The Mechanism of Anti-Tumor Activity of 6-Morpholino- and 6-Amino-9-Sulfonylpurine Derivatives on Human Leukemia Cells
Abstract
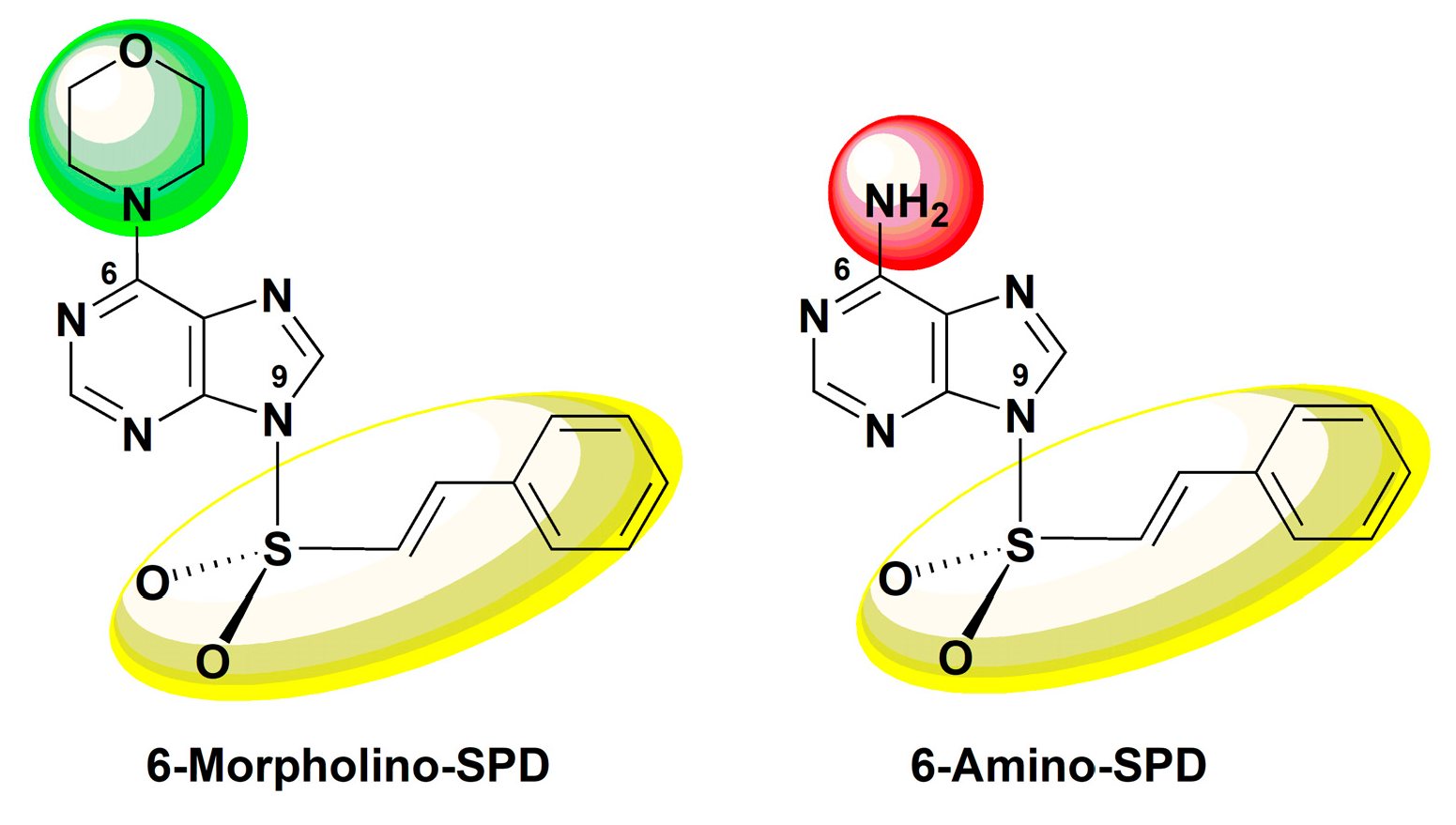
:1. Introduction
2. Results
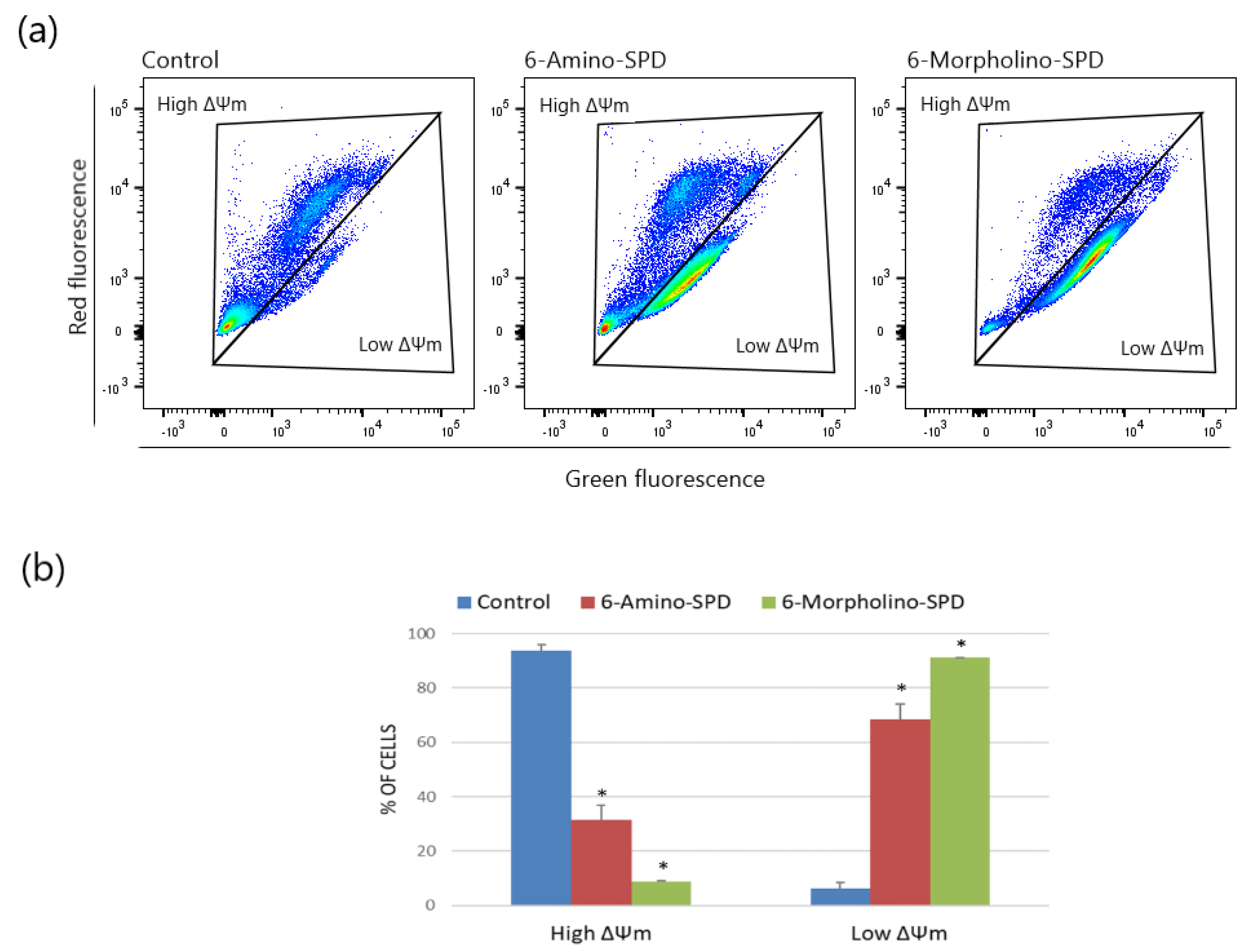
2.1. Effects of 6-Amino-SPD and 6-Morpholino-SPD on Apoptosis Induction and on Mitochondrial Membrane Potential (ΔΨm) in Leukemia Cells
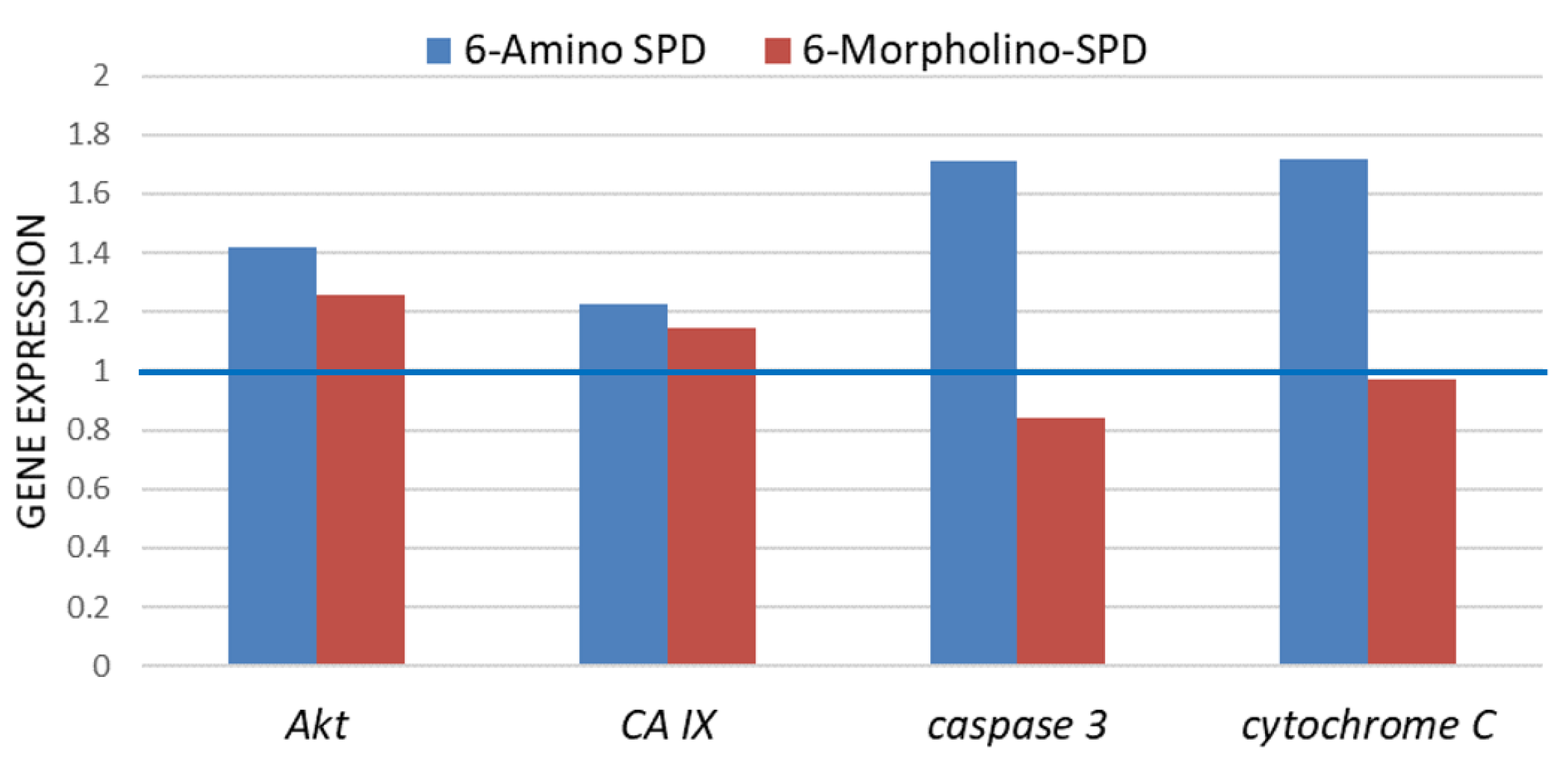
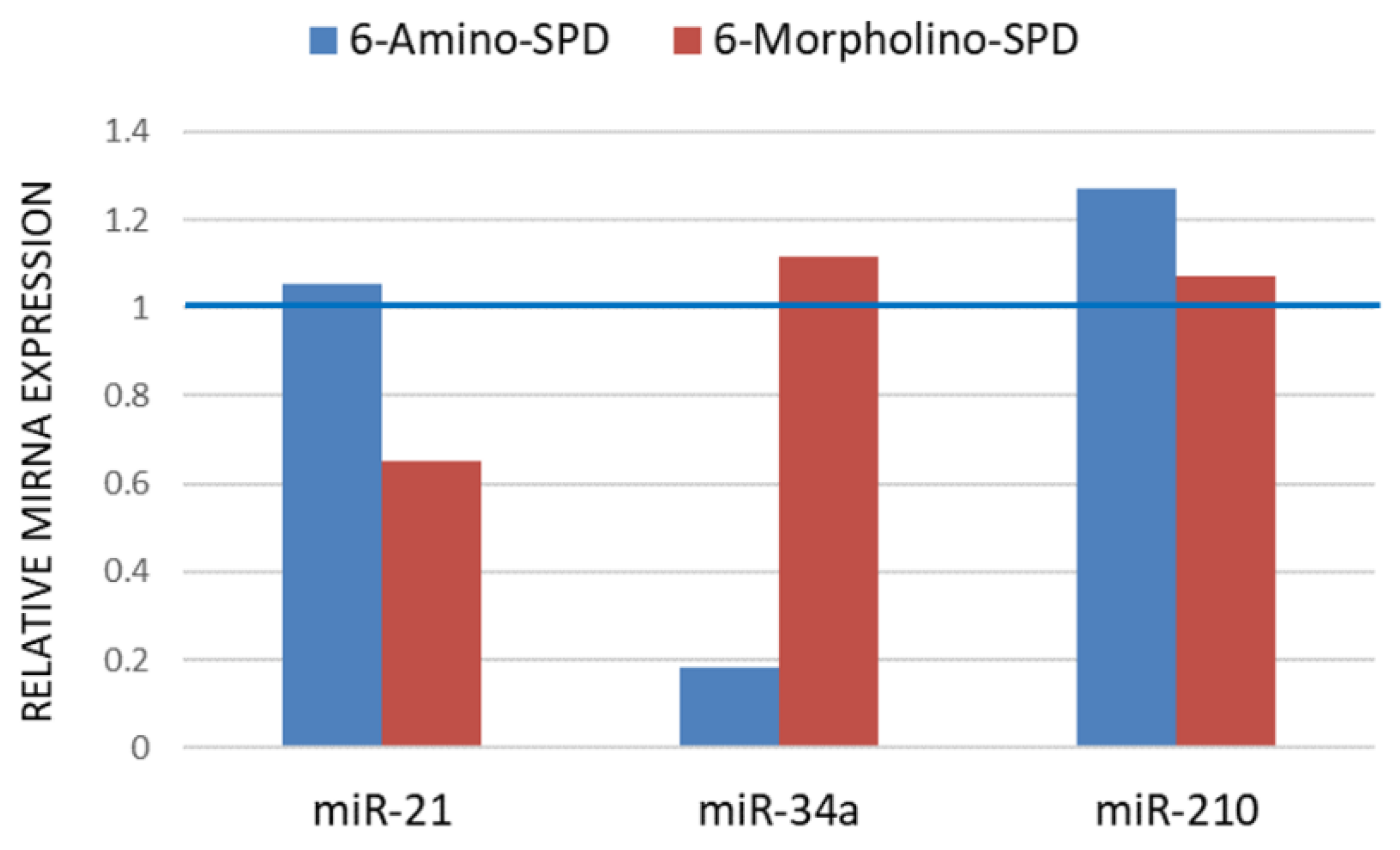
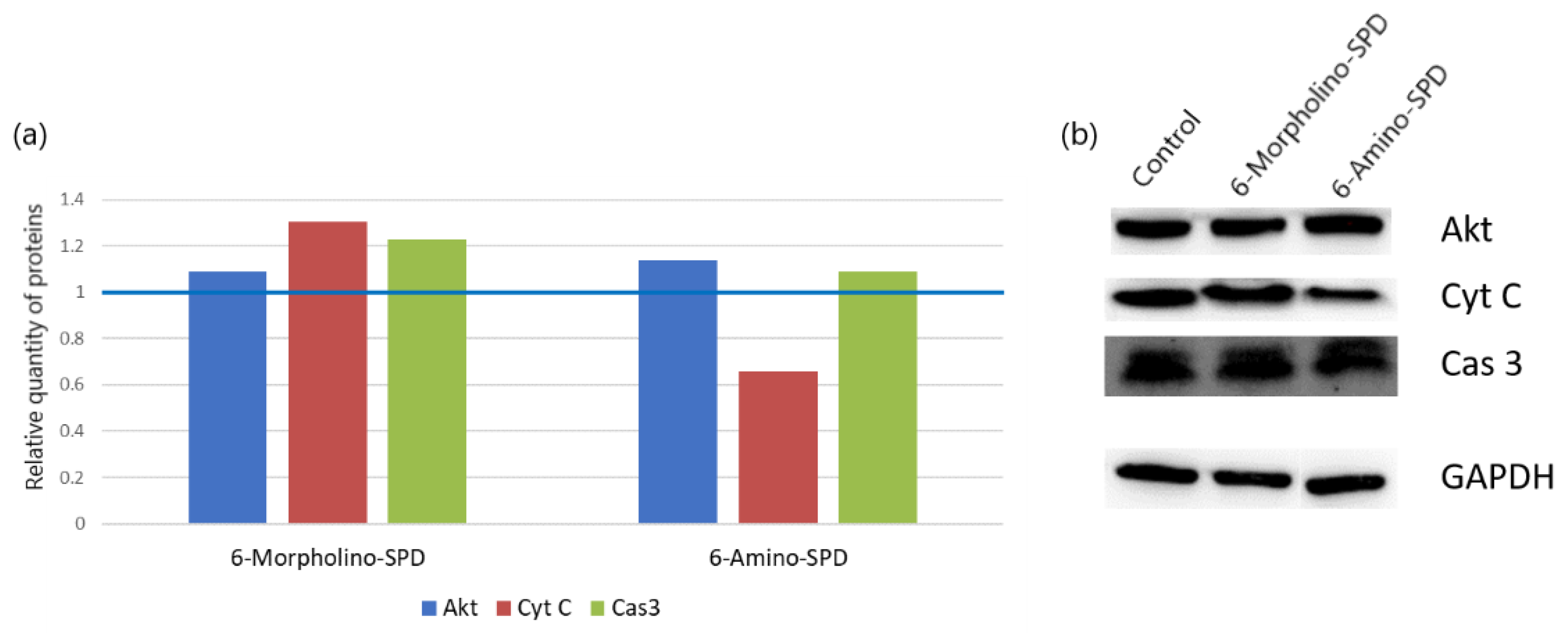
2.2. Effects of 6-Amino-SPD and 6-Morpholino-SPD on Cytochrome c, Caspase 3, Akt, and CA IX Genes, and microRNA (miRNA) Expression Profiles
2.3. Changes in Protein Synthesis
2.4. ADMET Properties
3. Discussion
4. Materials and Methods
4.1. Cell Culturing
4.2. Annexin V/PI Assay
4.3. Measurement of ΔΨm
4.4. Determination of Intracellular Reactive Oxygen Species (ROS)
4.5. Real-Time Quantitative PCR (qPCR)
4.6. miRNA Expression
4.7. Western Blot
4.8. ADMET Properties Prediction
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Nocentini, A.; Bua, S.; Lomelino, C.L.; McKenna, R.; Menicatti, M.; Bartolucci, G.; Tenci, B.; Di Cesare Mannelli, L.; Ghelardini, C.; Gratteri, P.; et al. Discovery of New Sulfonamide Carbonic Anhydrase IX Inhibitors Incorporating Nitrogenous Bases. ACS Med. Chem. Lett. 2017, 8, 1314–1319. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.A.; Tüzün, B.; Alsaif, N.A.; Khan, A.A.; Naglah, A.M. Synthesis, characterization, molecular modeling against EGFR target and ADME/T analysis of novel purine derivatives of sulfonamides. J. Mol. Struct. 2022, 1257, 132600. [Google Scholar] [CrossRef]
- Pastewska, M.; Żołnowska, B.; Kovačević, S.; Kapica, H.; Gromelski, M.; Stoliński, F.; Sławiński, J.; Sawicki, W.; Ciura, K. Modeling of Anticancer Sulfonamide Derivatives Lipophilicity by Chemometric and Quantitative Structure-Retention Relationships Approaches. Molecules 2022, 27, 3965. [Google Scholar] [CrossRef] [PubMed]
- Abas, M.; Bahadur, A.; Ashraf, Z.; Iqbal, S.; Riaz Rajoka, M.S.; Rashid, S.G.; Jabeen, E.; Iqbal, Z.; Abbas, Q.; Bais, A.; et al. Designing novel anticancer sulfonamide based 2,5-disubstituted-1,3,4-thiadiazole derivatives as potential carbonic anhydrase inhibitor. J. Mol. Struct. 2021, 1246, 131145. [Google Scholar] [CrossRef]
- Nitulescu, G.M.; Margina, D.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Saloustros, E.; Fenga, C.; Spandidos, D.A.; Libra, M.; Tsatsakis, A.M. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use. Int. J. Oncol. 2016, 48, 869–885. [Google Scholar] [CrossRef]
- Al-Rawashde, F.A.; Al-Wajeeh, A.S.; Vishkaei, M.N.; Saad, H.K.M.; Johan, M.F.; Taib, W.R.W.; Ismail, I.; Al-Jamal, H.A.N. Thymoquinone Inhibits JAK/STAT and PI3K/Akt/mTOR Signaling Pathways in MV4-11 and K562 Myeloid Leukemia Cells. Pharmaceuticals 2022, 15, 1123. [Google Scholar] [CrossRef]
- Gilmore, A.; King, L. Emerging approaches to target mitochondrial apoptosis in cancer cells. F1000Research 2019, 8, 1–11. [Google Scholar] [CrossRef]
- Markowicz-Piasecka, M.; Huttunen, J.; Zajda, A.; Sikora, J.; Huttunen, K.M. Sulfonamide metformin derivatives induce mitochondrial-associated apoptosis and cell cycle arrest in breast cancer cells. Chem. Biol. Interact. 2022, 352, 109795. [Google Scholar] [CrossRef]
- King, L.E.; Rodriguez-Enriquez, R.; Pedley, R.; Mellor, C.E.; Wang, P.; Zindy, E.; White, M.R.; Brennan, K.; Gilmore, A.P. Apoptotic priming is defined by the dynamic exchange of Bcl-2 proteins between mitochondria and cytosol. Cell Death Differ. 2022, 29, 2262–2274. [Google Scholar] [CrossRef]
- Koyuncu, I.; Tülüce, Y.; Slahaddin Qadir, H.; Durgun, M.; Supuran, C.T. Evaluation of the anticancer potential of a sulphonamide carbonic anhydrase IX inhibitor on cervical cancer cells. J. Enzym. Inhib. Med. Chem. 2019, 34, 703–711. [Google Scholar] [CrossRef]
- Petreni, A.; Bonardi, A.; Lomelino, C.; Osman, S.M.; ALOthman, Z.A.; Eldehna, W.M.; El-Haggar, R.; McKenna, R.; Nocentini, A.; Supuran, C.T. Inclusion of a 5-fluorouracil moiety in nitrogenous bases derivatives as human carbonic anhydrase IX and XII inhibitors produced a targeted action against MDA-MB-231 and T47D breast cancer cells. Eur. J. Med. Chem. 2020, 15, 112112. [Google Scholar] [CrossRef] [PubMed]
- Combs, J.; Bozdag, M.; Cravey, L.D.; Kota, A.; McKenna, R.; Angeli, A.; Carta, F.; Supuran, C.T. New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors. Molecules 2023, 28, 890. [Google Scholar] [CrossRef] [PubMed]
- Gaál, Z. Implication of microRNAs in Carcinogenesis with Emphasis on Hematological Malignancies and Clinical Translation. Int. J. Mol. Sci. 2022, 23, 5838. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, D.; Brown, E.; Javadala, J.; Uysal-Onganer, P.; Guinn, B. microRNA expression in acute myeloid leukaemia: New targets for therapy? EJHaem 2022, 3, 596–608. [Google Scholar] [CrossRef]
- Schmidt, M.F. Drug target miRNAs: Chances and challenges. Trends Biotechnol. 2014, 32, 578–585. [Google Scholar] [CrossRef]
- Otmani, K.; Lewalle, P. Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front. Oncol. 2021, 11, 708765. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Chen, X.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Matić, J.; Jukić, M.; Ismaili, H.; Saftić, D.; Ban, Ž.; Tandarić, T.; Vianello, R.; Opačak-Bernardi, T.; Glavaš-Obrovac, L.; Žinić, B. 6-Morpholino- and 6-amino-9-sulfonylpurine derivatives. Synthesis, computational analysis, and biological activity. Nucleosides Nucleotides Nucleic Acids 2021, 40, 470–503. [Google Scholar] [CrossRef]
- Žinić, B.; Žinić, M.; Krizmanić, I. Synthesis of the Sulfonypyrimidine Derivatives with Anticancer Activity, EP 0 977 022 B1. 2004. Available online: https://www.bib.irb.hr/162702?rad=162702 (accessed on 21 March 2023).
- Saftić, D.; Vianello, R.; Žinić, B. 5-Triazolyluracils and Their N1-Sulfonyl Derivatives: Intriguing Reactivity Differences in the Sulfonation of Triazole N1′-Substituted and N1′-Unsubstituted Uracil Molecules. Eur. J. Org. Chem. 2015, 35, 7695–7704. [Google Scholar] [CrossRef]
- Matić, J.; Nekola, I.; Višnjevac, A.; Kobetić, R.; Martin-Kleiner, I.; Kralj, M.; Žinić, B. C5-Morpholinomethylation of N1-sulfonylcytosines by a one-pot microwave assisted Mannich reaction. Org. Biomol. Chem. 2018, 16, 2678–2687. [Google Scholar] [CrossRef] [PubMed]
- Glavaš-Obrovac, L.; Karner, I.; Žinić, B.; Pavelić, K. Antineoplastic activity of novel N-1-sulfonypyrimidine derivatives. Anticancer. Res. 2001, 21, 1979–1986. Available online: https://pubmed.ncbi.nlm.nih.gov/11497287/ (accessed on 21 March 2023). [PubMed]
- Oprea, T.I. Property distribution of drug-related chemical databases. J. Comput. Aided Mol. Des. 2000, 14, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Burkner, G.T.; Dias, D.A.; Souza, K.F.S.D.; Araújo, A.J.P.D.; Basilio, D.C.L.S.; Jacobsen, F.T.; Moraes, A.C.R.D.; Silva-Filho, S.E.; Cavalcante, M.F.D.O.; Moraes, C.A.D.O.; et al. Selenylated Imidazo[1,2-a]pyridine Induces Cell Senescence and Oxidative Stress in Chronic Myeloid Leukemia Cells. Molecules 2023, 28, 893. [Google Scholar] [CrossRef] [PubMed]
- Haneef, J.; Parvathy, M.M.P.; Thankayyan, R.S.K.; Sithul, H.; Sreeharshan, S. Bax translocation mediated mitochondrial apoptosis and caspase dependent photosensitizing effect of Ficus religiosa on cancer cells. PLoS ONE 2012, 7, e40055. [Google Scholar] [CrossRef]
- Pogacar, Z.; Johnson, J.L.; Krenning, L.; De Conti, G.; Jochems, F.; Lieftink, C.; Velds, A.; Wardak, L.; Groot, K.; Schepers, A.; et al. Indisulam synergizes with palbociclib to induce senescence through inhibition of CDK2 kinase activity. PLoS ONE 2022, 17, e0273182. [Google Scholar] [CrossRef]
- Nijhuis, A.; Sikka, A.; Yogev, O.; Herendi, L.; Eckold, C.; Valbuena, G.; Liu, Y.; Kouloura, E.; Poon, E.; Martins da Costa, B.; et al. Indisulam targets RNA splicing and metabolism to serve as a therapeutic strategy for high-risk neuroblastoma. Nat. Commun. 2022, 13, 1380. [Google Scholar] [CrossRef]
- Monção, C.C.D.; Scrideli, C.A.; Andrade, A.F.; Viapiano, M.S.; Carlotti, C.G.; Moreno, D.A.; Baroni, M.; Tone, L.G.; Teixeira, S.A. Indisulam Reduces Viability and Regulates Apoptotic Gene Expression in Pediatric High-Grade Glioma Cells. Biomedicines 2023, 11, 68. [Google Scholar] [CrossRef]
- Salas, C.; Zarate, A.M.; Kryštof, V.; Mella, J.; Faundez, M.; Brea, J.; Loza, M.I.; Brito, I.; Hendrychová, D.; Jorda, R.; et al. Promising 2,6,9-Trisubstituted Purine Derivatives for Anticancer Compounds: Synthesis, 3D-QSAR, and Preliminary Biological Assays. Int. J. Mol. Sci. 2020, 21, 161. [Google Scholar] [CrossRef]
- Ricci, J.-E.; Muñoz-Pinedo, C.; Fitzgerald, P.; Bailly-Maitre, B.; Perkins, G.A.; Yadava, N.; Scheffler, I.E.; Ellisman, M.H.; Gree, D.R. Disruption of Mitochondrial Function during Apoptosis Is Mediated by Caspase Cleavage of the p75 Subunit of Complex I of the Electron Transport Chain. Cell 2004, 117, 773–786. [Google Scholar] [CrossRef]
- Bhadra, K. A Mini Review on Molecules Inducing Caspase-Independent Cell Death: A New Route to Cancer Therapy. Molecules 2022, 27, 6401. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Liu, Y.; Li, Z.; Zhu, P.; Zhao, M. Mitochondria Related Cell Death Modalities and Disease. Front. Cell Dev. Biol. 2022, 10, 832356. [Google Scholar] [CrossRef] [PubMed]
- Song, L.L.; Tu, Y.Y.; Xia, L.; Wang, W.W.; Wei, W.; Ma, C.M.; Wen, D.H.; Lei, H.; Xu, H.Z.; Wu, Y.L. Targeting Catalase but Not Peroxiredoxins Enhances Arsenic Trioxide-Induced Apoptosis in K562 Cells. PLoS ONE 2014, 9, e104985. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Mu, T.; Wang, G.; Jiang, X. Mitochondria-mediated apoptosis in mammals. Protein Cell 2014, 5, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadis, T.; Pissas, G.; Liakopoulos, V.; Stefanidis, I. Cytochrome c as a Potentially Clinical Useful Marker of Mitochondrial and Cellular Damage. Front. Immunol. 2016, 7, 279. [Google Scholar] [CrossRef]
- Chandra, D.; Liu, J.W.; Tang, D.G. Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J. Biol. Chem. 2002, 277, 50842–50854. [Google Scholar] [CrossRef]
- Liu, G.; Zou, H.; Luo, T.; Long, M.; Bian, J.; Liu, X.; Gu, J.; Yuan, Y.; Song, R.; Wang, Y.; et al. Caspase-Dependent and Caspase-Independent Pathways Are Involved in Cadmium-Induced Apoptosis in Primary Rat Proximal Tubular Cell Culture. PLoS ONE 2016, 11, e0166823. [Google Scholar] [CrossRef]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Elias, M.H.; Syed Mohamad, S.F.; Abdul Hamid, N. A Systematic Review of Candidate miRNAs, Its Targeted Genes and Pathways in Chronic Myeloid Leukemia—An Integrated Bioinformatical Analysis. Front. Oncol. 2022, 12, 848199. [Google Scholar] [CrossRef]
- Alves, R.; Gonçalves, A.C.; Jorge, J.; Marques, M.; Luís, D.; Ribeiro, A.B.; Freitas-Tavares, P.; Oliveiros, B.; Almeida, A.M.; Sarmento-Ribeiro, A.B. MicroRNA signature refine response prediction in CML. Sci. Rep. 2019, 9, 9666. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Liu, R.; Kasinski, A.L.; Shen, H.; Slack, F.J.; Tang, D.G. MicroRNA-34a: Potent Tumor Suppressor, Cancer Stem Cell Inhibitor, and Potential Anticancer Therapeutic. Front. Cell Dev. Biol. 2021, 9, 640587. [Google Scholar] [CrossRef]
- Usuda, J.; Inomata, M.; Fukumoto, H.; Iwamoto, Y.; Suzuki, T.; Kuh, H.J.; Fukuoka, K.; Kato, H.; Saijo, N.; Nishio, K. Restoration of p53 gene function in 12-O-tetradecanoylphorbor 13-acetate-resistant human leukemia K562/TPA cells. Int. J. Oncol. 2003, 22, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Law, J.C.; Ritke, M.K.; Yalowich, J.C.; Leder, G.H.; Ferrell, R.E. Mutational inactivation of the p53 gene in the human erythroid leukemic K562 cell line. Leuk. Res. 1993, 12, 1045–1050. [Google Scholar] [CrossRef]
- McDonald, P.C.; Winum, J.Y.; Supuran, C.T.; Dedhar, S. Recent developments in targeting carbonic anhydrase IX for cancer therapeutics. Oncotarget 2012, 3, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yao, L.; Yang, J.; Wang, Z.; Du, G. PI3K/Akt and HIF-1 signaling pathway in hypoxia-ischemia. Mol. Med. Rep. 2018, 18, 3547–3554. [Google Scholar] [CrossRef] [PubMed]
- Khalilian, S.; Bijanvand, A.; Abedinlou, H.; Ghafouri-Fard, S. A review on the role of miR-210 in human disorders. Pathol. Res. Pract. 2023, 241, 154244. [Google Scholar] [CrossRef]
- Tang, X.; Chen, L.; Yan, X.; Li, Y.; Xiong, Y.; Zhou, X. Overexpression of miR-210 is Associated with Poor Prognosis of Acute Myeloid Leukemia. Med. Sci. Monit. 2015, 21, 3427–3433. [Google Scholar] [CrossRef]
- Hassan, N.M.; Refaat, L.A.; Ismail, G.N.; Abdellateif, M.; Fadel, S.A.; AbdelAziz, R.S. Diagnostic, prognostic and predictive values of miR-100 and miR-210 in pediatric acute lymphoblastic Leukemia. Hematology 2020, 25, 405–413. [Google Scholar] [CrossRef]
- Drobna, M.; Szarzyńska-Zawadzka, B.; Daca-Roszak, P.; Kosmalska, M.; Jaksik, R.; Witt, M.; Dawidowska, M. Identification of Endogenous Control miRNAs for RT-qPCR in T-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2018, 19, 2858. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

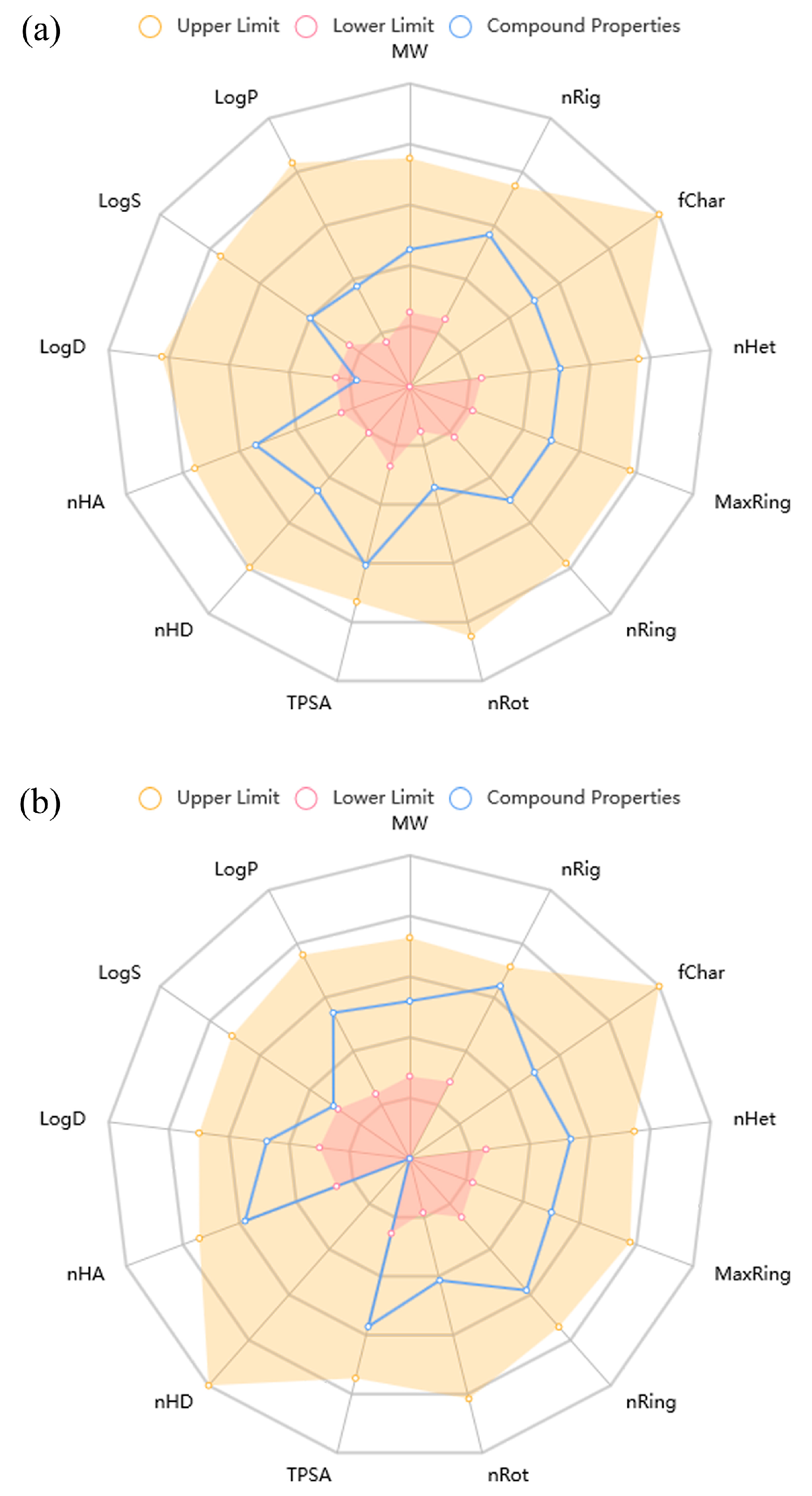
| Amino-SPD | Morpholino-SPD | Results Interpretation | |
|---|---|---|---|
| Medicinal Chemistry | |||
| QED | 0.885 | 0.687 | >0.67 attractive compounds; ≤0.67 poor |
| Synthetic accessibility score | 3.736 | 2.709 | ≤6 excellent; >6 poor |
| Fsp3 | 0.077 | 0.235 | ≥0.42 excellent; <0.42 poor |
| Lipinski rule | Accepted | Accepted | |
| Absorption | |||
| Caco-2 permeability | −5.009 | −4.479 | >−5.15 excellent; <−5.15 poor |
| MDCK permeability | 6.5 × 10−6 | 1.70 × 10−5 | >2 × 10−6 cm/s excellent; <2 × 10−6 cm/s poor |
| F30% | 0.996 | 0.813 | 0–0.3 excellent; 0.3–0.7 medium; 0.7–1.0 poor |
| Distribution | |||
| Plasma protein binding | 17.01% | 46.09% | ≤90% excellent; >90% poor |
| Volume distribution | 1.121 | 0.812 | 0.04–20 excellent; otherwise poor |
| BBB penetration | 0.359 | 0.923 | 0–0.3 excellent; 0.3–0.7 medium; 0.7–1.0 poor |
| Fraction unbound in plasma (Fu) | 82.10% | 61.47% | >20% high Fu; 5–20% medium Fu; <5% low Fu |
| Metabolism | |||
| CYP2C9 substrate | 0.636 | 0.848 | probability of being substrate 0–1 |
| Excretion | |||
| Clearance of a drug | 4.683 | 6.78 | ≥5 excellent; <5 poor |
| Half-life of a drug (T1/2) | 0.657 | 0.456 | 0–0.3 excellent; 0.3–0.7 medium; 0.7–1.0 poor |
| Toxicity | |||
| hERG blockers | 0.037 | 0.025 | 0–0.3 non-toxic; 0.3–0.7 medium; 0.7–1.0 toxic |
| Human hepatotoxicity | 0.488 | 0.806 | probability of being toxic 0–1 |
| Drug-induced liver injury | 0.975 | 0.993 | probability of being toxic 0–1 |
| AMES toxicity | 0.216 | 0.796 | probability of being toxic 0–1 |
| Rat oral acute toxicity | 0.630 | 0.141 | probability of being toxic 0–1 |
| Skin sensitization | 0.259 | 0.936 | probability of being toxic 0–1 |
| Carcinogenicity | 0.364 | 0.372 | probability of being toxic 0–1 |
| Eye irritation | 0.029 | 0.095 | probability of being toxic 0–1 |
| Respiratory toxicity | 0.921 | 0.378 | probability of being toxic 0–1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leventić, M.; Opačak-Bernardi, T.; Rastija, V.; Matić, J.; Pavlović Saftić, D.; Ban, Ž.; Žinić, B.; Glavaš-Obrovac, L. The Mechanism of Anti-Tumor Activity of 6-Morpholino- and 6-Amino-9-Sulfonylpurine Derivatives on Human Leukemia Cells. Molecules 2023, 28, 6136. https://doi.org/10.3390/molecules28166136
Leventić M, Opačak-Bernardi T, Rastija V, Matić J, Pavlović Saftić D, Ban Ž, Žinić B, Glavaš-Obrovac L. The Mechanism of Anti-Tumor Activity of 6-Morpholino- and 6-Amino-9-Sulfonylpurine Derivatives on Human Leukemia Cells. Molecules. 2023; 28(16):6136. https://doi.org/10.3390/molecules28166136
Chicago/Turabian StyleLeventić, Marijana, Teuta Opačak-Bernardi, Vesna Rastija, Josipa Matić, Dijana Pavlović Saftić, Željka Ban, Biserka Žinić, and Ljubica Glavaš-Obrovac. 2023. "The Mechanism of Anti-Tumor Activity of 6-Morpholino- and 6-Amino-9-Sulfonylpurine Derivatives on Human Leukemia Cells" Molecules 28, no. 16: 6136. https://doi.org/10.3390/molecules28166136
APA StyleLeventić, M., Opačak-Bernardi, T., Rastija, V., Matić, J., Pavlović Saftić, D., Ban, Ž., Žinić, B., & Glavaš-Obrovac, L. (2023). The Mechanism of Anti-Tumor Activity of 6-Morpholino- and 6-Amino-9-Sulfonylpurine Derivatives on Human Leukemia Cells. Molecules, 28(16), 6136. https://doi.org/10.3390/molecules28166136








