Structure Activity Relationship and Molecular Docking of Some Quinazolines Bearing Sulfamerazine Moiety as New 3CLpro, cPLA2, sPLA2 Inhibitors
Abstract
1. Introduction
2. Results
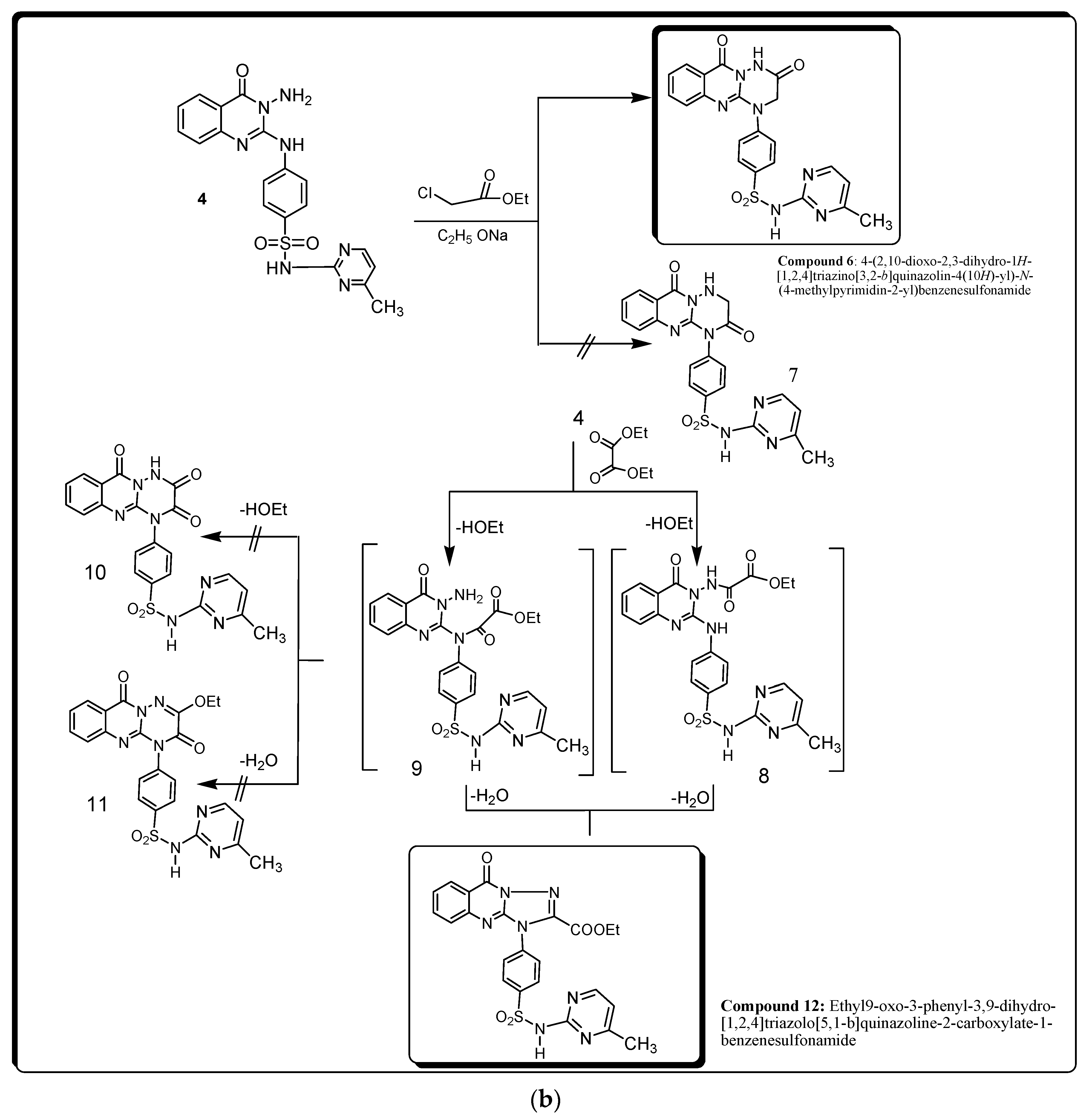
2.1. Chemistry
2.2. Biological Studies
2.2.1. CLpro Inhibitory Activity of Synthesized Compounds 4–6 and 12
2.2.2. sPLA2 Inhibitory Activity of Synthesized Compounds 4–6, and 12
2.2.3. cPLA2 Inhibitory Activity of Synthesized Compounds 4–6 and 12
2.2.4. Effects of Compounds 4–6, and 12 as well as Baicalein and Ivermectin on sPLA2, cPLA2, IL-8, TNF-α, and NO in Isolated Lung Cells Treated with LPS
2.2.5. In Silico Studies
Drug Likeliness
ADMET Prediction
3. Discussion
3.1. Chemistry
3.2. Biological Studies
Effects of Compounds 4–6 and 12 as well as Baicalein and Ivermectin on sPLA2, cPLA2, IL-8, TNF-α, and NO in Isolated Lung Cells Treated with LPS
3.3. Molecular Docking (MD) Simulations
ADMET Prediction
4. Materials and Methods
4.1. Chemistry
4.1.1. Synthesis and Characterization of Methyl-2-Isothiocyanato Benzoic Acid
4.1.2. Synthesis and Characterization of Compound 3
4.1.3. Synthesis and Characterization of Compound 4
4.1.4. Synthesis and Characterization of Compound 5
4.1.5. Synthesis and Characterization of Compounds 6 and 12
4.2. Biological Testing
4.2.1. 3CLpro Enzyme Inhibition Assay
4.2.2. sPLA2 Enzyme Inhibition Assay
4.2.3. cPLA2 Enzyme Inhibition Assay
4.2.4. Inhibition by Synthesized Compounds (4–6 and 12) as well as Baicalein and Ivermectin against cPLA2 and sPLA2 in LPS-Treated Mouse Lung Cells
4.3. Molecular Docking
4.3.1. Molecular Docking (MD) Simulations
4.3.2. ADMET Prediction
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- WHO. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 13 July 2023).
- Kneller, D.W.; Phillips, G.; O’Neill, H.M.; Jedrzejczak, R.; Stols, L.; Langan, P.; Joachimiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef]
- Kneller, D.W.; Galanie, S.; Phillips, G.; O’Neill, H.M.; Coates, L.; Kovalevsky, A. Malleability of the SARS-CoV-2 3CL Mpro Active-Site Cavity Facilitates Binding of Clinical Antivirals. Structure 2020, 28, 1313–1320.e3. [Google Scholar] [CrossRef]
- Hussein, M.A. Administration of exogenous surfactant and cytosolic phospholipase A2α Inhibitors may help COVID-19 infected patients with chronic diseases. Coronaviruses 2021, 2, e080921192222. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel coronavirus from wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Talluri, S. Computational protein design of bacteriocins based on structural scaffold of aureocin A53. Int. J. Bioinform. Res. 2019, 15, 129–143. [Google Scholar] [CrossRef]
- Mosaad, Y.O.; Baraka, M.A.; Warda, A.E.A.; Ateyya, H.; Hussein, M.A.; Gaber, S. Plasma lipid profile: A predictive marker of disease severity among COVID-19 patients-an opportunity for low-income countries. Drugs. Ther. Perspect. 2022, 38, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.A.; Ismail, N.E.M.; Mohamed, A.H.; Borik, R.M.; Ali, A.A.; Mosaad, Y.O. Plasma Phospholipids: A Promising Simple Biochemical Parameter to Evaluate COVID-19 Infection Severity. Bioinform. Biol. Insights 2021, 15, 11779322211055891. [Google Scholar] [CrossRef]
- Ghorab, M.M.; Ismail, Z.H.; Abdalla, M. Synthesis and biological activities of some novel triazoloquinazolines and triazinoquinazolines containing benzenesulfonamide moieties. Arzneim. Forsch. Drug Res. 2010, 60, 87–95. [Google Scholar] [CrossRef]
- Abdel-Gawad, S.M.; Ghorab, M.M.; El-Sharief, A.M.; El-Telbany, F.A.; Abdel-Alla, M. Design, synthesis, and antimicrobial activity of some new pyrazolo[3,4-d]pyrimidines. Heteroat. Chem. 2003, 14, 530–534. [Google Scholar] [CrossRef]
- Borik, R.M.; Hussein, M.A. Synthesis, Molecular Docking, Biological Potentials and Structure Activity Relationship of New Quinazoline and Quinazoline-4-one Derivatives. Asian J. Chem. 2021, 33, 423–438. [Google Scholar] [CrossRef]
- Borik, R.M.; Hussein, M.A. A Novel Quinazoline-4-one Derivatives as a Promising Cytokine Inhibitors: Synthesis, Molecular Docking, and Structure-activity Relationship. Curr. Pharm. Biotechnol. 2022, 23, 1179–1203. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Cheng, H.; Xiong, R.; Zhang, G.; Du, R.; Anantpadma, M.; Davey, R.A.; Rong, L. Identification of DiarylQuinoline Compounds as Entry Inhibitors of Ebola Virus. Viruses 2018, 10, 678. [Google Scholar] [CrossRef]
- Carta, A.; Briguglio, I.; Piras, S.; Corona, P.; Boatto, G.; Nieddu, M.; Giunchedi, P.; Marongiu, M.E.; Giliberti, G.; Iuliano, F.; et al. Quinoline tricyclic derivatives. Design, synthesis and evaluation of the antiviral activity of three new classes of RNA-dependent RNA polymerase inhibitors. Bioorg. Med. Chem. 2011, 19, 7070–7084. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, G.; Wang, Y.; Wang, J.; Zhu, M.; Cen, S.; Wang, Y. Design, synthesis and anti-influenza A virus activity of novel 2,4-disubstituted quinazoline derivatives. Bioorg. Med. Chem. Lett. 2020, 30, 127143. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, G.; Zhao, J.; Cheng, N.; Wang, Y.; Fu, Y.; Zheng, Y.; Wang, J.; Zhu, M.; Cen, S.; et al. Synthesis and antiviral activity of a series of novel quinoline derivatives as anti-RSV or anti-IAV agents. Eur. J. Med. Chem. 2011, 214, 113208. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.H.; Zhang, Q.; Ma, Y.B.; Huang, X.Y.; Luo, J.; Wang, L.J.; Geng, C.A.; Zhang, X.M.; Zhou, J.; Jiang, Z.Y.; et al. Synthesis and biological assay of 4-aryl-6-chloro-quinoline derivatives as novel non-nucleoside anti-HBV agents. Bioorg. Med. Chem. 2011, 19, 1400–1408. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, S.; Yi, D.; Li, Q.; Ma, L.; Zhang, Y.; Wang, J.; Li, X.; Guo, F.; Lin, R.; et al. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antivir. Res. 2021, 190, 105078. [Google Scholar] [CrossRef]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Hoult, J.R. Pharmacological and biochemical actions of sulphasalazine. Drugs 1986, 32 (Suppl. S1), 18–26. [Google Scholar] [CrossRef]
- Nielsen OH, Bukhave K, Elmgreen J, Ahnfelt-Ronne I: Inhibition of 5-lipoxygenase pathway of arachidonic acid metabolism in human neutrophils by sulfasalazine and 5-aminosalicylic acid. Dig. Dis. Sci. 1987, 32, 577–582. [CrossRef]
- Generini, S.; Fiori, G.; Matucci Cerinic, M. Therapy of spondylarthropathy in inflammatory bowel disease. Clin. Exp. Rheumatol. 2002, 20 (Suppl. S28), S88–S94. [Google Scholar] [PubMed]
- Allgayer, H. Review article: Mechanisms of action of mesalazine in preventing colorectal carcinoma in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2003, 18 (Suppl. S2), 10–14. [Google Scholar] [PubMed]
- Pruzanski, W.; Stefanski, E.; Vadas, P.; Ramamurthy, N.S. Inhibition of extracellular release of proinflammatory secretory phospholipase A2 (sPLA2) by sulfasalazine: A novel mechanism of anti-inflammatory activity. Biochem. Pharmacol. 1997, 53, 1901–1907. [Google Scholar]
- Weber, C.K.; Liptay, S.; Wirth, T.; Adler, G.; Schmid, R.M. Suppression of NF-kappaB activity by sulfasalazine is mediated by direct inhibition of IkappaB kinases alpha and beta. Gastroenterology 2000, 119, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, C.; Lefebvre, B.; Dubuquoy, L.; Lefebvre, P.; Romano, O.; Auwerx, J.; Metzger, D.; Wahli, W.; Desvergne, B.; Naccari, G.C.; et al. Desreumaux P: Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. J. Exp. Med. 2005, 201, 1205–1215. [Google Scholar] [CrossRef]
- Cevallos, S.A.; Lee, J.Y.; Velazquez, E.M.; Foegeding, N.J.; Shelton, C.D.; Tiffany, C.R.; Parry, B.H.; Stull-Lane, A.R.; Olsan, E.E.; Savage, H.P.; et al. Baumler AJ: 5-Aminosalicylic Acid Ameliorates Colitis and Checks Dysbiotic Escherichia coli Expansion by Activating PPAR-gamma Signaling in the Intestinal Epithelium. mBio 2021, 12, e03227-20. [Google Scholar] [CrossRef]
- Wahl, C.; Liptay, S.; Adler, G.; Schmid, R.M. Sulfasalazine: A potent specific inhibitor of nuclear factor kappa B. J. Clin. Investig. 1998, 101, 1163–1174. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Leeson, P.D.; Springthorpe, B. The Influence of Drug-like Concepts on Decision-Making in Medicinal Chemistry. Nat. Rev. Drug Discov. 2007, 6, 881–890. [Google Scholar] [CrossRef]
- Clark, D.E.; Pickett, S.D. Computational Methods for the Prediction of ‘Drug-Likeness’. Drug Discov. Today 2000, 5, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Herbertz, T.; Hudkins, R.L.; Dorsey, B.D.; Mallamo, J.P. Knowledge-Based, Central Nervous System (CNS) Lead Selection and Lead Optimization for CNS Drug Discovery. ACS Chem. Neurosci. 2012, 3, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, W.; Zhang, W. As the SARS-CoV-2 virus evolves, should Omicron subvariant BA.2 be subjected to quarantine, or should we learn to live with it? Front. Public Health 2022, 10, 1039123. [Google Scholar] [CrossRef]
- Murakami, M.; Taketomi, Y.; Miki, Y.; Sato, H.; Yamamoto, K.; Lambeau, G. Emerging roles of secreted phospholipase A2 enzymes: The 3rd edition. Biochimie 2014, 107 Pt A, 105–113. [Google Scholar]
- Murakami, M. Novel functions of phospholipase A(2)s: Overview. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 763–765. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Zhang, C.; Zhang, C.; Yang, H.; Gao, R.; Tong, Z. Lung fluid biomarkers for acute respiratory distress syndrome: A systematic review and meta-analysis. Crit. Care 2019, 23, 43. [Google Scholar]
- Murakami, M.; Yamamoto, K.; Miki, Y.; Murase, R.; Sato, H.; Taketomi, Y. The roles of the secreted phospholipase A(2) gene family in immunology. Adv. Immunol. 2016, 132, 91–134. [Google Scholar]
- van Hensbergen, V.P.; Wu, Y.; van Sorge, N.M.; Touqui, L. Type IIA secreted phospholipase A2 in host defense against bacterial infections. Trends Immunol. 2020, 41, 313–326. [Google Scholar] [CrossRef]
- Papadopoulos, S.; Kazepidou, E.; Antonelou, M.H.; Leondaritis, G.; Tsapinou, A.; Koulouras, V.P.; Avgeropoulos, A.; Nakos, G.; Lekka, M.E. Secretory phospholipase A(2)-IIA protein and mRNA pools in extracellular vesicles of bronchoalveolar lavage fluid from patients with early acute respiratory distress syndrome: A new perception in the dissemination of inflammation? Pharmaceuticals 2020, 13, 415. [Google Scholar] [PubMed]
- Vijay, R.; Hua, X.; Meyerholz, D.K.; Miki, Y.; Yamamoto, K.; Gelb, M.; Murakami, M.; Perlman, S. Critical role of phospholipase A2 group IID in age-related susceptibility to severe acute respiratory syndrome-CoV infection. J. Exp. Med. 2015, 212, 1851–1868. [Google Scholar] [CrossRef]
- El-Sayed, N.N.E.; Almaneai, N.M.; Ben Bacha, A.; El-Ashrey, M.K.; Al-Zaben, M.I.; Almarhoon, Z.M. Biological Evaluation, Molecular Docking Analyses, and ADME Profiling of Certain New Quinazolinones as Anti-colorectal Agents. ACS Omega 2022, 7, 18443–18458. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Islam, M.S.; Al-Majid, A.M.; Ghabbour, H.A.; Fun, H.K.; Javed, K.; Imad, R.; Yousuf, S.; Choudhary, M.I.; Wadood, A. Synthesis, in vitro biological activities and in silico study of dihydropyrimidines derivatives. Bioorg. Med. Chem. 2015, 23, 6740–6748. [Google Scholar] [CrossRef] [PubMed]
- Ibezim, A.; Onoabedje, E.A.; Adaka, I.C.; Omeje, K.O.; Onoabedje, U.S.; Obi, B.C. Carboxamides bearing sulfonamide functionality as potential novel phospholipase A2 inhibitors. Chem. Sel. 2020, 5, 14416–14421. [Google Scholar]
- Gandhirajan, R.K.; Meng, S.; Chandramoorthy, H.C.; Mallilankaraman, K.; Mancarella, S.; Gao, H.; Razmpour, R.; Yang, X.F.; Houser, S.R.; Chen, J.; et al. Blockade of NOX2 and STIM1 signaling limits lipopolysaccharide-induced vascular inflammation. J. Clin. Investig. 2013, 123, 887–902. [Google Scholar] [CrossRef]
- Lee, I.; Dodia, C.; Chatterjee, S.; Zagorski, J.; Mesaros, C.; Blair, I.A.; Feinstein, S.I.; Jain, M.; Fisher, A.B. A novel nontoxic inhibitor of the activation of NADPH oxidase reduces reactive oxygen species production in mouse lung. J. Pharmacol. Exp. Ther. 2013, 345, 284–296. [Google Scholar] [CrossRef]
- Sato, H.; Taketomi, Y.; Murakami, M. Metabolic regulation by secreted phospholipase A2. Inflamm Regen. 2016, 36, 7–18. [Google Scholar] [CrossRef]
- Kelvin, A.A.; Degousee, N.; Banner, D.; Stefanski, E.; Leόn, A.J.; Angoulvant, D.; Paquette, S.G.; Huang, S.S.; Danesh, A.; Robbins, C.S.; et al. Lack of group X secreted phospholipase A2 increases survival following pandemic H1N1 influenza infection. Virology 2014, 454–455, 78–92. [Google Scholar] [CrossRef][Green Version]
- Jawarkar, R.D.; Bakal, R.L.; Zaki, M.E.A.; Al-Hussain, S.; Ghosh, A.; Gandhi, A.; Mukerjee, N.; Samad, A.; Masand, V.H.; Lewaa, I. QSAR based virtual screening derived identification of a novel hit as a SARS CoV-229E 3CLpro Inhibitor: GA-MLR QSAR modeling supported by molecular Docking, molecular dynamics simulation and MMGBSA calculation approaches. Arab. J. Chem. 2022, 15, 103499. [Google Scholar] [CrossRef]
- Gupta, Y.; Kumar, S.; Zak, S.E.; Jones, K.A.; Upadhyay, C.; Sharma, N.; Azizi, S.A.; Kathayat, R.S.; Poonam Herbert, A.S.; Durvasula, R.; et al. Antiviral evaluation of hydroxyethylamine analogs: Inhibitors of SARS-CoV-2 main protease (3CLpro), a virtual screening and simulation approach. Bioorg. Med. Chem. 2021, 47, 116393. [Google Scholar] [CrossRef]
- Jo, S.; Kim, S.; Kim, D.Y.; Kim, M.S.; Shin, D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme. Inhib. Med. Chem. 2020, 35, 1539–1544. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, H.; Lin, Q.; Lyu, J.; Lu, L.; Chen, H.; Zhang, X.; Zhang, Y.; Chen, K. Progress on SARS-CoV-2 3CLpro Inhibitors: Inspiration from SARS-CoV 3CLpro Peptidomimetics and Small-Molecule Anti-Inflammatory Compounds. Drug Des. Devel. Ther. 2022, 16, 1067–1082. [Google Scholar] [CrossRef]
- Su, H.; Yao, S.; Zhao, W.; Li, M.; Liu, J.; Shang, W.; Xie, H.; Ke, C.; Hu, H.; Gao, M.; et al. Anti-SARS-CoV-2 Activities in Vitro of Shuanghuanglian Preparations and Bioactive Ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef]
- Conterno, L.O.; Turchi, M.D.; Corrêa, I.; Monteiro de Barros Almeida, R.A. Anthelmintic drugs for treating ascariasis. Cochrane Database Syst. Rev. 2020, 1, CD010599. [Google Scholar] [CrossRef]
- Calligari, P.; Bobone, S.; Ricci, G.; Bocedi, A. Molecular Investigation of SARS-CoV-2 Proteins and Their Interactions with Antiviral Drugs. Viruses 2020, 12, 445. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.; Baker, S.; Baric, R.; de Groot, R.; Drosten, C.; Gulyaeva, A. The species severe acute respiratory syndrome related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Dömling, A.; Gao, L. Chemistry and biology of SARS-CoV-2. Chem 2020, 6, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Koonin, E.V.; Donchenko, A.P.; Blinov, V.M. Coronavirus genome: Prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989, 17, 4847–4861. [Google Scholar] [CrossRef] [PubMed]
- Ziebuhr, J.; Snijder, E.J.; Gorbalenya, A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000, 81, 853–879. [Google Scholar] [CrossRef]
- Lee, H.J.; Shieh, C.K.; Gorbalenya, A.E.; Koonin, E.; La Monica, N.; Tuler, J. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 1991, 180, 567–582. [Google Scholar] [CrossRef]
- Osvaldo, Y.; Manuel, I.O.; Eugenio, U.; Carlos, A.; William, T.; José, M. Pérez-Donoso, Olimpo García-Beltrán8 and Fernando González-Nilo, In Silico Study of Coumarins and Quinolines Derivatives as Potent Inhibitors of SARS-CoV-2 Main Protease. Front. Chem. 2021, 8, 595097. [Google Scholar] [CrossRef]
- Anger, K.E.; Degrado, J.R.; Greenwood, B.C.; Cohen, S.A.; Szumita, P.M. Evaluation of recombinant activated protein C for severe sepsis at a tertiary academic medical center. Ther. Clin. Risk. Manag. 2013, 9, 277–284. [Google Scholar] [PubMed]
- Wang, J.; Niu, N.; Xu, S.; Jin, Z.G. A simple protocol for isolating mouse lung endothelial cells. Sci. Rep. 2019, 9, 1458. [Google Scholar] [CrossRef] [PubMed]
- Kitsiouli, E.; Antoniou, G.; Gotzou, H.; Karagiannopoulos, M.; Basagiannis, D.; Christoforidis, S.; Nakos, G.; Lekka, M.E. Effect of azithromycin on the LPS-induced production and secretion of phospholipase A2 in lung cells. Biochim. Biophys. Acta 2015, 1852, 1288–1297. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar]
- Schevitz, R.W.; Bach, N.J.; Carlson, D.G.; Chirgadze, N.Y.; Clawson, D.K.; Dillard, R.; Draheim, S.D.; Hartley, L.W.; Jones, N.D.; Mihelich, E.D.; et al. Structure-based design of the first potent and selective inhibitor of human non-pancreatic secretory phospholipase A2. Nat. Struct. Biol. 1995, 2, 458–465. [Google Scholar] [CrossRef]
- Dessen, A.; Tang, J.; Schmidt, H.; Stahl, M.; Clark, J.D.; Seehra, J.; Somers, W.S. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 1999, 97, 349–360. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web toolto evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]

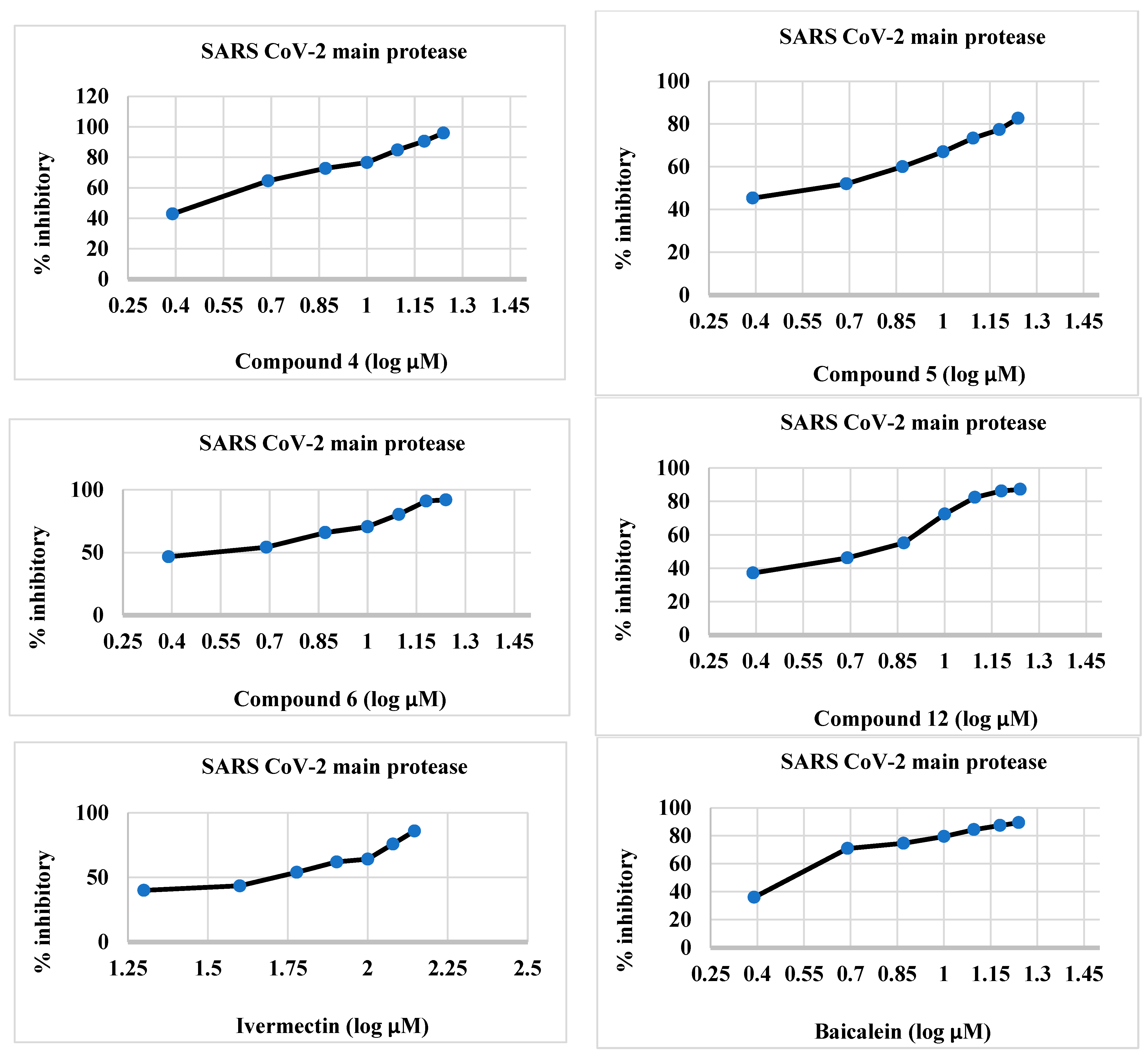

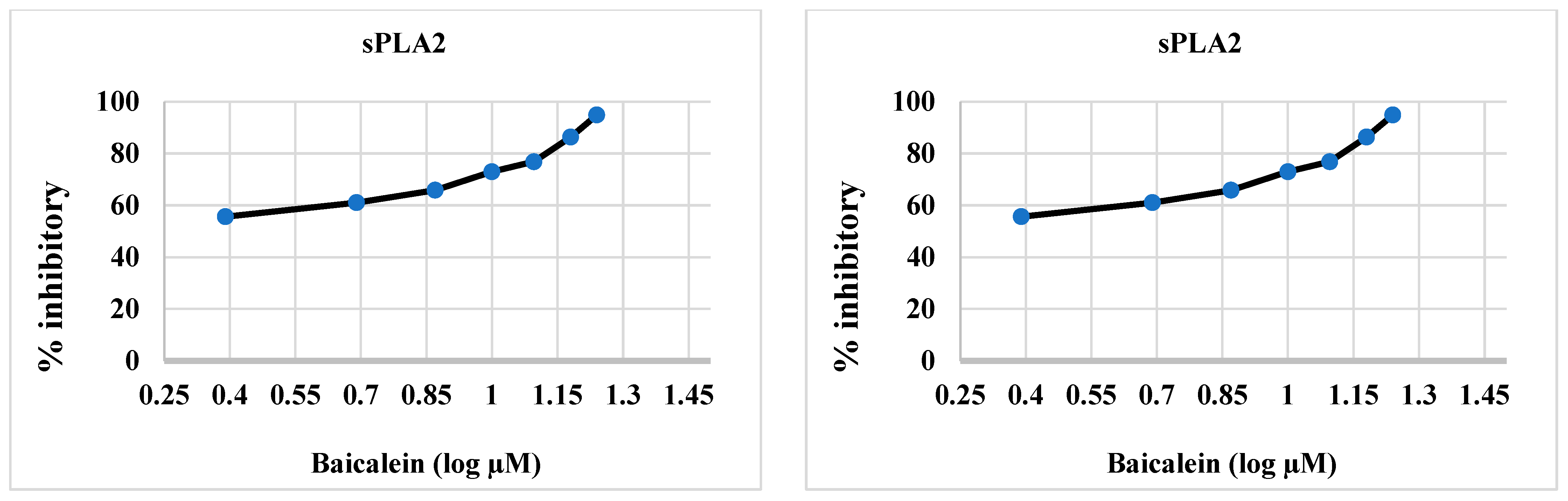

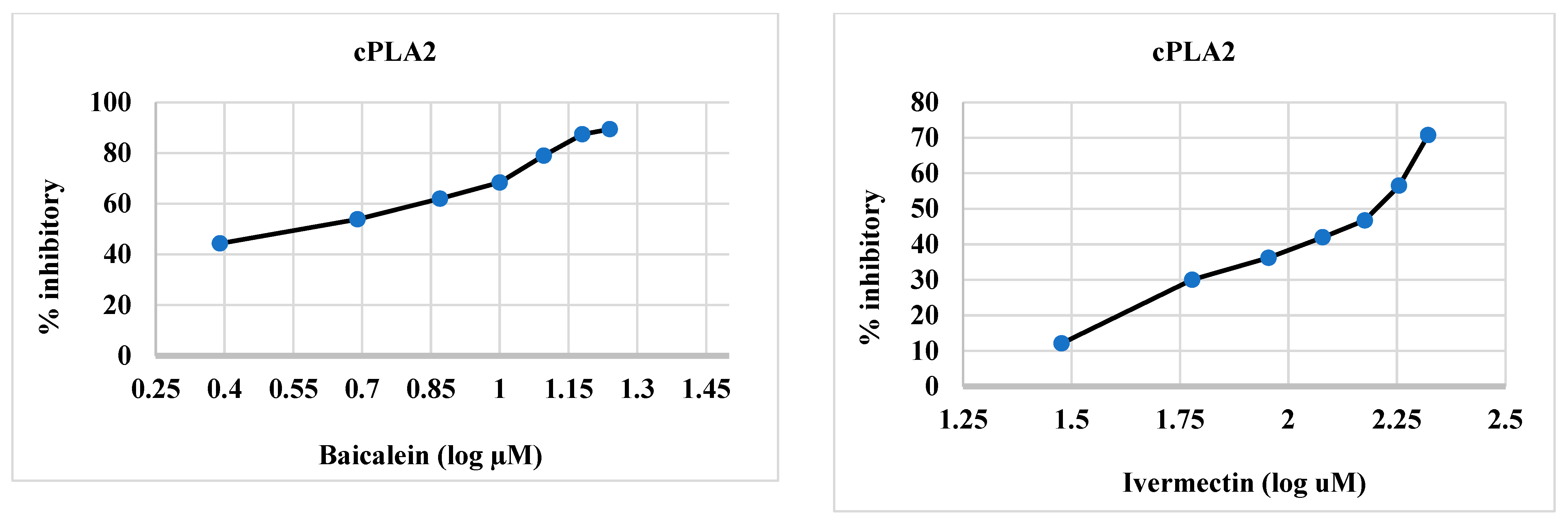
| Synthesized Compounds/Drugs | SARS-CoV-2 Main Protease (µM) | sPLA2 (µM) | cPLA2 (µM) |
|---|---|---|---|
| Compound 4 | 2.012 ± 0.004 c | 2.84 ± 0.026 c | 1.44 ± 0.009 b |
| Compound 5 | 3.68 ± 2.35 d | 2.73 ± 0.008 c | 2.08 ± 0.016 c |
| Compound 6 | 1.180 ± 0.025 a | 1.016 ± 0.039 b | 0.5 ± 0.008 a |
| Compound 12 | 5.47 ± 0.018 e | 4.45 ± 0.007 d | 2.39 ± 0.046 d |
| Baicalein | 1.72 ± 0.006 b | 0.89 ± 0.041 a | 3.88 ± 0.013 e |
| Ivermectin | 42.39 ± 2.50 f | 109.6 ± 3.27 e | 138.0 ± 1.54 f |
| Group No. | Synthesized Compounds/Drugs | Doses | sPLA2 (pg/mL) | cPLA2 (dpm/mL) | IL-8 (ng/mL) | TNF-α (pg/mL) | NO (µmol/L) |
|---|---|---|---|---|---|---|---|
| I | Negative control sample | 0 μg/ML | 12.59 ± 0.46 a | 474.22 ± 25.21 a | 0.82 ± 0.08 a | 476.97 ± 32.77 a | 17.79 ± 2.16 a |
| II | Positive control (LPS) | (1 μg/mL) | 129.58 ± 6.34 n | 2817.86 ± 40.51 l | 5.74 ± 0.47 h | 980.27 ± 75.96 l | 68.59 ± 5.82 j |
| III | LPS (1 μg/mL) + Compound 4 | 1.44 µM | 57.93 ± 2.92 i | 1077.77 ± 68.61 i | 2.55 ± 0.26 c | 654.20 ± 22.28 h | 37.27 ± 3.00 c |
| 2.84 µM | 29.10 ± 2.52 d | 671.05 ± 29.17 d | 2.17 ± 0.07 d | 481.36 ± 39.63 d | 25.18 ± 2.98 d | ||
| IV | LPS (1 μg/mL) + Compound 5 | 2.08 µM | 72.13 ± 3.94 k | 1258.99 ± 35.57 k | 2.74 ± 0.32 c | 671.91 ± 23.11 i | 45.75 ± 2.87 g |
| 3.68 µM | 34.17 ± 1.73 e | 719.32 ± 37.15 e | 2.40 ±0.09 e | 524.98 ± 19.11 e | 29.37 ± 2.21 f | ||
| V | LPS (1 μg/mL) + Compound 6 | 0.5 µM | 43.44 ± 2.74 g | 830.11 ± 34.00 f | 1.79 ± 0.14 d | 582.31 ± 52.25 e | 24.67 ± 2.70 e |
| 1.18 µM | 18.29 ± 2.35 b | 536.93 ± 23.50 b | 1.17 ± 0.06 b | 424.25 ± 36.86 f | 19.02 ± 1.5 b | ||
| VI | LPS (1 μg/mL) + Compound 12 | 2.39 µM | 85.23 ± 6.51 l | 1348.21 ± 50.26 | 3.84 ± 0.29 g | 748.98 ± 31.22 j | 50.36 ± 3.96 h |
| 5.47 µM | 40.61 ± 4.19 f | 722.24 ± 30.40 e | 3.21 ± 0.21 f | 551.52 ± 24.65 f | 30.94 ± 3.76 f | ||
| VII | LPS (1 μg/mL) + Baicalein | 0.89 µM | 54.27 ± 3.46 h | 903.43 ± 35.71 g | 2.06 ± 0.23 g | 639.55 ± 37.84 g | 32.01 ± 4.81 f |
| 3.88 µM | 24.77 ± 2.73 c | 616.34 ± 28.34 c | 1.42 ± 0.13 c | 431.21 ± 32.88 c | 20.53 ± 1.89 c | ||
| VIII | LPS (1 μg/mL) + Ivermectin | 131.01 µM | 106.13 ± 9.81 m | 1172.50 ± 72.55 j | 3.77 ± 0.62 g | 870.22 ± 40.62 k | 62.01 ± 4.81 i |
| 149.39 µM | 62.31 ± 1.41 j | 1002.65 ± 90.98 h | 3.10 ± 0.1 f | 714.29 ± 28.09 g | 50.74 ± 3.34 h |
| CPDS | SARS-CoV-2 Main Protease (3CLpro) (PDB = 5RFS) # | Phospholipase A2 (sPLA2) (PDB = 1DCY) | Cytosolic Phospholipase A2 (cPLA2) (PDB = 1CJY) | |||
|---|---|---|---|---|---|---|
| Binding Energy (Kcal/mol) | Binding Interactions | Binding Energy (Kcal/mol) | Binding Interactions | Binding Energy (Kcal/mol) | Binding Interactions | |
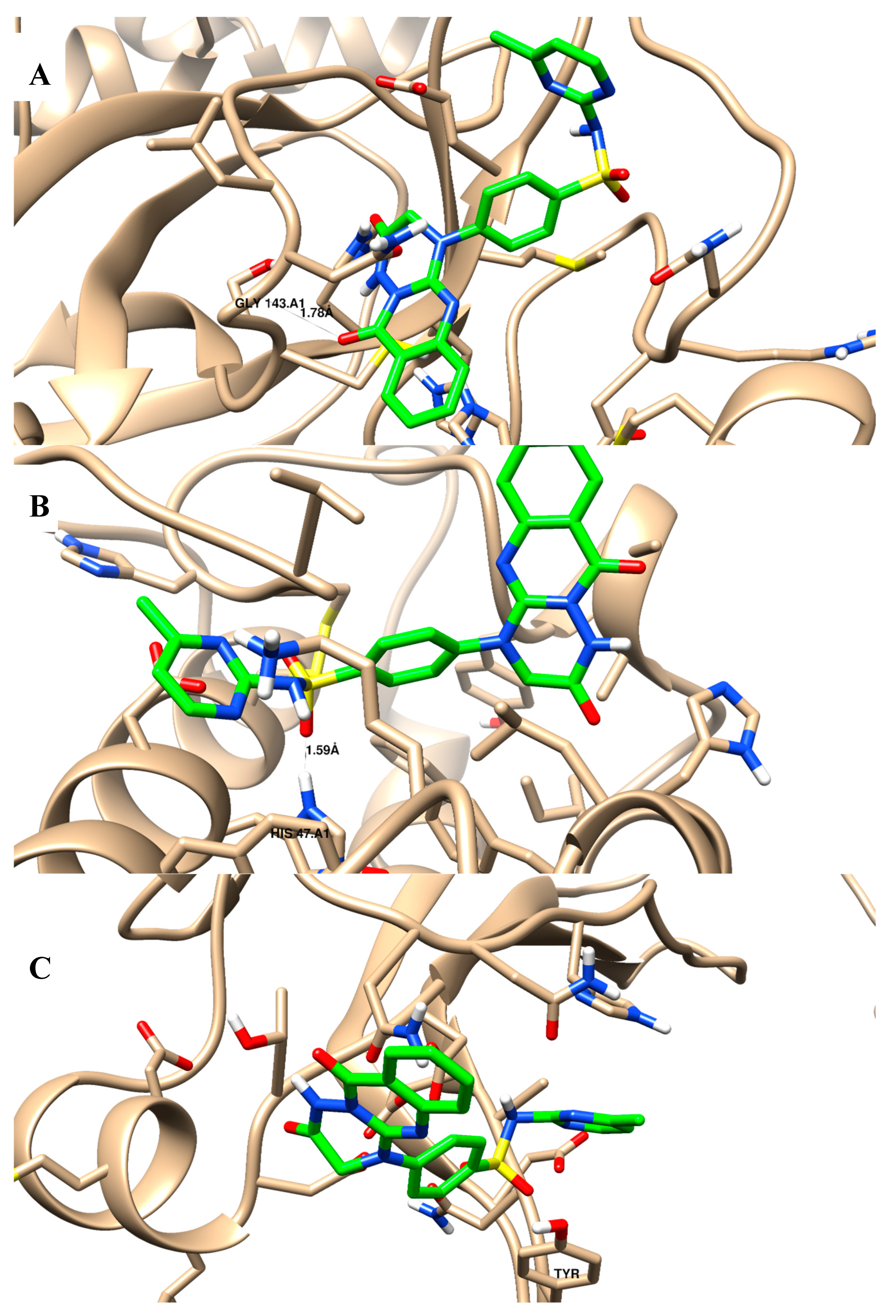
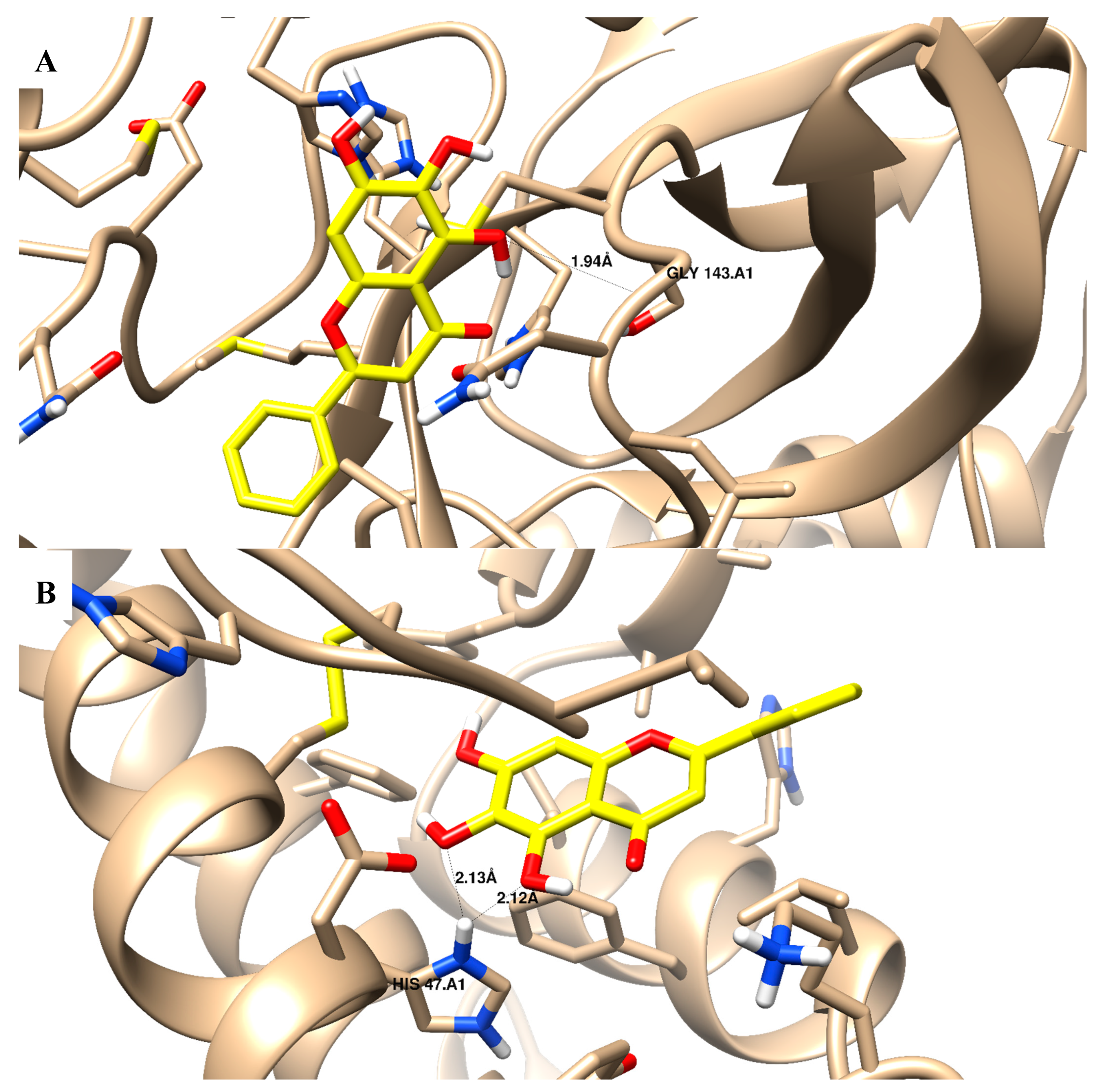
| 2 | −12.92 | 1 H-bond with Gly 143 pi–pi interaction with His 41 | −13.19 | 1 H-bond with His 47 | −7.22 | Cation–arene interaction with Tyr 96 |
| 3 | −19.91 | 1 H-bond with Gly 143 | −11.43 | 1 H-bond with His 47 | −11.17 | pi–pi interaction with Tyr 96 |
| 4 | −26.06 | 1 H-bond with Gly 143 | −22.33 | 1 H-bond with His 47 | −17.82 | pi–pi interaction with Tyr 96 |
| 5 | −25.83 | 1 H-bond with Gly 143 | −23.57 | 1 H-bond with His 47 | −13.77 | pi–pi interaction with Tyr 96 |
| 6 | −23.14 | 1 H-bond with Gly 143 | −18.38 | 1 H-bond with His 47 | −9.99 | pi–pi interaction with Tyr 96 |
| 12 | −23.28 | 1 H-bond with Gly 143 pi–pi interaction with His 41 | −23.49 | 1 H-bond with His 47 | −12.84 | pi–pi interaction with Tyr 96 |
| Ivermectin | −23.37 | -- | −6.09 | -- | −16.63 | -- |
| Baicalein | −15.07 | 1 H-bond with Gly 143 | −18.24 | 2 H-bond with His 47 | −15.54 | -- |
| Comp. No | MW g/mol | Log p | HBA | HBD | TPSA Å2 | MR | nRB | No. Lipinski Violations |
|---|---|---|---|---|---|---|---|---|
| 2 | 193.22 | 2.88 | 3 | 0 | 70.75 | 52.41 | 3 | 0 |
| 3 | 457.53 | 2.35 | 6 | 3 | 162.78 | 121.31 | 9 | 0 |
| 4 | 423.45 | 1.18 | 6 | 3 | 153.27 | 113.36 | 5 | 0 |
| 5 | 557.58 | 2.90 | 9 | 3 | 169.07 | 151.17 | 8 | 2 |
| 6 | 463.47 | 1.19 | 7 | 2 | 147.56 | 127.12 | 4 | 1 |
| 12 | 505.51 | 2.09 | 9 | 1 | 158.82 | 129.85 | 7 | 2 |
| Models | 2 | 3 | 4 | 5 | 6 | 12 |
|---|---|---|---|---|---|---|
| Blood–Brain Barrier | BBB+ | BBB− | BBB+ | BBB− | BBB+ | BBB− |
| Human Intestinal Absorption | HIA+ | HIA+ | HIA+ | HIA+ | HIA+ | HIA+ |
| Caco-2 Permeability | Caco-2- | Caco-2- | Caco-2- | Caco-2- | Caco-2- | Caco-2- |
| Aqueous solubility (Log S) | −2.8928 | −3.2434 | −3.1843 | −3.3822 | −3.5571 | −3.9130 |
| P-glycoprotein Substrate | Non-substrate | Non-substrate | Non-substrate | Non-substrate | Substrate | Non-substrate |
| P-glycoprotein Inhibitor I | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| P-glycoprotein Inhibitor II | Non-inhibitor | Non-inhibitor | Non-inhibitor | Inhibitor | Non-inhibitor | Non-inhibitor |
| Renal Organic Cation Transporter | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| Subcellular localization | Mitochondria | Mitochondria | Mitochondria | Mitochondria | Mitochondria | Mitochondria |
| CYP450 2C9 Substrate | Non-Substrate | Non-Substrate | Non-Substrate | Non-Substrate | Non-Substrate | Non-Substrate |
| CYP450 2D6 Substrate | Non-Substrate | Non-Substrate | Non-Substrate | Non-Substrate | Non-Substrate | Non-Substrate |
| CYP450 3A4 Substrate | Non-Substrate | Non-substrate | Non-substrate | Non-substrate | Substrate | Non-substrate |
| CYP450 1A2 Inhibitor | Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| CYP450 2D6 Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| CYP450 3A4 Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Inhibitor |
| CYP450 2C19 Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| Human Ether-a-go-go-Related Gene Inhibition I | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| Human Ether-a-go-go Related Gene Inhibition II | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| AMES Toxicity | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic |
| Carcinogens | Non-carcinogen | Non-carcinogen | Non-carcinogen | Non-carcinogen | Non-carcinogen | Non-carcinogen |
| Honeybee toxicity | High HBT | Low HBT | Low HBT | Low HBT | Low HBT | Low HBT |
| Biodegradation | Not readily biodegradable | Not readily biodegradable | Not readily biodegradable | Not readily biodegradable | Not readily biodegradable | Not readily biodegradable |
| Acute oral toxicity | III | III | III | III | III | III |
| Carcinogenicity (three class) | Non-required | Non-required | Non-required | Non-required | Non-required | Non-required |
| Rat Acute Toxicity (LD50, mol/kg) | 2.2363 | 2.1081 | 2.0649 | 2.2374 | 2.2163 | 2.4056 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, M.A.; Borik, R.M.; Nafie, M.S.; Abo-Salem, H.M.; Boshra, S.A.; Mohamed, Z.N. Structure Activity Relationship and Molecular Docking of Some Quinazolines Bearing Sulfamerazine Moiety as New 3CLpro, cPLA2, sPLA2 Inhibitors. Molecules 2023, 28, 6052. https://doi.org/10.3390/molecules28166052
Hussein MA, Borik RM, Nafie MS, Abo-Salem HM, Boshra SA, Mohamed ZN. Structure Activity Relationship and Molecular Docking of Some Quinazolines Bearing Sulfamerazine Moiety as New 3CLpro, cPLA2, sPLA2 Inhibitors. Molecules. 2023; 28(16):6052. https://doi.org/10.3390/molecules28166052
Chicago/Turabian StyleHussein, Mohammed Abdalla, Rita M. Borik, Mohamed S. Nafie, Heba M. Abo-Salem, Sylvia A. Boshra, and Zahraa N. Mohamed. 2023. "Structure Activity Relationship and Molecular Docking of Some Quinazolines Bearing Sulfamerazine Moiety as New 3CLpro, cPLA2, sPLA2 Inhibitors" Molecules 28, no. 16: 6052. https://doi.org/10.3390/molecules28166052
APA StyleHussein, M. A., Borik, R. M., Nafie, M. S., Abo-Salem, H. M., Boshra, S. A., & Mohamed, Z. N. (2023). Structure Activity Relationship and Molecular Docking of Some Quinazolines Bearing Sulfamerazine Moiety as New 3CLpro, cPLA2, sPLA2 Inhibitors. Molecules, 28(16), 6052. https://doi.org/10.3390/molecules28166052








