The Homoleptic Curcumin–Copper Single Crystal (ML2): A Long Awaited Breakthrough in the Field of Curcumin Metal Complexes
Abstract
1. Introduction
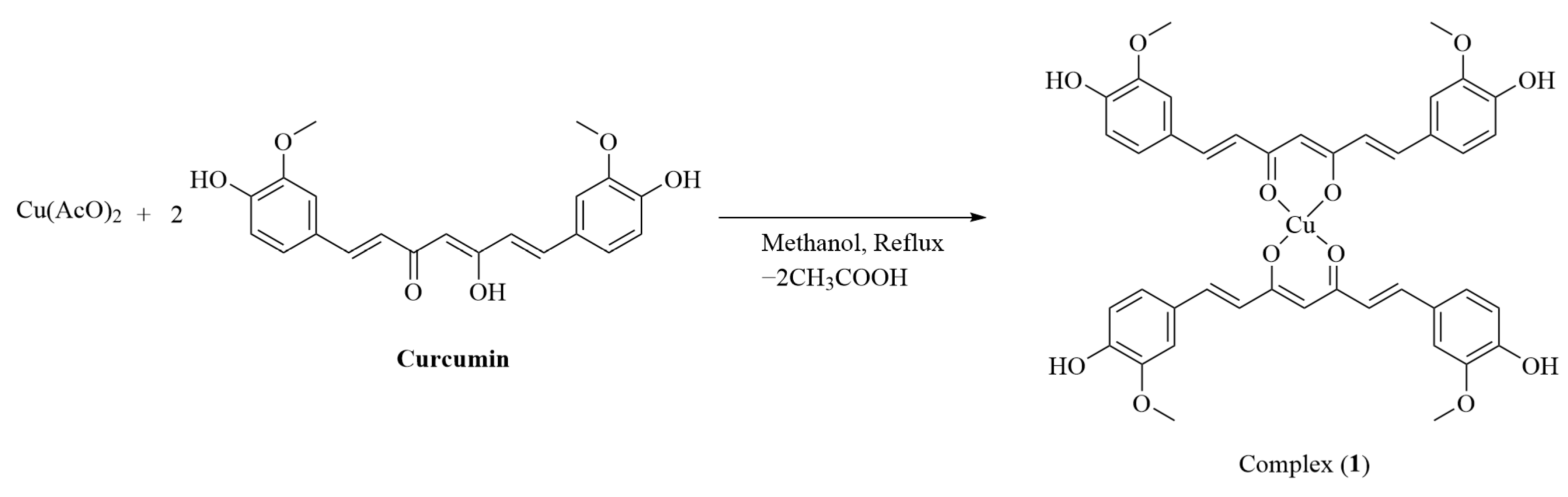
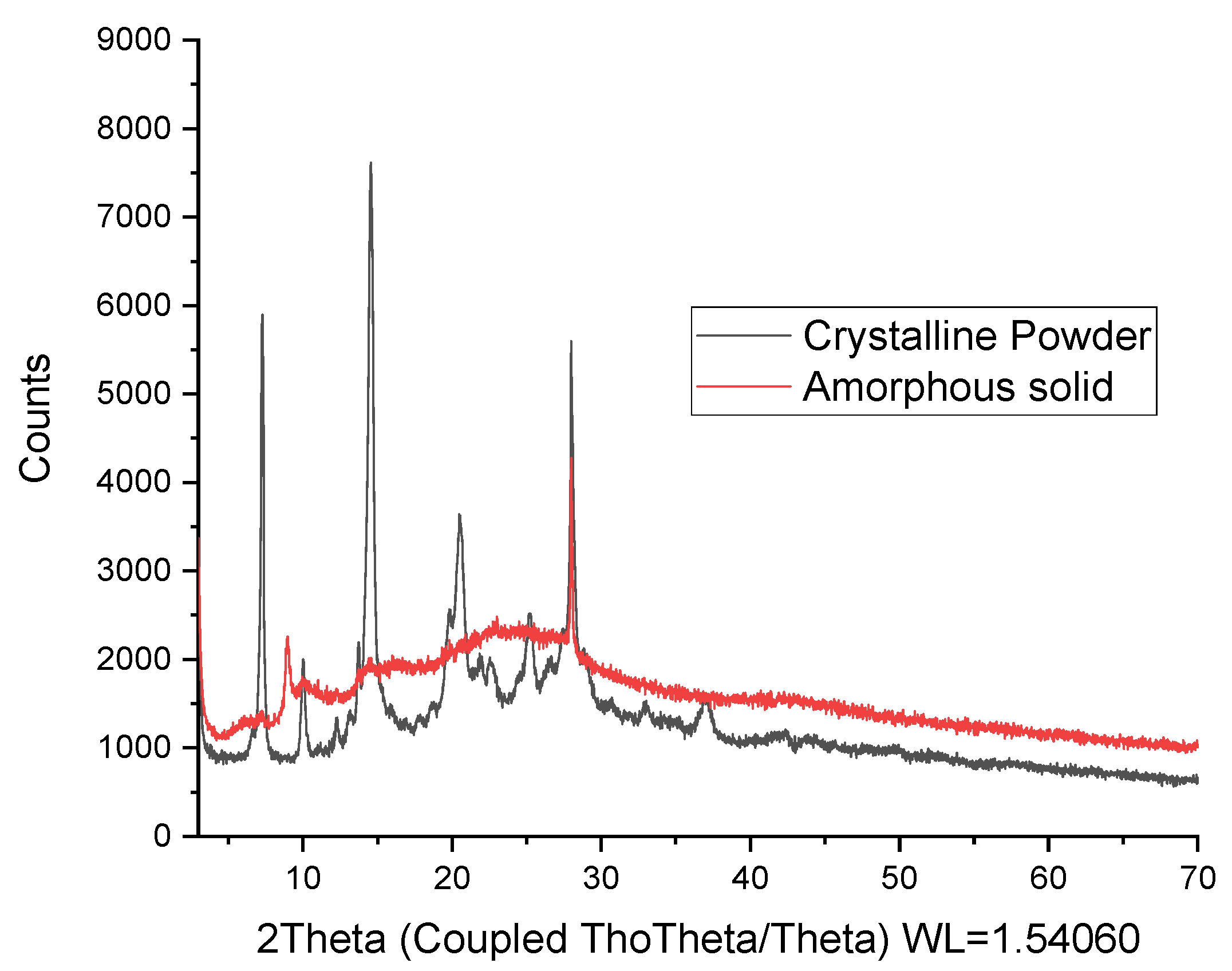
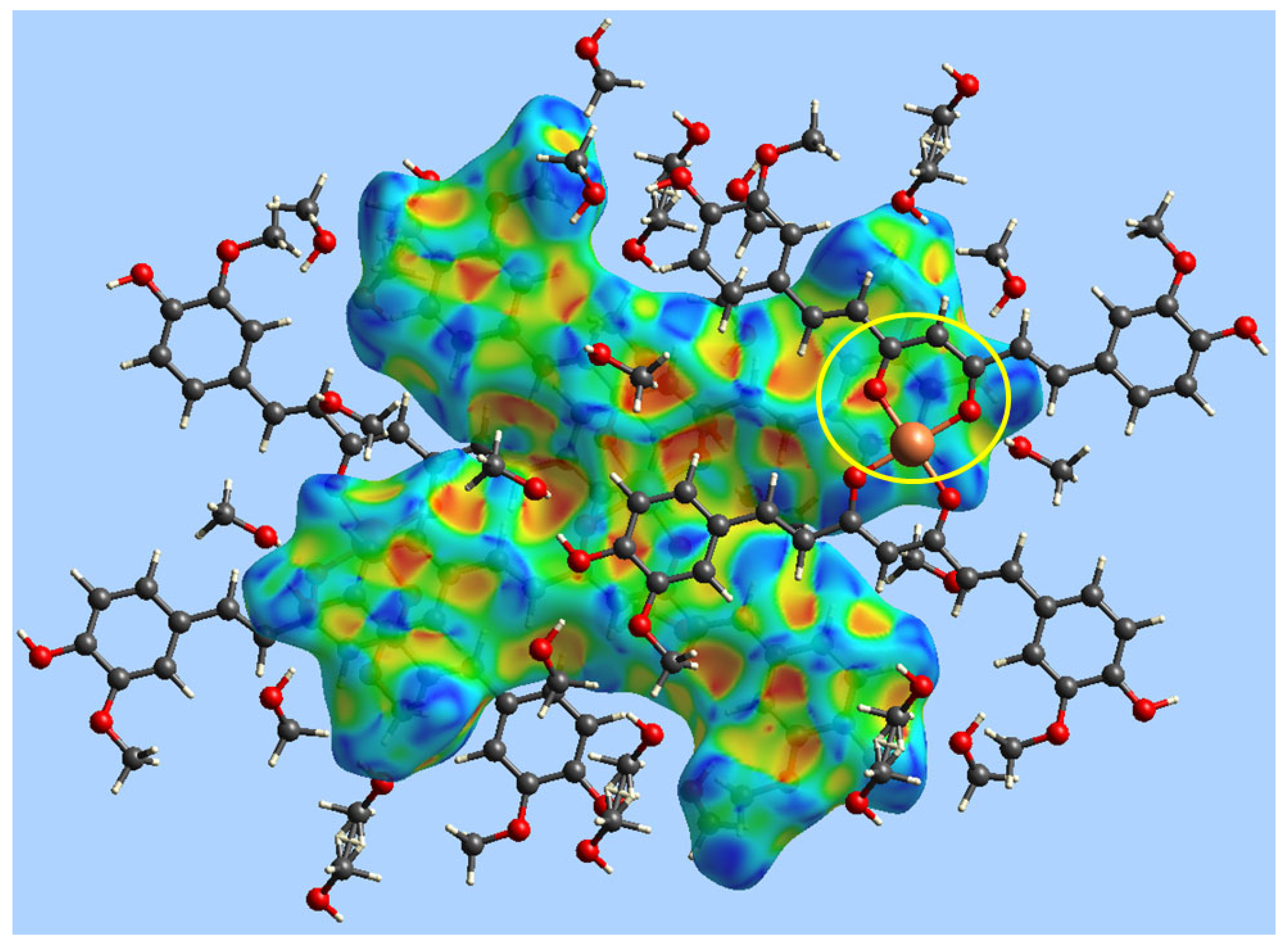
2. Results
3. Discussion
4. Materials and Methods
4.1. Physical Measurements
4.2. Spectroscopic Determinations
4.3. Biological Assays
4.4. Determination of Partition Coefficients
4.5. Synthesis of Complex 1
4.6. Powder and Single-Crystal X-ray Diffraction Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hewlings, S.J.; Kalman, D.S. Curcumin: A Review of Its Effects on Human Health. Foods 2017, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Dubourdieu, D.; Srivastava, A.; Kumar, P.; Lall, R. Metal–Curcumin Complexes in Therapeutics: An Approach to Enhance Pharmacological Effects of Curcumin. Int. J. Mol. Sci. 2021, 22, 7094. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Palanivelu, K. The Effect of Curcumin (Turmeric) on Alzheimer′s Disease: An Overview. Ann. Indian. Acad. Neurol. 2008, 11, 13. [Google Scholar] [CrossRef]
- Boarescu, P.M.; Boarescu, I.; Bocșan, I.C.; Gheban, D.; Bulboacă, A.E.; Nicula, C.; Pop, R.M.; Râjnoveanu, R.M.; Bolboacă, S.D. Antioxidant and Anti-Inflammatory Effects of Curcumin Nanoparticles on Drug-Induced Acute Myocardial Infarction in Diabetic Rats. Antioxidants 2019, 8, 504. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.; Ahmad, S.; Shah, S.W.A.; Ullah, A.; Rahman, S.U.; Ahmad, M.; Almehmadi, M.; Abdulaziz, O.; Allahyani, M.; Alsaiari, A.A.; et al. Synthetic Mono-Carbonyl Curcumin Analogues Attenuate Oxidative Stress in Mouse Models. Biomedicines 2022, 10, 2597. [Google Scholar] [CrossRef]
- Witika, B.A.; Makoni, P.A.; Matafwali, S.K.; Mweetwa, L.L.; Shandele, G.C.; Walker, R.B. Enhancement of Biological and Pharmacological Properties of an Encapsulated Polyphenol: Curcumin. Molecules 2021, 26, 4244. [Google Scholar] [CrossRef]
- Ghosh, S.; Banerjee, S.; Sil, P.C. The Beneficial Role of Curcumin on Inflammation, Diabetes and Neurodegenerative Disease: A Recent Update. Food Chem. Toxicol. 2015, 83, 111–124. [Google Scholar] [CrossRef]
- Chauhan, G.; Rath, G.; Goyal, A.K. In-Vitro Anti-Viral Screening and Cytotoxicity Evaluation of Copper-Curcumin Complex. Artif. Cells Nanomed. Biotechnol. 2013, 41, 276–281. [Google Scholar] [CrossRef]
- Reddy, R.C.; Vatsala, P.G.; Keshamouni, V.G.; Padmanaban, G.; Rangarajan, P.N. Curcumin for Malaria Therapy. Biochem. Biophys. Res. Commun. 2005, 326, 472–474. [Google Scholar] [CrossRef]
- Mahady, G.B.; Pendland, S.L.; Yun, G.; Lu, Z.Z. Turmeric (Curcuma Longa) and Curcumin Inhibit the Growth of Helicobacter Pylori, a Group 1 Carcinogen. Anticancer Res. 2002, 22, 4179–4181. [Google Scholar] [PubMed]
- Ghorbani, Z.; Hekmatdoost, A.; Mirmiran, P. Anti-Hyperglycemic and Insulin Sensitizer Effects of Turmeric and Its Principle Constituent Curcumin. Int. J. Endocrinol. Metab. 2014, 12, e18081. [Google Scholar] [CrossRef]
- Quispe, C.; Herrera-Bravo, J.; Javed, Z.; Khan, K.; Raza, S.; Gulsunoglu-Konuskan, Z.; Daştan, S.D.; Sytar, O.; Martorell, M.; Sharifi-Rad, J.; et al. Therapeutic Applications of Curcumin in Diabetes: A Review and Perspective. BioMed Res. Int. 2022, 2022, 1375892. [Google Scholar] [CrossRef] [PubMed]
- Lei, F.; Li, P.; Chen, T.; Wang, Q.; Wang, C.; Liu, Y.; Deng, Y.; Zhang, Z.; Xu, M.; Tian, J.; et al. Recent Advances in Curcumin-Loaded Biomimetic Nanomedicines for Targeted Therapies. J. Drug Deliv. Sci. Technol. 2023, 80, 104200. [Google Scholar] [CrossRef]
- de Waure, C.; Bertola, C.; Baccarini, G.; Chiavarini, M.; Mancuso, C. Exploring the Contribution of Curcumin to Cancer Therapy: A Systematic Review of Randomized Controlled Trials. Pharmaceutics 2023, 15, 1275. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.-Y.; Ngai, S.C.; Goh, B.-H.; Lee, L.-H.; Htar, T.-T.; Chuah, L.-H. Is Curcumin the Answer to Future Chemotherapy Cocktail? Molecules 2021, 26, 4329. [Google Scholar] [CrossRef]
- Zoi, V.; Galani, V.; Lianos, G.D.; Voulgaris, S.; Kyritsis, A.P.; Alexiou, G.A. The Role of Curcumin in Cancer Treatment. Biomedicines 2021, 9, 1086. [Google Scholar] [CrossRef]
- Jalili-Nik, M.; Soltani, A.; Moussavi, S.; Ghayour-Mobarhan, M.; Ferns, G.A.; Hassanian, S.M.; Avan, A. Current Status and Future Prospective of Curcumin as a Potential Therapeutic Agent in the Treatment of Colorectal Cancer. J. Cell Physiol. 2018, 233, 6337–6345. [Google Scholar] [CrossRef]
- Devassy, J.G.; Nwachukwu, I.D.; Jones, P.J.H. Curcumin and Cancer: Barriers to Obtaining a Health Claim. Nutr. Rev. 2015, 73, 155–165. [Google Scholar] [CrossRef]
- Ravindran, J.; Prasad, S.; Aggarwal, B.B. Curcumin and Cancer Cells: How Many Ways Can Curry Kill Tumor Cells Selectively? AAPS J. 2009, 11, 495–510. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of Curcumin: Problems and Promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Prasad, S.; Tyagi, A.K.; Aggarwal, B.B. Recent Developments in Delivery, Bioavailability, Absorption and Metabolism of Curcumin: The Golden Pigment from Golden Spice. Cancer Res. Treat. 2014, 46, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.-I.; Priya, A.; Balasubramaniam, B.; Muthuramalingam, P.; Sivasankar, C.; Selvaraj, A.; Valliammai, A.; Jothi, R.; Pandian, S. Biomedical Applications and Bioavailability of Curcumin—An Updated Overview. Pharmaceutics 2021, 13, 2102. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Ji, H.-F. The Pharmacology of Curcumin: Is It the Degradation Products? Trends Mol. Med. 2012, 18, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wei, D.; Jiang, B.; Liu, T.; Ni, J.; Zhou, S. Two Copper(II) Complexes of Curcumin Derivatives: Synthesis, Crystal Structure and in Vitro Antitumor Activity. Transit. Metal Chem. 2014, 39, 553–558. [Google Scholar] [CrossRef]
- Basile, V.; Ferrari, E.; Lazzari, S.; Belluti, S.; Pignedoli, F.; Imbriano, C. Curcumin Derivatives: Molecular Basis of Their Anti-Cancer Activity. Biochem. Pharmacol. 2009, 78, 1305–1315. [Google Scholar] [CrossRef]
- Medigue, N.E.H.; Bouakouk-Chitti, Z.; Bechohra, L.L.; Kellou-Taïri, S. Theoretical Study of the Impact of Metal Complexation on the Reactivity Properties of Curcumin and Its Diacetylated Derivative as Antioxidant Agents. J. Mol. Model. 2021, 27, 192. [Google Scholar] [CrossRef]
- Wanninger, S.; Lorenz, V.; Subhan, A.; Edelmann, F.T. Metal Complexes of Curcumin—Synthetic Strategies, Structures and Medicinal Applications. Chem. Soc. Rev. 2015, 44, 4986–5002. [Google Scholar] [CrossRef]
- Barik, A.; Mishra, B.; Kunwar, A.; Kadam, R.M.; Shen, L.; Dutta, S.; Padhye, S.; Satpati, A.K.; Zhang, H.Y.; Indira Priyadarsini, K. Comparative Study of Copper(II)-Curcumin Complexes as Superoxide Dismutase Mimics and Free Radical Scavengers. Eur. J. Med. Chem. 2007, 42, 431–439. [Google Scholar] [CrossRef]
- Leung Mandy, H.M.; Harada, T.; Kee Tak, W. Delivery of Curcumin and Medicinal Effects of the Copper(II)-Curcumin Complexes. Curr. Pharm. Des. 2013, 19, 2070–2083. [Google Scholar] [CrossRef]
- Vajragupta, O. Manganese Complexes of Curcumin and Its Derivatives: Evaluation for the Radical Scavenging Ability and Neuroprotective Activity. Free Radic. Biol. Med. 2003, 35, 1632–1644. [Google Scholar] [CrossRef]
- Khalil, M.I.; Al-Zahem, A.M.; Al-Qunaibit, M.H. Synthesis, Characterization, Mössbauer Parameters, and Antitumor Activity of Fe(III) Curcumin Complex. Bioinorg. Chem. Appl. 2013, 2013, 982423. [Google Scholar] [CrossRef] [PubMed]
- Al-Noor, T.H.; Ali, A.M.; Al-Sarray, A.J.A.; Al-Obaidi, O.H.; Obeidat, A.I.M.; Habash, R.R. A Short Review: Chemistry of Curcumin and Its Metal Complex Derivatives. J. Univ. Anbar Pure Sci. 2022, 16, 20–26. [Google Scholar] [CrossRef]
- Portolés-Gil, N.; Lanza, A.; Aliaga-Alcalde, N.; Ayllón, J.A.; Gemmi, M.; Mugnaioli, E.; López-Periago, A.M.; Domingo, C. Crystalline Curcumin BioMOF Obtained by Precipitation in Supercritical CO2 and Structural Determination by Electron Diffraction Tomography. ACS Sustain. Chem. Eng. 2018, 6, 12309–12319. [Google Scholar] [CrossRef]
- Su, H.; Sun, F.; Jia, J.; He, H.; Wang, A.; Zhu, G. A Highly Porous Medical Metal–Organic Framework Constructed from Bioactive Curcumin. Chem. Commun. 2015, 51, 5774–5777. [Google Scholar] [CrossRef] [PubMed]
- Wazir, S.M.; Ghobrial, I. Copper Deficiency, a New Triad: Anemia, Leucopenia, and Myeloneuropathy. J. Community Hosp. Intern. Med. Perspect. 2017, 7, 265–268. [Google Scholar] [CrossRef]
- Sierpinska, T.; Konstantynowicz, J.; Orywal, K.; Golebiewska, M.; Szmitkowski, M. Copper Deficit as a Potential Pathogenic Factor of Reduced Bone Mineral Density and Severe Tooth Wear. Osteoporos. Int. 2014, 25, 447–454. [Google Scholar] [CrossRef]
- Percival, S. Copper and Immunity. Am. J. Clin. Nutr. 1998, 67, 1064S–1068S. [Google Scholar] [CrossRef]
- Collins, J.F.; Klevay, L.M. Copper. Adv. Nutr. 2011, 2, 520–522. [Google Scholar] [CrossRef]
- Desai, V.; Kaler, S.G. Role of Copper in Human Neurological Disorders. Am. J. Clin. Nutr. 2008, 88, 855S–858S. [Google Scholar] [CrossRef]
- Xu, G.; Wang, J.; Liu, T.; Wang, M.; Zhou, S.; Wu, B.; Jiang, M. Synthesis and Crystal Structure of a Novel Copper(II) Complex of Curcumin-Type and Its Application in in Vitro and in Vivo Imaging. J. Mater. Chem. B 2014, 2, 3659–3666. [Google Scholar] [CrossRef] [PubMed]
- Pi, Z.; Wang, J.; Jiang, B.; Cheng, G.; Zhou, S. A Curcumin-Based TPA Four-Branched Copper(II) Complex Probe for in Vivo Early Tumor Detection. Mater. Sci. Eng. C 2015, 46, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, C.; Shi, H.; Yang, M.; Liu, Y.; Ji, P.; Chen, H.; Tan, R.X.; Li, E. Curcumin Is a Biologically Active Copper Chelator with Antitumor Activity. Phytomedicine 2016, 23, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Angulo, J.; Delgado-Villanueva, J. Preparación y Caracterización de Complejos de Curcumina Con Zinc(II), Níquel(II), Magnesio(II), Cobre(II) y Su Evaluación Frente a Bacterias Grampositiva y Gramnegativa. Rev. Politécnica 2023, 51, 63–72. [Google Scholar] [CrossRef]
- Su, H.; He, H.; Tian, Y.; Zhao, N.; Sun, F.; Zhang, X.; Jiang, Q.; Zhu, G. Syntheses and Characterizations of Two Curcumin-Based Cocrystals. Inorg. Chem. Commun. 2015, 55, 92–95. [Google Scholar] [CrossRef]
- Matlinska, M.A.; Wasylishen, R.E.; Bernard, G.M.; Terskikh, V.V.; Brinkmann, A.; Michaelis, V.K. Capturing Elusive Polymorphs of Curcumin: A Structural Characterization and Computational Study. Cryst. Growth Des. 2018, 18, 5556–5563. [Google Scholar] [CrossRef]
- Rajesh, J.; Gubendran, A.; Rajagopal, G.; Athappan, P. Synthesis, Spectra and DNA Interactions of Certain Mononuclear Transition Metal(II) Complexes of Macrocyclic Tetraaza Diacetyl Curcumin Ligand. J. Mol. Struct. 2012, 1010, 169–178. [Google Scholar] [CrossRef]
- Barik, A.; Mishra, B.; Shen, L.; Mohan, H.; Kadam, R.M.; Dutta, S.; Zhang, H.-Y.; Priyadarsini, K.I. Evaluation of a New Copper(II)–Curcumin Complex as Superoxide Dismutase Mimic and Its Free Radical Reactions. Free Radic. Biol. Med. 2005, 39, 811–822. [Google Scholar] [CrossRef]
- Sumi, M.; Nevaditha, N.T.; Sindhu Kumari, B. Synthesis, Spectroscopic Investigation and Bioactivities of Metal Complexes from Curcuma Longa Derivative. Inorganica Chim. Acta 2023, 549, 121397. [Google Scholar] [CrossRef]
- Shakeel Nawaz, S.; Manjunatha, K.B.; Ranganatha, S.; Supriya, S.; Ramakrishna, D. Z-Scan and Degenerate Four Wave Mixing Studies on Newly Synthesized Copper Complexes of Curcumin. Mater. Today Commun. 2023, 36, 106601. [Google Scholar] [CrossRef]
- Subhan, M.A.; Alam, K.; Rahaman, M.S.; Rahman, M.A.; Awal, R. Synthesis and Characterization of Metal Complexes Containing Curcumin (C21H20O6) and Study of Their Anti-Microbial Activities and DNA-Binding Properties. J. Sci. Res. 2013, 6, 97–109. [Google Scholar] [CrossRef]
- Leung, M.H.M.; Pham, D.-T.; Lincoln, S.F.; Kee, T.W. Femtosecond Transient Absorption Spectroscopy of Copper(Ii)–Curcumin Complexes. Phys. Chem. Chem. Phys. 2012, 14, 13580. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards Quantitative Analysis of Intermolecular Interactions with Hirshfeld Surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef] [PubMed]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting Intermolecular Interactions in Molecular Crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Allen, F.H. The Cambridge Structural Database: A Quarter of a Million Crystal Structures and Rising. Acta Crystallogr. B 2002, 58, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Caruso, F.; Rossi, M.; Benson, A.; Opazo, C.; Freedman, D.; Monti, E.; Gariboldi, M.B.; Shaulky, J.; Marchetti, F.; Pettinari, R.; et al. Ruthenium-Arene Complexes of Curcumin: X-ray and Density Functional Theory Structure, Synthesis, and Spectroscopic Characterization, in Vitro Antitumor Activity, and DNA Docking Studies of (p-Cymene)Ru(Curcuminato)Chloro. J. Med. Chem. 2012, 55, 1072–1081. [Google Scholar] [CrossRef]
- Pettinari, R.; Marchetti, F.; Di Nicola, C.; Pettinari, C.; Cuccioloni, M.; Bonfili, L.; Eleuteri, A.M.; Therrien, B.; Batchelor, L.K.; Dyson, P.J. Novel Osmium(II)–Cymene Complexes Containing Curcumin and Bisdemethoxycurcumin Ligands. Inorg. Chem. Front. 2019, 6, 2448–2457. [Google Scholar] [CrossRef]
- Pettinari, R.; Petrini, A.; Marchetti, F.; Di Nicola, C.; Scopelliti, R.; Riedel, T.; Pittet, L.D.; Galindo, A.; Dyson., P.J. Influence of Functionalized η6-Arene Rings on Ruthenium(II) Curcuminoids Complexes. ChemistrySelect 2018, 3, 6696–6700. [Google Scholar] [CrossRef]
- da Silva, B.A.; Pitasse-Santos, P.; Sueth-Santiago, V.; Monteiro, A.R.M.; Marra, R.K.F.; Guedes, G.P.; Ribeiro, R.R.; de Lima, M.E.F.; Decoté-Ricardo, D.; Neves, A.P. Effects of Cu(II) and Zn(II) Coordination on the Trypanocidal Activities of Curcuminoid-Based Ligands. Inorganica Chim. Acta 2020, 501, 119237. [Google Scholar] [CrossRef]
- Hussain, A.; Somyajit, K.; Banik, B.; Banerjee, S.; Nagaraju, G.; Chakravarty, A.R. Enhancing the Photocytotoxic Potential of Curcumin on Terpyridyl Lanthanide(III) Complex Formation. Dalton Trans. 2013, 42, 182–195. [Google Scholar] [CrossRef]
- Banerjee, S.; Prasad, P.; Hussain, A.; Khan, I.; Kondaiah, P.; Chakravarty, A.R. Remarkable Photocytotoxicity of Curcumin in HeLa Cells in Visible Light and Arresting Its Degradation on Oxovanadium(Iv) Complex Formation. Chem. Commun. 2012, 48, 7702. [Google Scholar] [CrossRef]
- Halevas, E.; Pekou, A.; Papi, R.; Mavroidi, B.; Hatzidimitriou, A.G.; Zahariou, G.; Litsardakis, G.; Sagnou, M.; Pelecanou, M.; Pantazaki, A.A. Synthesis, Physicochemical Characterization and Biological Properties of Two Novel Cu(II) Complexes Based on Natural Products Curcumin and Quercetin. J. Inorg. Biochem. 2020, 208, 111083. [Google Scholar] [CrossRef] [PubMed]
- Triantis, C.; Tsotakos, T.; Tsoukalas, C.; Sagnou, M.; Raptopoulou, C.; Terzis, A.; Psycharis, V.; Pelecanou, M.; Pirmettis, I.; Papadopoulos, M. Synthesis and Characterization of fac-[M(CO)3(P)(OO)] and cis-trans-[M(CO)2(P)2(OO)] Complexes (M = Re, 99mTc) with Acetylacetone and Curcumin as OO Donor Bidentate Ligands. Inorg. Chem. 2013, 52, 12995–13003. [Google Scholar] [CrossRef] [PubMed]
- Rohlíček, J.; Skořepová, E.; Babor, M.; Čejka, J. CrystalCMP: An Easy-to-Use Tool for Fast Comparison of Molecular Packing. J. Appl. Crystallogr. 2016, 49, 2172–2183. [Google Scholar] [CrossRef]
- Yan, F.-S.; Sun, J.-L.; Xie, W.-H.; Shen, L.; Ji, H.-F. Neuroprotective Effects and Mechanisms of Curcumin–Cu(II) and –Zn(II) Complexes Systems and Their Pharmacological Implications. Nutrients 2017, 10, 28. [Google Scholar] [CrossRef]
- Zebib, B.; Mouloungui, Z.; Noirot, V. Stabilization of Curcumin by Complexation with Divalent Cations in Glycerol/Water System. Bioinorg. Chem. Appl. 2010, 2010, 292760. [Google Scholar] [CrossRef]
- Khorasani, M.Y.; Langari, H.; Sany, S.B.T.; Rezayi, M.; Sahebkar, A. The Role of Curcumin and Its Derivatives in Sensory Applications. Mater. Sci. Eng. C 2019, 103, 109792. [Google Scholar] [CrossRef]
- Shakeri, A.; Panahi, Y.; Johnston, T.P.; Sahebkar, A. Biological Properties of Metal Complexes of Curcumin. BioFactors 2019, 45, 304–317. [Google Scholar] [CrossRef]
- Shahabadi, N.; Falsafi, M.; Moghadam, N.H. DNA Interaction Studies of a Novel Cu(II) Complex as an Intercalator Containing Curcumin and Bathophenanthroline Ligands. J. Photochem. Photobiol. B 2013, 122, 45–51. [Google Scholar] [CrossRef]
- Meza-Morales, W.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Arenaza-Corona, A.; Ramírez-Apan, M.T.; Toscano, R.A.; Poveda-Jaramillo, J.C.; Enríquez, R.G. Three New Coordination Geometries of Homoleptic Zn Complexes of Curcuminoids and Their High Antiproliferative Potential. RSC Adv. 2023, 13, 8577–8585. [Google Scholar] [CrossRef]
- Priyadarsini, K. The Chemistry of Curcumin: From Extraction to Therapeutic Agent. Molecules 2014, 19, 20091–20112. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar Rao, A.; Prasad, E.; Deepthi, S.S.; Ansari, I.A. Synthesis and Biological Evaluation of Glucosyl Curcuminoids. Arch. Pharm. 2014, 347, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Priya, R.S.; Balachandran, S.; Daisy, J.; Mohanan, P.V. Reactive Centers of Curcumin and the Possible Role of Metal Complexes of Curcumin as Antioxidants. Univers. J. Phys. Appl. 2015, 9, 6–16. [Google Scholar] [CrossRef]
- Jakubczyk, K.; Drużga, A.; Katarzyna, J.; Skonieczna-Żydecka, K. Antioxidant Potential of Curcumin—A Meta-Analysis of Randomized Clinical Trials. Antioxidants 2020, 9, 1092. [Google Scholar] [CrossRef]
- Obregón-Mendoza, M.A.; Meza-Morales, W.; Alvarez-Ricardo, Y.; Estévez-Carmona, M.M.; Enríquez, R.G. High Yield Synthesis of Curcumin and Symmetric Curcuminoids: A “Click” and “Unclick” Chemistry Approach. Molecules 2022, 28, 289. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, F.; Wang, Z.T. Antioxidative Activity of Natural Products from Plants. Life Sci. 2000, 66, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Meza-Morales, W.; Alejo-Osorio, Y.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Machado-Rodriguez, J.C.; Arenaza-Corona, A.; Toscano, R.A.; Ramírez-Apan, M.T.; Enríquez, R.G. Homoleptic Complexes of Heterocyclic Curcuminoids with Mg(II) and Cu(II): First Conformationally Heteroleptic Case, Crystal Structures, and Biological Properties. Molecules 2023, 28, 1434. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for Lipid Peroxides in Animal Tissues by Thiobarbituric Acid Reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Obregón-Mendoza, M.A.; Arias-Olguín, I.I.; Estévez-Carmona, M.M.; Meza-Morales, W.; Alvarez-Ricardo, Y.; Toscano, R.A.; Arenas-Huertero, F.; Cassani, J.; Enríquez, R.G. Non-Cytotoxic Dibenzyl and Difluoroborate Curcuminoid Fluorophores Allow Visualization of Nucleus or Cytoplasm in Bioimaging. Molecules 2020, 25, 3205. [Google Scholar] [CrossRef]
- Andrés, A.; Rosés, M.; Ràfols, C.; Bosch, E.; Espinosa, S.; Segarra, V.; Huerta, J.M. Setup and Validation of Shake-Flask Procedures for the Determination of Partition Coefficients (LogD) from Low Drug Amounts. Eur. J. Pharm. Sci. 2015, 76, 181–191. [Google Scholar] [CrossRef]
- Roisnel, T.; Rodriguez-Carvajal, J. WinPLOTR: A windows tool for powder diffraction patterns analysis materials science forum. In Proceedings of the Seventh European Powder Diffraction Conference (EPDIC 7), Barcelona, Spain, 20–23 May 2000; pp. 118–123. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt Graphical User Interface for SHELXL. J. Appl. Crystallogr. 2011, 44, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From Visualization to Analysis, Design and Prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed]
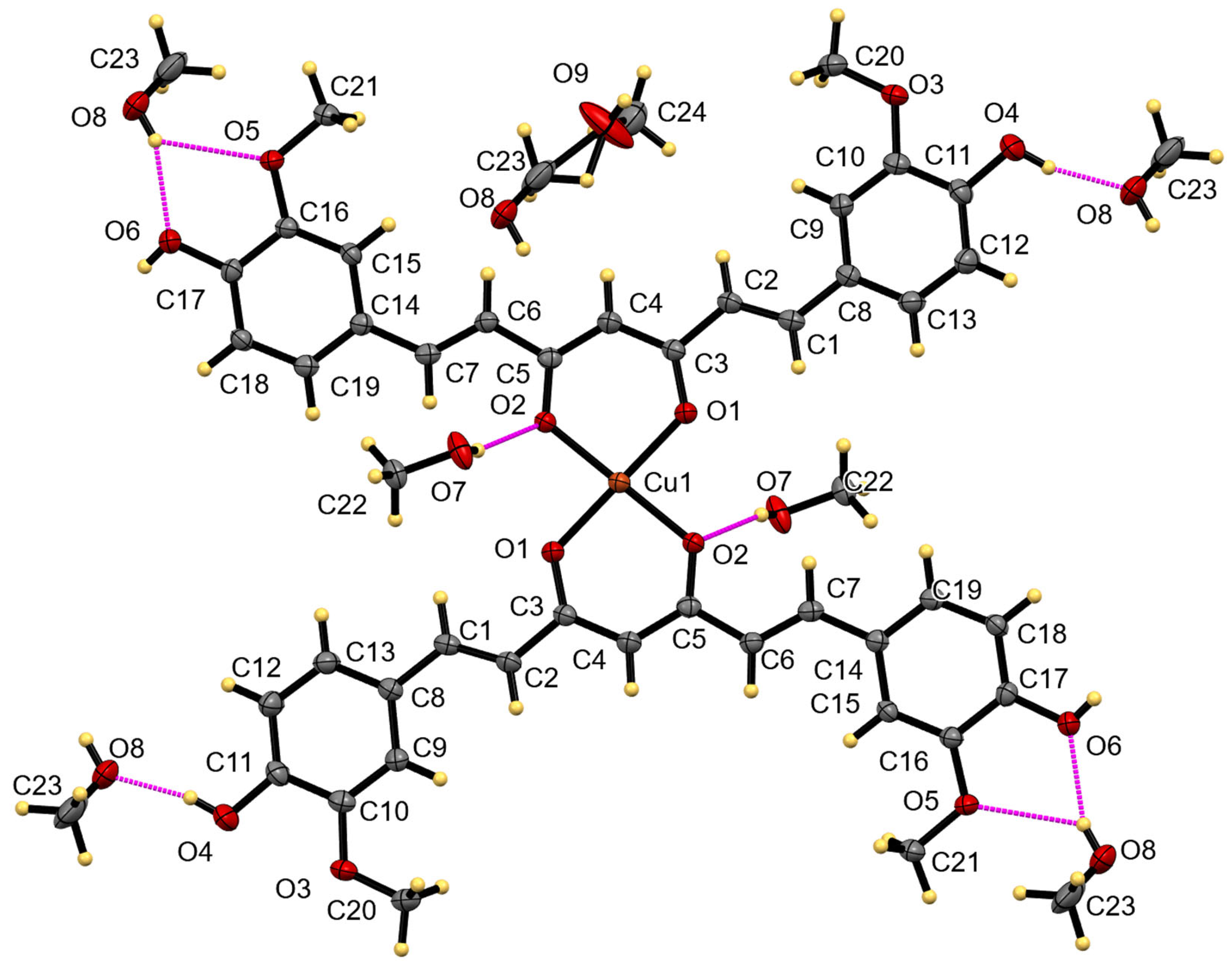


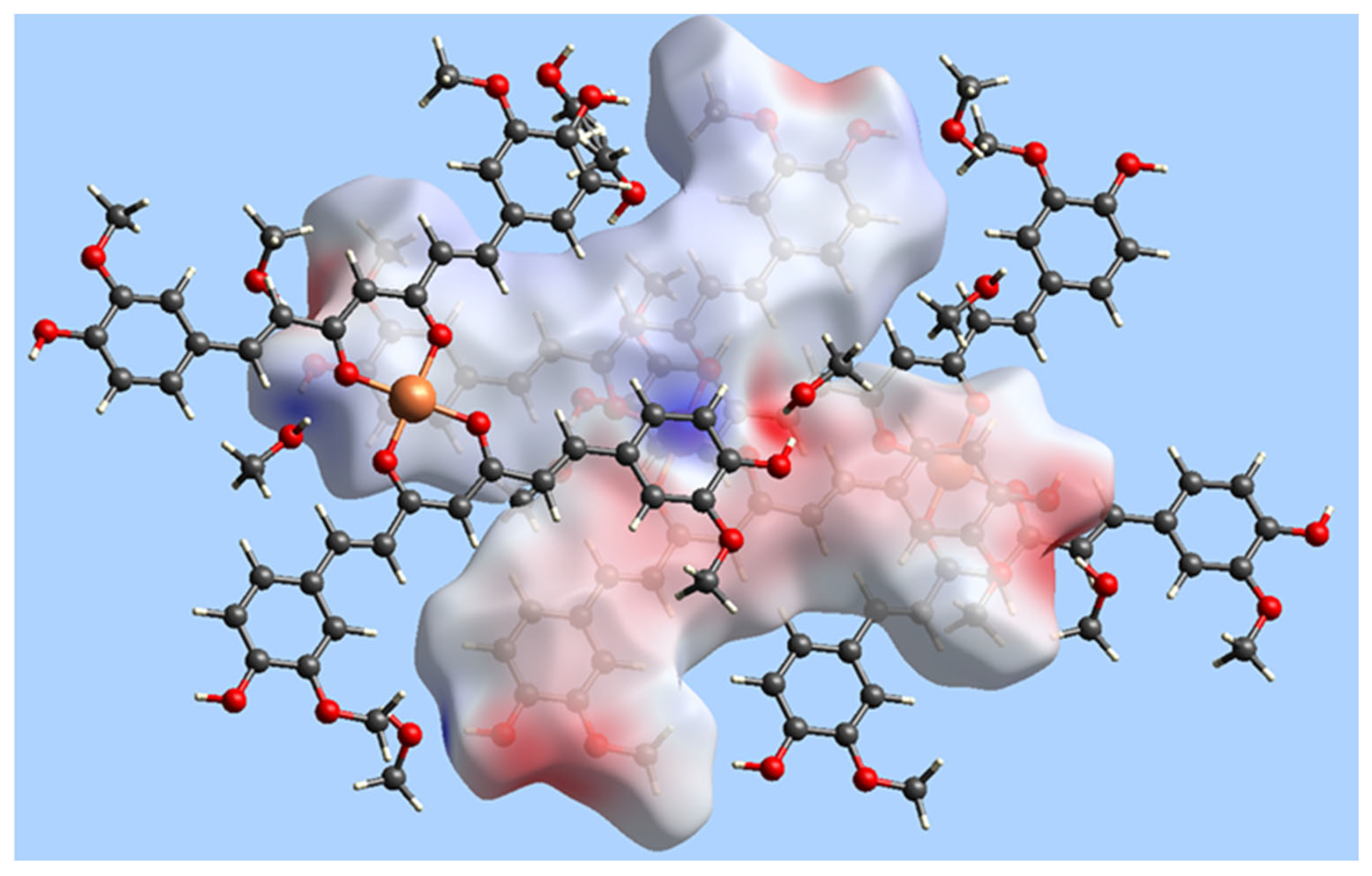
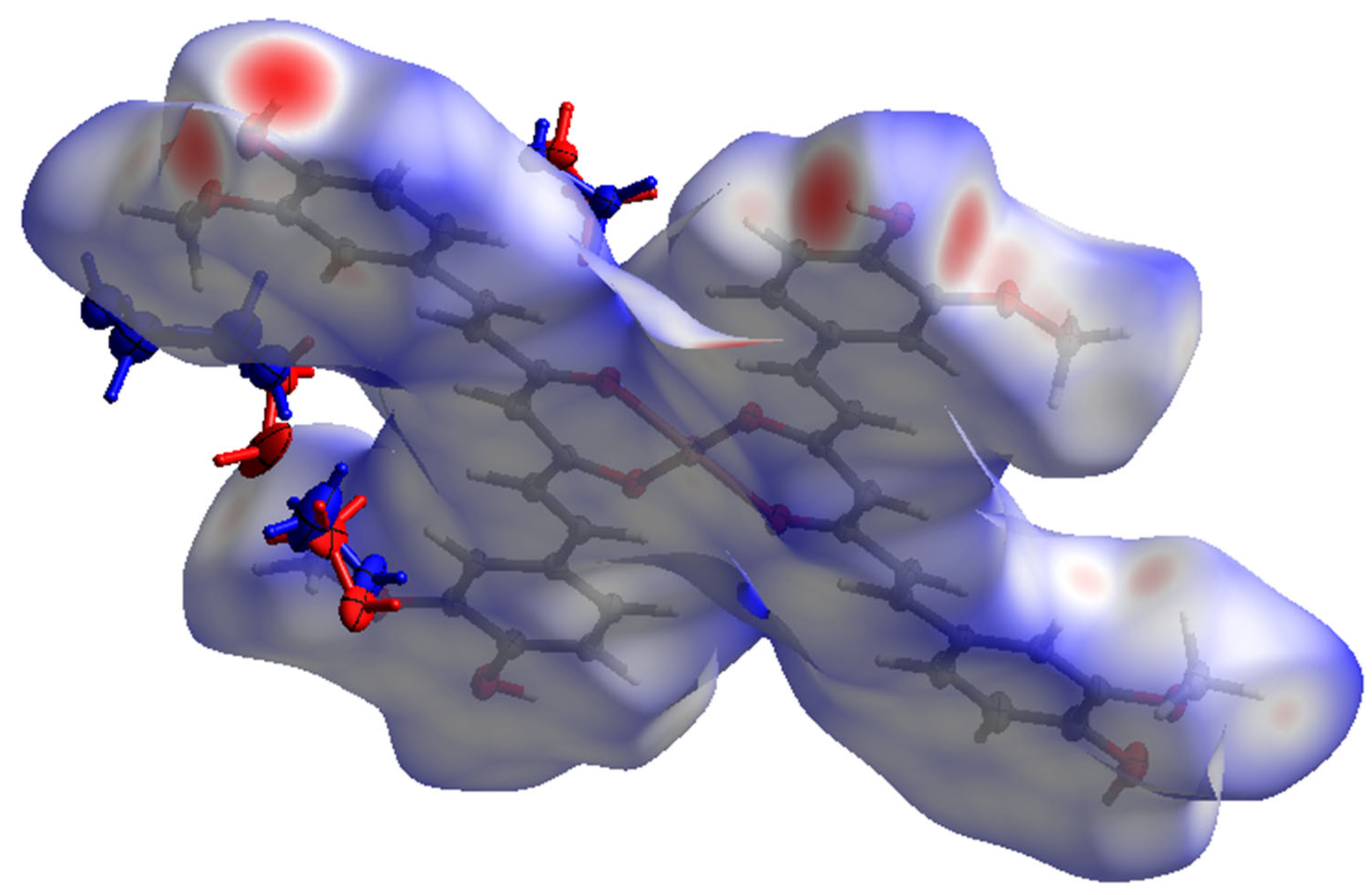
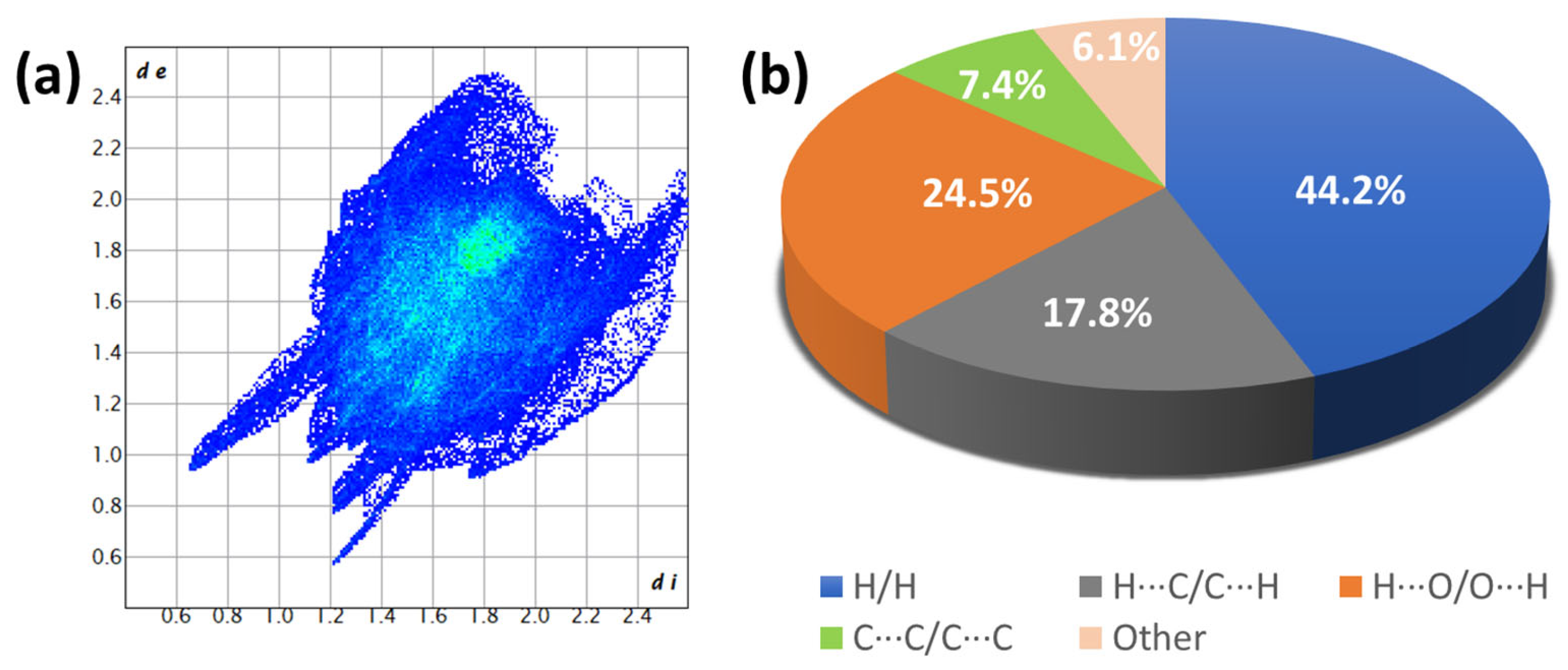
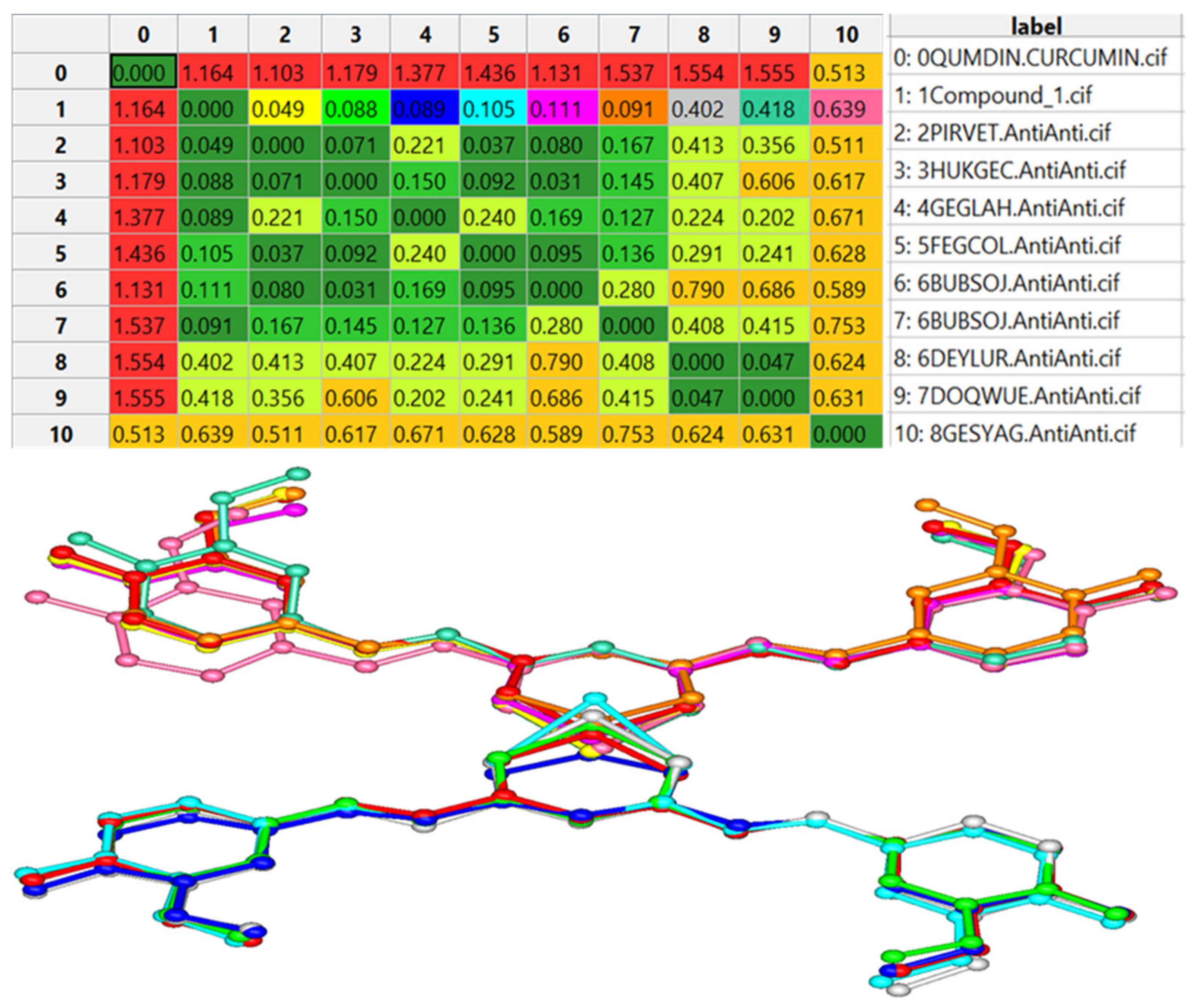
| Bond | Length | Bond | Length |
|---|---|---|---|
| Cu(1)-O(1) | 1.9189(15) | C(3)-C(4) | 1.407(3) |
| Cu(1)-O(2) | 1.9191(14) | C(4)-C(5) | 1.389(3) |
| O(1)-C(3) | 1.280(2) | C(5)-C(6) | 1.469(3) |
| O(2)-C(5) | 1.292(2) | C(6)-C(7) | 1.344(3) |
| C(1)-C(2) | 1.340(3) | C(7)-C(14) | 1.457(3) |
| C(1)-C(8) | 1.458(3) | ||
| C(2)-C(3) | 1.466(3) | ||
| Bonds | Angle | Bonds | Angle |
| O(1)-Cu(1)-O(2)#1 | 85.99(6) | C(1)-C(2)-C(3)-O(1) | −10.6(3) |
| O(1)-Cu(1)-O(2) | 94.01(6) | C(1)-C(2)-C(3)-C(4) | 169.6(2) |
| C(2)-C(1)-C(8) | 126.4(2) | O(1)-C(3)-C(4)-C(5) | 7.7(4) |
| C(1)-C(2)-C(3) | 123.3(2) | C(2)-C(3)-C(4)-C(5) | −172.5(2) |
| O(1)-C(3)-C(4) | 124.7(2) | Cu(1)-O(2)-C(5)-C(4) | −5.3(3) |
| O(1)-C(3)-C(2) | 118.2(2) | Cu(1)-O(2)-C(5)-C(6) | 175.8(1) |
| C(4)-C(3)-C(2) | 117.1(2) | C(3)-C(4)-C(5)-O(2) | 0.0(4) |
| C(5)-C(4)-C(3) | 125.2(2) | C(3)-C(4)-C(5)-C(6) | 178.9(2) |
| O(2)-C(5)-C(4) | 124.3(2) | O(2)-C(5)-C(6)-C(7) | −9.4(3) |
| O(2)-C(5)-C(6) | 117.0(2) | C(4)-C(5)-C(6)-C(7) | 171.6(2) |
| C(4)-C(5)-C(6) | 118.6(2) | C(5)-C(6)-C(7)-C(14) | 175.3(2) |
| C(7)-C(6)-C(5) | 122.2(2) | C(2)-C(1)-C(8)-C(9) | 8.5(3) |
| C(6)-C(7)-C(14) | 127.6(2) | C(6)-C(7)-C(14)-C(15) | −2.2(3) |
| D-H···A | d(D-H) | d(H···A) | d(D···A) | <(DHA) |
|---|---|---|---|---|
| O(4)-H(4A)...O(8) #1 | 0.83 | 1.76 | 2.552(12) | 160.9 |
| O(4)-H(4A)...O(8B) #1 | 0.83 | 1.88 | 2.694(12) | 166.7 |
| O(6)-H(6A)...O(7) #2 | 0.76 | 2.02 | 2.767(12) | 170.1 |
| O(6)-H(6A)...O(7B) #2 | 0.76 | 1.82 | 2.574(10) | 174.9 |
| C(9)-H(9)...O(9B) | 0.95 | 2.52 | 3.431(9) | 161.0 |
| C(18)-H(18)...O(7) #2 | 0.95 | 2.53 | 3.197(12) | 127.4 |
| C(20)-H(20A)...O(9B) | 0.98 | 2.41 | 3.208(9) | 138.4 |
| C(21)-H(21A)...O(4) #3 | 0.98 | 2.64 | 3.580(3) | 161.1 |
| O(7)-H(7A)...O(2) #4 | 0.84 | 1.96 | 2.734(15) | 153.2 |
| O(8)-H(8)...O(5) #5 | 0.80 | 2.42 | 2.988(12) | 129.3 |
| O(8)-H(8)...O(6) #5 | 0.80 | 2.04 | 2.801(12) | 159.7 |
| C(23)-H(23C)...O(5) #6 | 0.98 | 2.46 | 3.129(10) | 125.1 |
| O(7B)-H(7B)...O(2) #4 | 0.84 | 1.99 | 2.786(14) | 158.5 |
| O(8B)-H(8B)...O(5) #5 | 0.84 | 2.39 | 3.070(11) | 138.5 |
| O(8B)-H(8B)...O(6) #5 | 0.84 | 2.11 | 2.861(11) | 148.8 |
| O(9)-H(9A)...O(4) #6 | 0.83 | 1.94 | 2.768(9) | 179.3 |
| C(24B)-H(24F)...O(4) #7 | 0.98 | 2.62 | 3.303(13) | 126.9 |
| Complex | U-251 (IC50 = µM) | K562 (IC50 = µM) |
|---|---|---|
| Complex (1) | 5.2 ± 0.2 | 9.5 ± 0.3 |
| Curcumin * | 20.5 ± 1.7 | 16.4 ± 0.04 |
| Complex | (IC50 = µM) |
|---|---|
| Complex (1) | 1.24 ± 0.08 |
| Curcumin | 3.03 ± 0.15 |
| BHT | 1.22 ± 0.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arenaza-Corona, A.; Obregón-Mendoza, M.A.; Meza-Morales, W.; Ramírez-Apan, M.T.; Nieto-Camacho, A.; Toscano, R.A.; Pérez-González, L.L.; Sánchez-Obregón, R.; Enríquez, R.G. The Homoleptic Curcumin–Copper Single Crystal (ML2): A Long Awaited Breakthrough in the Field of Curcumin Metal Complexes. Molecules 2023, 28, 6033. https://doi.org/10.3390/molecules28166033
Arenaza-Corona A, Obregón-Mendoza MA, Meza-Morales W, Ramírez-Apan MT, Nieto-Camacho A, Toscano RA, Pérez-González LL, Sánchez-Obregón R, Enríquez RG. The Homoleptic Curcumin–Copper Single Crystal (ML2): A Long Awaited Breakthrough in the Field of Curcumin Metal Complexes. Molecules. 2023; 28(16):6033. https://doi.org/10.3390/molecules28166033
Chicago/Turabian StyleArenaza-Corona, Antonino, Marco A. Obregón-Mendoza, William Meza-Morales, María Teresa Ramírez-Apan, Antonio Nieto-Camacho, Rubén A. Toscano, Leidys L. Pérez-González, Rubén Sánchez-Obregón, and Raúl G. Enríquez. 2023. "The Homoleptic Curcumin–Copper Single Crystal (ML2): A Long Awaited Breakthrough in the Field of Curcumin Metal Complexes" Molecules 28, no. 16: 6033. https://doi.org/10.3390/molecules28166033
APA StyleArenaza-Corona, A., Obregón-Mendoza, M. A., Meza-Morales, W., Ramírez-Apan, M. T., Nieto-Camacho, A., Toscano, R. A., Pérez-González, L. L., Sánchez-Obregón, R., & Enríquez, R. G. (2023). The Homoleptic Curcumin–Copper Single Crystal (ML2): A Long Awaited Breakthrough in the Field of Curcumin Metal Complexes. Molecules, 28(16), 6033. https://doi.org/10.3390/molecules28166033






