison with Other Cystatins. Plant Cell 2009, 21, 861–875. [Google Scholar] [CrossRef]
- Stiekema, W.J.; Heidekamp, F.; Dirkse, W.G.; van Beckum, J.; de Haan, P.; ten Bosch, C.; Louwerse, J.D. Molecular Cloning and Analysis of Four Potato Tuber MRNAs. Plant Mol. Biol. 1988, 11, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Ohta, S.; Matsuoka, K.; Hattori, T.; Nakamura, K. A Family of Potato Genes That Encode Kunitz-Type Proteinase Inhibitors: Structural Comparisons and Differential Expression. Plant Cell Physiol. 1994, 35, 303–312. [Google Scholar] [CrossRef]
- Fettke, J.; Poeste, S.; Eckermann, N.; Tiessen, A.; Pauly, M.; Geigenberger, P.; Steup, M. Analysis of Cytosolic Heteroglycans from Leaves of Transgenic Potato (Solanum tuberosum, L.) Plants That under- or Overexpress the Pho 2 Phosphorylase Isozyme. Plant Cell Physiol. 2005, 46, 1987–2004. [Google Scholar] [CrossRef]
- Mahlow, S.; Hejazi, M.; Kuhnert, F.; Garz, A.; Brust, H.; Baumann, O.; Fettke, J. Phosphorylation of Transitory Starch by α-Glucan, Water Dikinase during Starch Turnover Affects the Surface Properties and Morphology of Starch Granules. New Phytol. 2014, 203, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, L.; Gruppen, H.; Piersma, S.R.; Van den Broek, L.A.M.; Van Koningsveld, G.A.; Voragen, A.G.J. Relative Abundance and Inhibitory Distribution of Protease Inhibitors in Potato Juice from Cv. Elkana. J. Agric. Food Chem. 2001, 49, 2864–2874. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, L.; Gruppen, H.; Van Koningsveld, G.A.; Van Den Broek, L.A.M.; Voragen, A.G.J. The Most Abundant Protease Inhibitor in Potato Tuber (Cv. Elkana) Is a Serine Protease Inhibitor from the Kunitz Family. J. Agric. Food Chem. 2003, 51, 5001–5005. [Google Scholar] [CrossRef]
- Jørgensen, M.; Bauw, G.; Welinder, K.G. Molecular Properties and Activities of Tuber Proteins from Starch Potato Cv. Kuras. J. Agric. Food Chem. 2006, 54, 9389–9397. [Google Scholar] [CrossRef]
- Ritte, G.; Eckermann, N.; Haebel, S.; Lorberth, R.; Steup, M. Compartmentation of the Starch-Related R1 Protein in Higher Plants. Starch/Staerke 2000, 52, 179–185. [Google Scholar] [CrossRef]
- Kötting, O.; Pusch, K.; Tiessen, A.; Geigenberger, P.; Steup, M.; Ritte, G. Identification of a Novel Enzyme Required for Starch Metabolism in Arabidopsis Leaves. The Phosphoglucan, Water Dikinase. Plant Physiol. 2005, 137, 242–252. [Google Scholar] [CrossRef]
- Webster, J.; Oxley, D. Protein identification by MALDI-TOF mass spectrometry. Methods Mol. Biol. 2012, 800, 227–240. [Google Scholar] [CrossRef]
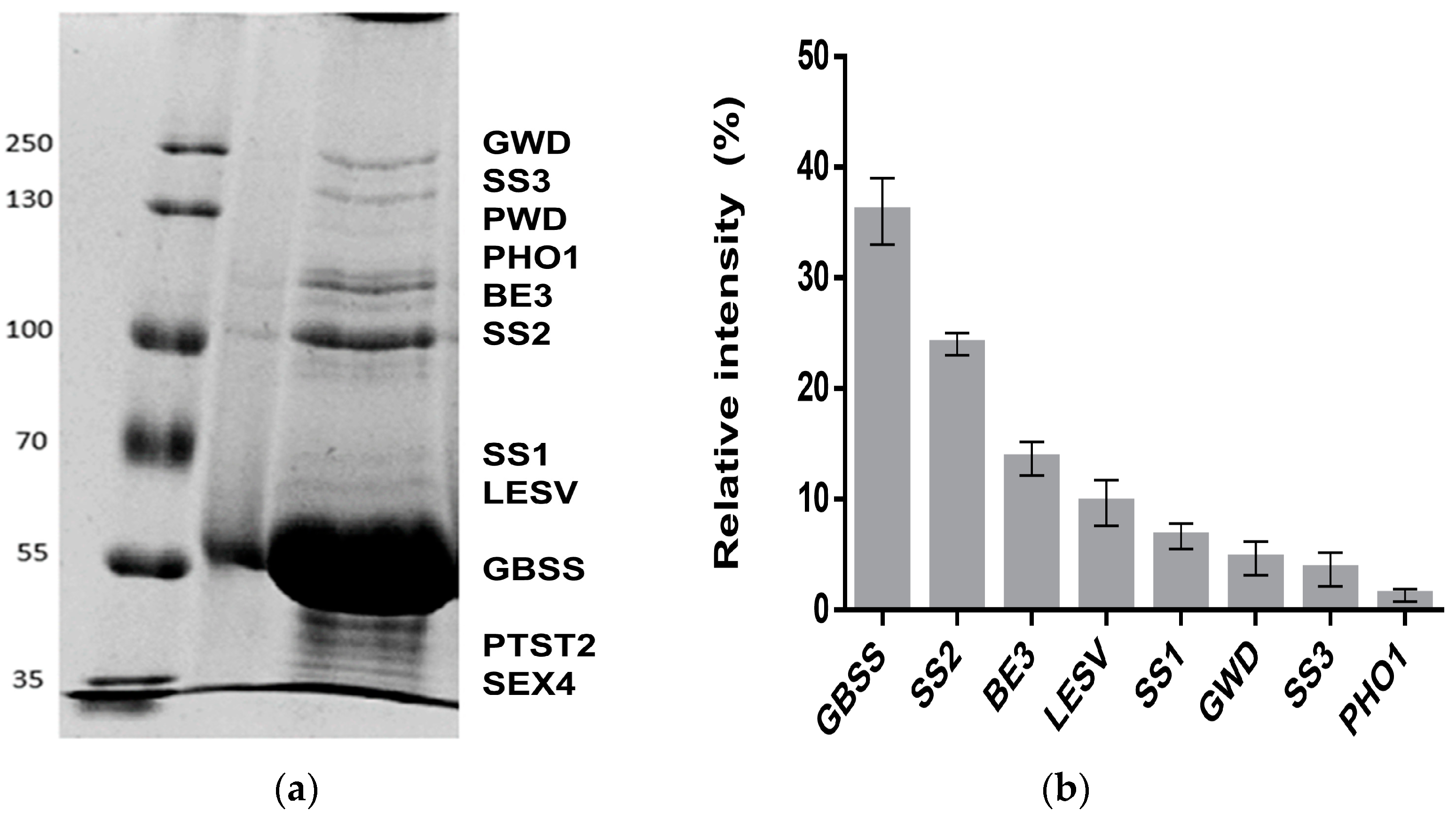

| Protein Name | Accession Number | Predicted MW (KDa) | Seq. Cov. (%) | Score | Identification Method | Cell Location |
|---|---|---|---|---|---|---|
| GWD | Q9AWA5 | 163.14 | 12 | 105 | MALDI-TOF | Chloro |
| SS3 | Q43846 | 139.02 | 18 | 80 | MALDI-TOF | Chloro |
| PWD | D2JRZ6 | 132.28 | N.A. | Immunodetected | Chloro | |
| BE3 | P30924 | 99.02 | 9 | 116 | MS/MS | Chloro |
| SS2 | Q43847 | 85.17 | 27 | 159 | MALDI-TOF | Chloro |
| SS1 | P93568 | 70.60 | 13 | 80 | MS/MS | Chloro |
| GBSS | Q00775 | 66.58 | 24 | 138 | MALDI-TOF | Chloro |
| LESV | Soltu.DM.06G014960.1 | 63.40 | 21 | 77 | MS/MS | Chloro |
| PTST2 | XP_006367350 | 47.26 | N.A. | Immunodetected | Chloro | |
| SEX4 | Soltu.DM.11G004900.2 | 41.57 | N.A. | Immuno-detected | Chloro |
| Protein Name | Accession Number | Predicted MW (KDa) | Seq. Cov. (%) | Score | Identification Method | Cell Location |
|---|---|---|---|---|---|---|
| PHO2 | P32811 | 95.05 | 23 | 108 | MALDI-TOF | Cytosol |
| LOX 13 Probable linoleate 9S-11 oxygenate | Q43189 | 96.98 | 11 | MALDI-TOF | Cytosol | |
| Multicystatin | P37842 | 86.71 | 16 | MALDI-TOF | N.D. | |
| Probable inactive patatin 3-Kuras PT3 | Q3YJS9 | 41.11 | 8 | MS/MS | Vacuole | |
| Kunitz type protease inhibitor | AAB32802.1 | 24.50 | 51 | MS/MS | Vacuole | |
| Cysteine protease inhibitor 10 (fragment) | O24383 | 20.96 | 37 | MALDI-TOF | Vacuole | |
| Cysteine protease inhibitor 8 (fragment) | O24384 | 24.69 | 31 | MALDI-TOF | Vacuole |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castellanos, J.F.; Khan, A.; Fettke, J. Gradual Analytics of Starch-Interacting Proteins Revealed the Involvement of Starch-Phosphorylating Enzymes during Synthesis of Storage Starch in Potato (Solanum tuberosum L.) Tubers. Molecules 2023, 28, 6219. https://doi.org/10.3390/molecules28176219
Castellanos JF, Khan A, Fettke J. Gradual Analytics of Starch-Interacting Proteins Revealed the Involvement of Starch-Phosphorylating Enzymes during Synthesis of Storage Starch in Potato (Solanum tuberosum L.) Tubers. Molecules. 2023; 28(17):6219. https://doi.org/10.3390/molecules28176219
Chicago/Turabian StyleCastellanos, Junio Flores, Arsalan Khan, and Joerg Fettke. 2023. "Gradual Analytics of Starch-Interacting Proteins Revealed the Involvement of Starch-Phosphorylating Enzymes during Synthesis of Storage Starch in Potato (Solanum tuberosum L.) Tubers" Molecules 28, no. 17: 6219. https://doi.org/10.3390/molecules28176219
APA StyleCastellanos, J. F., Khan, A., & Fettke, J. (2023). Gradual Analytics of Starch-Interacting Proteins Revealed the Involvement of Starch-Phosphorylating Enzymes during Synthesis of Storage Starch in Potato (Solanum tuberosum L.) Tubers. Molecules, 28(17), 6219. https://doi.org/10.3390/molecules28176219