General Synthetic Methods
Reagents and solvents were purchased from commercial suppliers and used without further purification unless otherwise stated. Toluene was dried by distillation first over phosphorus pentoxide then over calcium hydride and was then stored over activated 4 Å molecular sieves (MS). Solvents were dried by storage over activated MS for at least 24 h prior to use (dichloromethane 4 Å, acetonitrile, and DMF 3 Å). Residual moisture was determined by colorimetric titration on a Mitsubishi CA21 Karl Fischer apparatus and did not exceed 20 ppm. Reactions were monitored by TLC performed on silica gel 60 F254 HPTLC precoated glass plates with a 25 mm concentration zone (Merck, Darmstadt, Germany). Spots were visualized by dipping into a sulfuric acid—
p-anisaldehyde solution and subsequent charring at 250 °C. Solvents were removed under reduced pressure at ≤40 °C. Column chromatography was performed using silica gel 60 (0.040–0.063 mm). Size exclusion chromatography was performed using Bio-Beads S-X1 support (BioRad, Darmstadt, Germany). NMR spectra were recorded on a Bruker Avance III 600 spectrometer (
1H at 600.22 MHz;
13C at 150.93 MHz;
31P at 242.97 MHz) using standard Bruker NMR software. Chemical shifts are reported in ppm, where
1H NMR spectra recorded from samples in CDCl
3 were referenced to internal TMS and
13C spectra were referenced to the corresponding solvent signal (77.16 ppm for CDCl
3). NMR spectra recorded from samples in other solvents were referenced to residual solvent signals (for CD
3OD 3.31 and 49.00 ppm; for CD
2Cl
2 5.32 and 53.84 ppm; for DMSO-d
6 2.50 and 39.52 ppm; for
1H- and
13C-NMR, respectively). NMR spectra recorded in CDCl
3-MeOD (4:1,
v/
v) were referenced to residual solvent signals of CDCl
3 (7.26 ppm and 77.16 ppm;
1H- and
13C-NMR, respectively). NMR spectra recorded in CDCl
3:MeOD (1:1 to 4:1,
v/
v) were referenced to residual solvent signals of MeOD (3.31 and 49.00 ppm,
1H and
13C NMR, respectively).
31P-NMR spectra were referenced according to IUPAC recommendations from a referenced
1H-NMR spectrum. The NMR signals of the β-configured GlcN ring are indicated by primes. The NMR-spectra of all synthetic compounds can be found in the
Supporting Information file. High-resolution mass spectrometry (HRMS) was carried out on acetonitrile or acetonitrile-dichloromethane solutions via LC-TOF MS (Agilent 1200SL HPLC and Agilent 6210 ESI-TOF, Agilent Technologies, Santa Clara, CA, USA). Datasets were analysed using Agilent Mass Hunter Software. MALDI-TOF MS was performed in negative-ion mode using a Bruker Autoflex Speed instrument with 6-aza-2-thiothymine (ATT) as matrix and ammonium sulfate as additive. Optical rotation was measured on a PerkinElmer 243B polarimeter (PerkinElmer equipped with a Haake water circulation bath and a Haake D1 immersion circulator for temperature control or an Anton Paar MCP 100 polarimeter (Anton Paar, Graz, Austria) featuring integrated Peltier temperature control. All
values are reported in units of deg*dm
−1*cm
3*g
−1, the corresponding concentrations are reported in g/100 mL.
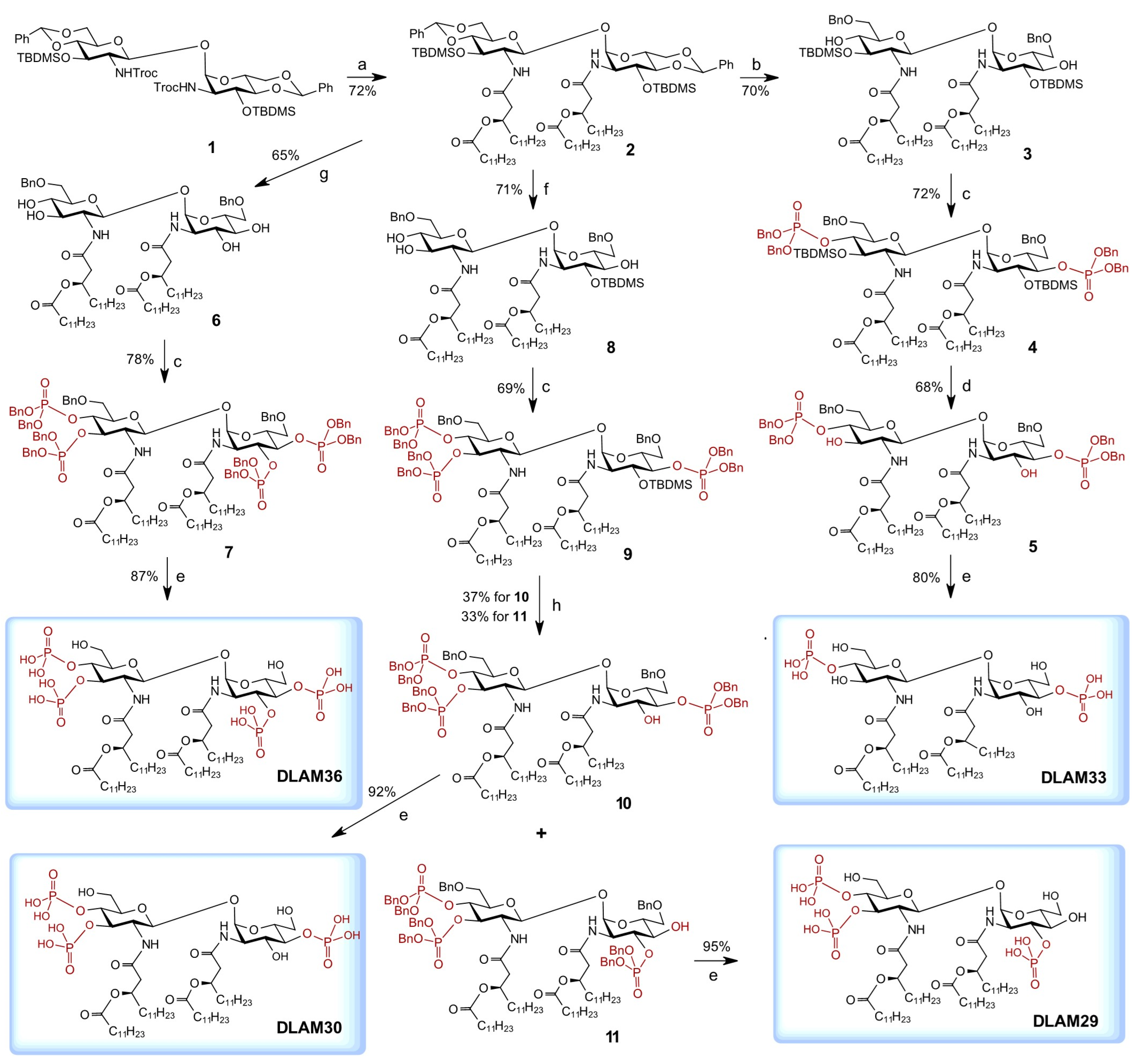
4,6-O-Benzylidene-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)-tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-4,6-O-benzylidene-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (2). To a cooled (0 °C) stirred solution of
1 (156 mg, 140 µmol) in DCM (2 mL), acetic acid (3 mL) and Zn powder (330 mg, 5 mmol) were added and the mixture was stirred for 20 h at 0 °C then diluted with DCM (10 mL), the solids were removed by filtration over a pad of Celite, then the filtrate was diluted with toluene (10 mL) and concentrated. The residue was repeatedly co-evaporated with toluene (3 × 10 mL), redissolved in chloroform (50 mL), and washed with satd. aq. NaHCO
3 (2 × 20 mL), water (20 mL), and brine (20 mL). The organic layer was dried over Na
2SO
4, filtered over cotton, and concentrated. The residue was taken up in dry DCM (5 mL) and solutions of (
R)-3-(dodecanoyloxy)tetradecanoic acid C
11H
23O(C
11H
23CO)CH
2COOH [
53] (149 mg, 350 µmol) in DCM (4 mL) and DIC (44 mg, 55 µL, 350 µmol) were added under stirring in the atmosphere of Ar. The reaction mixture was stirred for 20 h, then diluted with EtOAc (50 mL) and washed with satd. aq. NaHCO
3 (2 × 30 mL), water (30 mL), and brine (30 mL). The organic layer was dried over Na
2SO
4, filtered over cotton, and concentrated. The residue was purified by column chromatography on silica gel (toluene-EtOAc, 6:1) to give
2 (158 mg, 72%). R
f = 0.5 (toluene-EtOAc, 4:1);
= 9.0 (
c = 1, CHCl
3).
1H NMR (600 MHz, CDCl
3): δ 7.49–7.45 (m, 4H, Ph), 7.37–7.34 (m, 6H, Ph), 6.71 (d, 1H,
J2-NH = 7.9 Hz, NH′), 5.97 (d, 1H, NH), 5.48, 5.47 (2s, 2H, C
HPh), 5.11 (m, 1H, βC
HMyr), 5.03 (d, 1H,
J1′-2′ = 8.7 Hz, H-1′), 5.02 (d, 1H,
J1-2 = 1.5 Hz, H-1), 5.0 (m, 1H, βC
HMyr), 4.26 (dd, 1H,
J5′-6′b = 4.8 Hz, H-6′b), 4.21 (ddd, 1H,
J2-3 = 3.7,
J2-NH= 9.6 Hz, H-2), 4.19 (t, 1H,
J2′-3′ =
J3′-4′ = 8.7 Hz, H-3′), 4.15 (dd, 1H,
J6a-6b = 10.4 Hz, H-6b), 4.05 (ddd, 1H,
J4-5 =
J5-6a = 9.9 Hz,
J5-6b = 5.0 Hz, H-5), 3.86 (t, 1H, H-3), 3.75 (t, 1H,
J6′b-6′a =
J5′-6′a= 10.0 Hz, H-6′a), 3.69 (t, 1H, H-6a), 3.52 (dd, 1H,
J3-4 = 9.2 Hz, H-4), 3.51–3.47 (m, 2H, H-5′, H-2′), 3.45 (dd, 1H,
J4′-5′ = 8.9 Hz, H-4′), 2.60–2.25 (m, 8H, 2×αC
H2Myr, 2×αC
H2Lau), 1.68–1.57 (m, 8H, 2×βC
H2Lau, 2×γC
H2Myr), 1.36–1.20 (m, 68H, C
H2Myr,Lau), 0.89–0.87 (m, 12H, C
H3Myr,Lau), 0.84 and 0.80 (2s, 18H,
tBuSi), 0.02, 0.00, −0.04, and −0.05 (4s, 12H, SiMe).
13C NMR (150.9 MHz, CDCl
3): δ 174.60, 173.81, 170.61, 169.75 (4C, 4×
C=O), 137.50, 137.44 (2C,
Cq-Ph), 129.33, 129.26, 128.38, 128.35, 126.62 (10C, Ph), 102.21, 102.17 (2C, 2Ph
CH), 100.18 (1C, C-1′), 99.15 (1C, C-1), 82.51 (1C, C-4), 82.24 (1C, C-4′), 72.49 (1C,
CH
Myr), 71.90 (1C, C-3′), 71.32 (1C,
CH
Myr), 71.07 (1C, C-3), 68.98 (2C, C-6, C-6′), 66.26 (1C, C-5′), 63.61 (1C, C-5), 59.68 (1C, C-2′), 54.41 (1C, C-2), 41.79 (4C, 2×α
CMyr, 2×α
CLau), 35.06, 35.02, 34.97, 34.86, 32.18, 29.95, 29.90, 29,87, 29.81, 29.79, 29.74, 29.67, 29.62, 29.52, 29.50 (
CH
2Myr,Lau)
, 26.05, 25.97 (6C,
tBuSi), 25.73, 25.64, 25.37, 25.27, 22.94, 22.51 (
CH
2Myr,Lau), 14.35 (4C,
CH
3Myr,Lau), −3.71, −3.77, −4.52, −4.57 (4C, SiMe). HRMS (ESI-TOF)
m/
z: found 1562.3742, calc. for [M + H]
+ C
90H
156N
2O
15Si
2 + H
+:
m/
z = 1562.3744.
6-O-Benzyl-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (3). To a stirred solution of 2 (97 mg, 62 µmol) in dry DCM (15 mL), molecular sieves 4 Å (100 mg) were added and the suspension was stirred for 2 h. at r.t. in an atmosphere of Ar. The mixture was cooled to −85 °C (hexane/liquid N2) and a solution of triethylsilane (25 µL, 150 µmol, 2.5 eq) in dry DCM (5% solution) was added under stirring. Next, a solution of trifluoromethanesulfonic acid (13 µL, 150 µmol, 2.5 eq) in dry DCM (5% solution) was added, and the stirring was continued for 4 h at −85 °C under atmosphere of Ar. The reaction mixture was quenched by addition of a solution of Et3N (30 µL, 220 µmol, 3.5 eq) in DCM (5% solution). The mixture was stirred for 15 min at −85 °C, then brought up to r.t. and diluted with DCM (50 mL). The solids were removed by filtration over a pad of Celite, the filtrate was washed with satd. aq. NaHCO3 (30 mL), water (30 mL), and brine (30 mL), dried over Na2SO4, filtered over cotton, and concentrated. The residue was purified by column chromatography on silica gel (toluene-EtOAc, 10:1 → 4:1) which gave 3 (68 mg, 70%). Rf = 0.3 (toluene-EtOAc, 3:1); = 15 (c = 1, CHCl3). 1H NMR (600 MHz, CDCl3): δ 7.36–7.28 (m, 10H, Ph), 6.99 (d, 1H, NH′), 5.73 (d, 1H, NH), 5.27 (d, 1H, J1′-2′ = 9.0 Hz, H-1′), 5.21 (m, 1H, βCHMyr), 5.16 (d, 1H, J1-2 = 3.5 Hz, H-1), 5.07 (m, 1H, βCHMyr), 4.57–4.50 (m, 4H, CH2-Bn), 4.35 (t, 1H, J3-4 = 9.0 Hz, H-3′), 4.11 (ddd, 1H, J4-5 = 10.1 Hz, J5-6b =2.4 Hz, J5-6a = 7.7 Hz, H-5), 4.08 (dt, 1H, J2-NH = J2-3 = 9.9 Hz, H-2), 3.85 (dd, 1H, H-6b), 3.71 (dd, 1H, J6a′-6b′ = 5.6 Hz, H-6a′), 3.66 (t, 1H, H-6b′), 3.65 (dd, 1H, J3-4 = 4.3 Hz, H-3), 3.55 (dt, 1H, J5′-6a′ = J4′-5′ = 9.9 Hz, J5′-6b′ = 5.1 Hz, H-5′), 3.47 (dd, 1H, J6a-6b = 9.8 Hz, H-6a), 3.38 (t, 1H, H-4′), 3.27 (ddd, 1H, H-4), 3.16 (dt, 1H, J2′-3′ = 9.0 Hz, J2′-NH′ = 7.2 Hz, H-2′), 2.56–2.20 (m, 8H, 2×αCH2Myr, 2×αCH2Lau), 1.64–1.52 (m, 8H, 2×βCH2Lau, 2×γCH2Myr), 1.30–1.21 (m, 68 H, CH2Myr,Lau), 0.91–0.83 (m, 30H, CH3Myr,Lau, tBuSi), 0.10, 0.09, 0.07, 0.05 (4s, 12H, SiMe). 13C NMR (150.9 MHz, CDCl3): δ 173.58, 173.29, 170.32, 169.55 (4C, 4×C=O), 137.79, 137.07 (2C, Cq-Ph), 128.62, 128.49, 128.10, 128.01, 127.81, 127.64 (10C, Ph), 96.50 (1C, C-1′), 93.50 (1C, C-1), 73.95 (1C, C-4′), 73.72 (1C, C-3), 73.7, 73.61 (2C, CH2-Bn), 73.59 (1C, C-5), 73.25 (1C, C-3′), 72.71 (1C, C-4), 71.40 (1C, CHMyr), 71.28 (1C, C-5), 70.89 (1C, CHMyr), 70.80 (1C, C-6′), 70.60 (1C, C-6), 58.90 (1C, C-2′), 52.95 (1C, C-2), 41.78, 41.08 (4C, 2×αCMyr, 2×αCLau), 34.95, 34.65, 34.61, 34.30, 31.92, 29.71, 29.67, 29-65, 29.64, 29.58, 29.56, 29.54, 29.49, 29.47, 29.36, 29.23, 29.21 (CH2Myr,Lau), 25.92, 25.78 (6C, tBuSi), 25.34, 25.22, 25.09, 25.07, 22.68 (CH2Myr,Lau), 14.10 (4C, CH3Myr,Lau), −4.02, −4.17, −4.32, −4.57 (4C, SiMe). HRMS (ESI-TOF) m/z: found 1566.1460, calc. for [M + H]+ C90H160N2O15Si2 + H+: m/z = 1566.1432.
6-O-Benzyl-4-O-[bis(benzyloxy)phosphoryl]-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-4-O-[bis(benzyloxy)phosphoryl]-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (4). To a stirred solution of 3 (40 mg, 26 µmol) in dry DCM (4 mL), bisbenzyloxy(diisopropylamino)phosphine (27 mg, 26 µL, 77 µmol) and a solution of 1H-tetrazol (16 mg, 155 µmol, 0.45 M in CH3CN) were added successively and the stirring was continued for 1 h at r.t. in an atmosphere of Ar. The reaction mixture was cooled to −78 °C, a solution of m-chloroperoxybenzoic acid (mCPBA) (33 mg, 116 µmol) in DCM (0.6 mL) was added, and the stirring was continued for 2 h. The reaction was quenched by addition of a solution of Et3N (50 µL) in DCM (0.5 mL) and the mixture was stirred for 10 min at −78 °C, then allowed to warm up to r.t. The mixture was diluted with DCM (50 mL) and washed with satd. aq. NaHCO3 (2 × 20 mL), water (20 mL), and brine (20 mL). The organic phase was dried over Na2SO4, filtered over cotton, and concentrated. The residue was purified by column chromatography on silica gel (toluene – EtOAc, 6:1 → 4:1) to give 4 (39 mg, 72%). Rf = 0.6 (toluene – EtOAc, 3:1); = 18 (c = 1, CHCl3). 1H-NMR (600 MHz, CDCl3): δ 7.32–7.19 (m, 30H, Ph), 6.92 (d, 1H, JNH′-2′ = 7.8 Hz, NH′), 6.04 (d, 1H, JNH-2 = 9.6 Hz, NH), 5.21–5.14 (m, 2H, 2×βCHMyr), 4.98 (d, 1H, J1-2 = 3.1 Hz, H-1), 4.97 (d, 1H, J1′-2′ = 8.3 Hz, H-1′), 4.95–4.81 (m, 8H, 2×OP(O)(OCH2Ph)2), 4.52–4.35 (m, 4H, 2×CH2-Ph), 4.33 (m, 1H, H-4′), 4.32 (m, 1H, H-3′), 4.23 (ddd, 1H, J3-4 = J4-5 = 9.5 Hz, J4-P = 9.0 Hz, H-4), 4.19 (ddd, 1H, J1-2 = 3.1 Hz, J2-3 = 6.7 Hz, J2-NH = 9.6 Hz, H-2), 4.12 (ddd, 1H, J5-6a = 5.5 Hz, J5-6b = 2.0 Hz, H-5), 3.90 (dd, 1H, H-3), 3.88 (ddd, 1H, J5′-6′a = 5.7 Hz, J5′-6′b = 2.0 Hz, J5′-4′= 9.1 Hz, H-5′), 3.80 (dd, 1H, H-6′a), 3.78 (dd, 1H, H-6a), 3.70 (dd, 1H, H-6b), 3.69 (dd, 1H, H-6′b), 3.58 (m, 1H, H-2′), 2.63–2.18 (m, 8H, 2×αCH2Myr, 2×αCH2Lau), 1.61–1.53 (m, 8H, 2×βCH2Lau, 2×γCH2Myr), 1.30–1.22 (m, 68 H, CH2Myr,Lau), 0.89–0.86 (m, 12H, CH3Myr,Lau), 0.84 and 0.82 (2s, 18H, tBuSi), 0.09, 0.07, 0.06 and 0.03 (4s, 12H, SiMe). 13C-NMR (150.9 MHz, CDCl3): δ 173.17, 172.94, 170.21, 169.79 (4C, 4×C=O), 138.26, 138.09, 135.88, 135.84, 135.48, 135.42 (6C, Cq-Ph), 128.68, 128.61, 128.59, 128.47, 128.41, 128.37, 128.28, 128.12, 128.10, 127.91, 127.49, 127.42 (30C, Ph), 98.68 (1C, C-1′), 96.77 (1C, C-1), 76.80 (C-3), 76.18 (C-4) 74.82 (1C, C-4′), 72.98, 72.89 (2C, 2×CH2-Ph), 71.56 (1C, C-3′), 71.29 (1C, C-5). 71.26 (1C, C-5′), 71.01 (1C, CHMyr), 70.60 (1C, CHMyr), 69.88, 69.85, 69.78, 69.75, 69.66, 69.62, 69.20, 69.16 (8C, 2×CH2Ph, 2×OP(O)(OCH2Ph)2, C-6, C-6′), 56.53 (1C, C-2), 53.15 (1C, C-2′), 41.49, 40.65 (4C, 2×αCMyr, 2×αCLau), 34.58, 34.52, 34.47, 34.42, 31.93, 29.72, 29.68, 29.65, 29.62, 29.57, 29.55, 29.50, 29.37, 29.28, 29.24 (CH2Myr,Lau), 25.91, 25.88 (6C, tBuSi), 25.39, 25.33, 25.06, 25.05, 22.68 (CH2Myr,Lau), 14.09 (4C, CH3Myr,Lau), −3.07, −4.02, −4.50, and −4.62 (4C, SiMe). 31P NMR (243 MHz, CDCl3): δ −1.70 −2.27. HRMS (ESI-TOF) m/z: found 2086.2548, calc. for [M + H + NH4]2+ C118H186N2O21P2Si2 + NH4+ + H+: m/z = 2086.2527.
6-O-Benzyl-4-O-[bis(benzyloxy)phosphoryl]-2-deoxy-2-[(R)-3-(dodecanoyloxy)-tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-4-O-[bis(benzyloxy)-phosphoryl]-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (5). To a cooled (0 °C) stirred solution of 4 (25 mg, 12 µmol) in dry THF (2 mL) in a Teflon tube, a solution of 3HF∙Py (25 µL, 120 µmol) in THF (70%) was added and the reaction mixture was stirred for 72 h at 0 °C. The mixture was then diluted with DCM (20 mL) and washed with satd. aq. NaHCO3 (pH 8, 3 × 5 mL) and water (2 × 5 mL). The organic phase was dried over Na2SO4, filtered over cotton, and concentrated. The residue was purified by HPLC (toluene–EtOAc, 2:1 → 1:4) to give 5 (15 mg, 68%). Rf = 0.4 (hexane-EtOAc, 2:1); = 18 (c = 1, CHCl3). 1H NMR (600 MHz, CDCl3): δ 7.34–7.15 (m, 30H, Ph), 6.43 (d, JNH′-2′ = 7.2 Hz, NH′), 6.42 (d, 1H, JNH-2 = 8.3 Hz, NH), 5.19 (m, 1H, βCHMyr), 5.13 (m, 1H, βCHMyr), 5.07 (d,1H, J1-2 = 3.8 Hz, H-1), 5.04–4.95 (m, 8H, 2×OP(O)(OCH2Ph)2), 4.79 (d, 1H, J1′-2′ = 8.6 Hz, H-1′), 4.70 (bs, 0.7H, OH-3,3′), 4.39–4.30 (m, 5H, 2×CH2-Ph, H-4), 4.22 (ddd, 1H, J4′-P′ = J3′-4′ = 8.6 Hz, J4′-5′ = 8.3 Hz, H-4′), 4.14 (ddd, 1H, J4-5 = 10.1 Hz, J5-6a = 5.2 Hz, J5-6b = 1.6 Hz, H-5), 4.09 (ddd, 1H, J2-3 = 10.4 Hz, H-2), 4.05 (t, 1H, J2′-3′ = 9.4 Hz, H-3′), 3.92 (dd, 1H, J3′-4′ = 8.9 Hz, H-3), 3.62 (dd, 1H, J6a-6b = 10.7 Hz, H-6b), 3.58–3.55 (m, 1H, H-2′), 3.55 (dd, 1H, J6a′-6b′ = 11.0 Hz, J5′-6b′ = 1.7 Hz, H-6b′), 3.51–3.50 (m, 1H, H-5′), 3.48 (dd, 1H, H-6a), 3.43 (dd, 1H, J5′-6a′ = 4.9 Hz, H-6a′), 2.56–2.36 (m, 4H, 2×αCH2Myr,), 2.26 (m, 4H, 2×αCH2Lau), 1.62–1.53 (m, 8H, βCH2Lau, γCH2Myr), 1.30–1.22 (m, 68H, CH2Myr,Lau), 0.88–0.86 (m, 12H, CH3Myr;Lau). 13C-NMR (150.9 MHz, CDCl3): δ 174.07, 173.77, 171.28, 171.23 (4C, 4×C=O), 137.86, 137.80, 135.84, 135.80, 135.38, 135.36 (6C, Cq-Ph), 128.74, 128.68, 128.64, 128.60, 128.49, 128.42, 128.36, 128.30, 128.03, 127.99, 127.63, 127.57, 127.47 (30C, Ph), 99.37 (1C, C-1′), 96.80 (1C, C-1), 76.98 (1C, C-4), 76.83 (1C, C-4′), 74.04 (1C, C-5′), 73.40, 73.34 (2C, CH2-Bn), 72.32 (1C, C-3), 71.94 (1C, C-3′), 71.33 (1C, CHMyr), 70.99 (1C, CHMyr), 70.31 (1C, C-5), 70.14, 70.10, 69.97, 69.93, 69.76, 69.72 (6C, 2×CH2Ph, 2×OP(O)(OCH2Ph)2), 68.54 (1C, C-6), 68.32 (1C, C-6′), 57.03 (1C, C-2′), 53.35 (1C, C-2), 42.23, 41.33 (4C, 2×αCMyr, 2×αCLau), 34.58, 34.41, 34.22, 31.92, 29.65, 29.54, 29.43, 29.35, 29.21, 25.37, 25.00, 24.96, 22.68 (CH2Myr,Lau), and 14.09 (4C, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3): 0.03 0.77. HRMS (ESI-TOF) m/z: found 1858.0924, calc. for [M + H]+ C106H158N2O21P2 + H+: m/z = 1858.0905.
6-O-Benzyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (6). To a stirred solution of 2 (90 mg, 60 µmol) in dry DCM (5 mL), 4 Å molecular sieves were added and the suspension was stirred for 2 h at r.t. under atmosphere of Ar. The mixture was then cooled to −70 °C and a solution of triethylsilane (74 µL, 460 µmol, 8 eq) in DCM (5% solution) and a solution of trifluoromethanesulfonic acid (41 µL, 460 µmol, 8 eq) in DCM (5%) were added successively. The stirring was continued for 3 h, the reaction was quenched by addition of a solution of Et3N in DCM (5%, 70 µL, 460 µmol, 8 eq), and the mixture was stirred for 15 min at −70 °C. The reaction mixture was brought up to r.t., then diluted with DCM (50 mL), and the solids were removed by filtration over a pad of Celite. The filtrate was washed with satd. aq. NaHCO3 (2 × 20 mL), water (20 mL), and brine (20 mL), dried over Na2SO4, filtered over cotton, and concentrated. The residue was purified by column chromatography on silica gel (toluene - EtOAc, 10:1 → 0:1) which gave 6 (52 mg, 65%). Rf = 0.3 (toluene-EtOAc, 1:1), = 14 (c = 1, CHCl3). 1H NMR (600 MHz, CDCl3): δ 7.34–7.26 (m, 10H, Ph), 6.69 (d, 1H, NH′, J2′,N′ = 6.7 Hz), 6.49 (d, 1H, NH, J2,N = 8.3 Hz), 5.21 (m, 1H, βCH Myr), 5.15 (m, 1H, βCH Myr), 5.07 (d, 1H, H-1, J1,2 = 3.5 Hz), 4.90 (d, 1H, H-1′, J1,2 = 8.4 Hz), 4.55–4.47 (m, 4H, CH2Ph), 4.02 (m, 3H, H-2, H-5, H-4′), 3.78 (dd, 1H, J6a,6b = 2.1 Hz, J5,6a = 10.3 Hz, H-6a), 3.69 (m, 2H, H-3, H-6a′), 3.63 (dd, 1H, J5′,6a′ = 10.4 Hz and J6a′,6b′ = 5.6 Hz, H-6b′), 3.54 (m, 2H, H-6b, H-5′), 3.45 (m, 3H, H-4, H-2′, H-3′), 2.89 (bs, 4H, 4×OH), 2.55-2.22 (m, 8H, m, 8H, 2×αCH2Myr, 2×αCH2Lau), 1.55 (m, 8H, βCH2Lau, γCH2Myr), 1.58–1.25 (m, 68H, CH2Myr,Lau), 0.88 (m, 12H, CH3Myr,Lau). 13C-NMR (150.9 MHz, CDCl3): δ 174.77, 174.36, 171.82 and 171.68 (4C, 4×C=O), 137.73 and 137.60 (2C, Cq-Ph), 128.50, 127.88, 127.84, 127.66 and 127.61 (10C, Ph), 98.46 (1C, C-1′), 95.53 (1C, C-1), 74.44 (1C, H-4′), 74.20 (1C, H-4), 73.59 and 73.45 (2C, CH2Ph), 72.69 (1C, C-3′), 72.08 (1C, C-3), 71.78 and 71.48 (2C, CHFA), 71.44 (1C, C-5′), 71.38 (1C, C-5), 70.32 (1C, C-6′), 69.86 (1C, C-6), 57.38 (1C, C-2′), 53.28 (1C, C-2), 42.78, 41.68, 34.81, 34.59, 34.43, 31.93, 29.68, 29.66, 29.63, 29.60, 29.58, 29.57, 29.54, 29.53, 29.41, 29.37, 29.35, 29.33, 29.27, 29.20, 29.16, 25.37, 25.36, 24.96, 24.94 22.69 (40C, CH2Myr,Lau), and 14.10 (4C, CH3Myr,Lau). HRMS (ESI-TOF) m/z: found 1337.9700, calc. for [M + H]+ C78H132N2O15P2 + H+: m/z = 1337.9614.
6-O-Benzyl-4-O-[bis(benzyloxy)phosphoryl]-3-O-[bis(benzyloxy)phosphoryl]-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-2-deoxy-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-4-O-[bis(benzyloxy)phosphoryl]-3-O-[bis(benzyloxy)phosphoryl]-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-2-deoxy-β-d-glucopyranoside (7). To a stirred solution of 6 (40 mg, 30 µmol) in dry DCM (4 mL) bisbenzyloxy(diisopropylamino)phosphine (52 mg, 51 µL, 150 µmol) and 1H-tetrazole (11 mg, 150 µmol, 0.45M in CH3CN) were added successively under atmosphere of Ar. The mixture was stirred for 1 h at r.t., then cooled to −78 °C, and a solution of mCPBA (21 mg, 122 µmol, 0.1 M in dry CH2Cl2) was added. The reaction mixture was stirred for 1 h at −78 °C, then quenched by addition of a solution of Et3N (30 µL) in MeOH (300 µL) and brought to r.t. The mixture was diluted with CH2Cl2 (30 mL) and washed with sat. aq. NaHCO3 (2 × 10 mL), water (10 mL), and brine (10 mL). The organic layer was dried over Na2SO4, filtered over cotton, and concentrated. The residue was purified by chromatography on silica gel (toluene – EtOAc, 2:3 → 0:1) and by gel permeation chromatography on Bio-Beads® S-X1 support (600 × 1.5 mm, toluene—CHCl3, 3:1) to give 7 (55 mg, 78%). = 11 (c = 1.2, CHCl3); Rf = 0.7 (hexane-EtOAc, 2:1). 1H NMR (600 MHz, CDCl3): δ 7.28–7.11 (m, 50H, Ph), 7.03 (d, 1H, NH′, J2′,N′ = 6.7 Hz), 6.75 (d, 1H, NH, J2,N = 8.5 Hz), 5.25 (m, 1H, βCHMyr), 5.13 (m, 1H, βCHMyr), 5.10 (d, 1H, J1,2 = 3.5 Hz, H-1,), 5.07–4.85 (m, 8H, CH2OP(O)(OBn)), 4.80 (d, 1H, H-1′, J1′,2′ = 8.2 Hz), 4.74 (m, 2H, H-3, H-3′), 4.60 (q, 1H, J3,4 = J4,5 = J4,P = 9.5 Hz, H-4), 4.52 (q, 1H, J3′,4′ = J4′,5′ = J4′,P′ = 9.4 Hz, H-4′), 4.43, 4.36, 4.30, 4.24 (d(x4), 4H, 2×CH2Ph), 4.30 (ddd, 1H, H-2), 4.25 (m, 1H, H-5), 3.73 (m, 1H, H-2′), 3.68 (m, 2H, H-6a and H-6a′), 3.64 (dd, 1H, J5,6b = 1.7 Hz, J6a,6b = 11.0 Hz, H-6b), 3.64 (dd, 1H, J5,6b = 1.7 Hz, J6a,6b = 11.0 Hz, H-6b), 3.59 (dd, 1H, J5′,6b′ = 4.9 Hz, J6a′,6b′ = 10.9 Hz, H-6′b), 3.53 (m, 1H, H-5), 2.58 (dd, 1H, αCH2Myr), 2.44 (dd, 1H, αCH2Myr), 2.33 (dd, 1H, αCH2Myr), 2.28–2.16 (m, 5HH, αCH2Myr, 2×αCH2Lau), 1.68–1.39 (m, 8H, γCH2Myr, βCH2Lau), 1.55–1.17 (m, 68H, CH2Myr,Lau), 0.87 (m, 12H, CH3Myr,Lau). 13C NMR (150.9 MHz, CDCl3): δ 173.18, 173.00, 171.62 and 170.43 (4C, 4xC=O), 138.29 and 138.11 (2C, Cq-Ph), 130.14, 129.79, 128.55, 128.48, 128.46, 128.42, 128.40, 128.36, 128.34, 128.31, 128.25, 128.23, 128.18, 128.13, 128.06, 128.04, 127.98, 127.95, 127.54 and 127.32 (30C, Ph), 99.68 (1C, C-1′), 98.12 (1C, C-1), 77.49 (1C, C-3), 77.05 (1C, C-3′), 74.20(1C, H-4′), 74.05 (1C, H-4), 73.81 (1C, C-5′), 73.13 and 72.99 (2C, CH2Ph), 71.08 (1C, C-5), 70.88 and 70.47 (2C, CHFA), 69.97, 69.77, 69.72 and 69.57 (4C, CH2PhOP(O)(OBn)), 68.19 (1C, C-6′), 67.86 (1C, C-6), 56.51 (1C, C-2′), 52.31 (1C, H-2), 34.56, 34.46, 31.94, 31.93, 29.73, 29.70, 29.66, 29.59, 29.50, 29.42, 29.39, 29.37, 29.30, 29.28, 25.30, 25.07, 25.04, 22.69 (40C, CH2Myr,Lau), 14.11 (4C, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3): −0.30, −0.33, −1.89 and −1.99. HRMS (ESI-TOF) m/z: found 2378.2011, calc. for [M + H]+ C134H184N2O27P4 + H+: m/z = 2378.2007.
6-O-Benzyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (8). To a stirred solution of 2 (105 mg, 67 µmol) in dry DCM (15 mL), molecular sieves 4 Å were added and the suspension was stirred for 2 h in an atmosphere of Ar. The mixture was then cooled to −65 °C and a solution of triethylsilane (20 mg, 27 µL, 170 µmol, 2.5 eq) in dry DCM (5% solution) followed by a solution of trifluoromethanesulfonic acid (25 mg, 15 µL, 170 µmol, 2.5 eq) in dry DCM (5% solution) were added. The reaction mixture was stirred at −65 °C for 3 h under atmosphere of Ar, and then quenched by addition of a solution of Et3N (30 µL, 200 µmol, 3 eq) in DCM (5% solution), and the stirring was continued for 15 min at −65 °C. The mixture was diluted with DCM (30 mL), the solids were removed by filtration over a pad of Celite, the filtrate was washed with satd. aq. NaHCO3 (30 mL), water (30 mL), and brine (30 mL), dried over Na2SO4, filtered, and concentrated. The residue was purified by column chromatography on silica gel (toluene-EtOAc, 10:1 → 4:1) which gave 8 (68 mg, 71%). Rf = 0.4 (toluene - EtOAc, 1:1), = 12 (c = 1, CHCl3). 1H NMR (600 MHz, CDCl3): δ 7.35–7.27 (m, 10H, Ph), 6.69 (d, 1H, NH′), 5.83 (d, 1H, NH), 5.19–5.14 (m, 1H, βCHMyr), 5.12 (d, 1H, H-1), 5.11–5.06 (m, 1H, βCHMyr), 4.97 (d, 1H, H-1′), 4.54–4.47 (m, 4H, CH2-Bn), 4.11 (t, 1H, J3′-4′ = 9.7 Hz, H-3′), 4.08 (dt, 1H, J2-3 = J2-NH= 9.6 Hz, J1-2 = 3.7 Hz, H-2), 4.04 (ddd, 1H, J4-5 = 9.6 Hz, J5-6b = 2.8 Hz, J5-6a = 6.4 Hz, H-5), 3.76 (dd, 1H, J5′-6a′ = 10.3 Hz, J6a′-6b′ = 3.0 Hz, H-6a′), 3.69–3.64 (m, 3H, H-6b′, H-6a, H-3), 3.56–3.54 (m, 2H, H-6b, H-5′), 3.45 (t, 1H, J4′-5′ = 9.2 Hz, H-4′), 3.40 (dd,1H, J4-3 = 8.9 Hz, H-4), 3.31 (ddd, 1H, J2′-3′ = 9.9 Hz, J2′-NH′ = 6.7 Hz, J1′-2′ = 8.6 Hz, H-2′), 2.55–2.20 (m, 8H, 2×αCH2Myr, 2×αCH2Lau), 1.69–1.52 (m, 8H, 2×βCH2Lau, 2×γCH2Myr), 1.32–1.16 (m, 68 H, CH2Myr,Lau), 0.91–0.84 (m, 21H, CH3Myr,Lau, tBuSi), 0.10, 0.06 (2s, 6H, SiMe). 13C NMR (150.9 MHz, CDCl3): δ 174.78, 173.29, 171.45, 169.60 (4C, 4×C=O), 137.85, 137.48 (2C, Cq-Ph), 128.56, 128.48, 128.00, 127.83, 127.76, 127.69, 127.62 (10C, Ph), 98.20 (1C, C-1′), 95.72 (1C, C-1), 74.32 (1C, C-5′), 73.62, 73.56 (2C, CH2-Bn), 72.62 (1C, C-3′), 72.56 (1C, C-4), 72.22 (1C, C-4′), 71.92 (1C, CHMyr), 71.42 (1C, CHMyr), 71.22 (1C, C-5), 70.42 (1C, C-6′), 70.34 (1C, C-6), 58.04 (1C, C-2′), 53.40 (1C, C-2), 42.83, 41.08 (4C, 2×αCMyr, 2×αCLau), 34.96, 34.63, 34.61, 34.37, 31.92, 29.62, 29.59, 29.58, 29.56, 29.54, 29.52, 29.45, 29.36, 29.35, 29.27, 29.24, 29.16 (CH2FA),25.86, 25.79 (3C, tBuSi), 25.35, 25.28, 25.08, 22.69 (CH2Myr,Lau), 14.09 (4C, CH3Myr,Lau), −4.00, −4.32 (2C, SiMe). HRMS (ESI-TOF) m/z: found 1452.0485, calc. for [M + H]+ C84H146N2O15Si + H+: m/z = 1452.0477.
6-O-benzyl-3,4-O-[bis(benzyloxy)phosphoryl]-2-deoxy-2-[(R)-3-(dodecanoyloxy)-tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-4-O-[bis(benzyloxy)phosphoryl]-3-O-tert-butyldimethylsilyl-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (9). To a stirred solution of 8 (28 mg, 20 µmol) in dry DCM (5 mL), bisbenzyloxy(diisopropylamino)phosphine (32 mg, 31 µL, 90 µmol) and a solution of 1H-tetrazol (16 µL, 116 µmol, 0.45M in CH3CN) were added successively and the stirring was continued for 2 h at r.t. in an atmosphere of Ar. The reaction mixture was cooled to −78 °C, a solution of mCPBA (15 mg, 80 µmol) in DCM (0.6 mL) was added, and the stirring was continued for 2h at −78 °C. The reaction mixture was neutralized by addition of Et3N (50 µL, 36 µmol, 18 eq), stirred for 10 min at −78 °C, then brought to r.t. The mixture was diluted with DCM (20 mL) and washed with satd. aq. NaHCO3 (2 × 5 mL), water (10 mL), and brine (10 mL). The organic phase was dried over Na2SO4, filtered over cotton, and concentrated. The residue was purified by column chromatography on silica gel (toluene/EtOAc, 8:1 → 1:1) to give 9 (30 mg, 69%). Rf = 0.7 (toluene-EtOAc, 1:1); = 16 (c = 1, CHCl3). 1H NMR (600 MHz, CDCl3): δ 7.28–7.11 (m, 40H, Ph), 7.10 (d, 1H, J2′-NH′ = 7.5 Hz, NH′), 6.17 (d, 1H, J2-NH = 10.2 Hz, NH), 5.27–5.21 (m, 2H, 2×βCHMyr), 5.05–4.82 (m, 12 H, 3×OP(O)(OCH2Ph)2), 4.90 (m, 1H, H-1), 4.58 (d, 1H, J1′-2′= 8.3 Hz, H-1′), 4.51 (m, 1H, H-3′), 4.4 (m, 1H, H-4′), 4.45 (d, 1H, OCH2Ph), 4.43 (d, 1H, OCH2Ph), 4.35 (dd, 1H, J3-4 = J4-5= 9.3 Hz, J4-P = 9.5 Hz, H-4), 4.30 (d, 1H, OCH2Ph),4.28 (d, 1H, OCH2Ph), 4.23 (ddd, 1H, J2-NH = J2-3 = 9.9 Hz, J1-2 = 3.1 Hz, H-2), 4.18 (ddd, 1H, J5-6a = 4.2 Hz, J5-6b = 2.0 Hz, H-5), 3.94 (dd, 1H, H-3), 3.85 (ddd, 1H, J2′-NH′ = 10.3 Hz, J2′-3′ = 6.8 Hz, H-2′), 3.79 (dd, 1H, J6a-6b = 10.7 Hz, H-6a), 3.70 (dd, 1H, J6a′-6b′ = 10.9 Hz, H-6b′), 3.68 (dd, 1H, H-6b), 3.61 (dd, 1H, H-6a′), 3.49 (ddd, 1H, J4′-5′ = 9.1 Hz, J5′-6b′ = 2.1 Hz, J5′-6a′ = 4.6 Hz, H-5′), 2.61–2.17 (m, 8H, 2×αCH2Myr, 2×αCH2Lau), 1.68–1.47 (m, 8H, 2×βCH2Lau, 2×γCH2Myr), 1.30–1.22 (m, 68H, CH2Myr,Lau), 0.89–0.86 (m, 12H, CH3Myr,Lau), 0.81 (s, 9H, tBuSi), 0.11, 0.08 (2s, 6H, SiMe). 13C NMR (150.9 MHz, CDCl3): δ 172.95, 172.86, 172.05, 169.50 (4C, 4×C=O), 138.43, 138.06, 135.94, 135.89, 135.62, 135.58, 135.36, 135.31 (8C, Cq-Ph), 128.60, 128.56, 128.51, 128.47, 128.45, 128.37, 128.35, 128.32, 128.18, 128.15, 128.06, 128.02, 127.96, 127.93, 127.56, 127.51, 127.44, 127.23 (40C, Ph), 100.53 (1C, C-1′), 98.86 (1C, C-1), 76.93 (1C, C-4′), 76.89 (1C, C-4), 74.00 (1C, C-5′), 73.86 (1C, C-3′), 73.14, 72.76 (2C, 2×CH2-Ph), 71.59 (1C, C-5), 71.32 (1C, C-3), 70.45 (2C, CHMyr), 70.11, 70.08, 70.05, 69.79, 69.76, 69.72, 69.68, 69.64, 69.19, 69.15 (6C, 3×OP(O)(OCH2Ph)2), 68.37, 68.25 (2C, C-6, C-6′), 56.05 (1C, C-2′), 52.80 (1C, C-2), 41.39, 40.68 (4C, 2×αCMyr, 2×αCLau), 34.78, 34.65, 34.59, 34.41, 31.93, 29.70, 29.68, 29.69, 29.62, 29.58, 29.54, 29.43, 29.37, 29.30 (CH2Myr,Lau), 25.92 (3C, tBuSi), 25.48, 25.38, 25.05, 22.68 (CH2Myr,Lau), 14.10 (4C, CH3Myr,Lau),−3,66, −4.04 (2C, SiMe). 31P NMR (243 MHz, CDCl3): δ 0.49, −1.80 and −1.84. HRMS (ESI-TOF) m/z: found 2232.2340, calc. for [M + H]+ C126H185N2O24P3Si + H+: m/z = 2232.2372.
6-O-Benzyl-3,4-di-O-[bis(benzyloxy)phosphoryl]-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-4-O-[bis(benzyloxy)phosphoryl]-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (10) and 6-O-Benzyl-3,4-di-O-[bis(benzyloxy)phosphoryl]-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-6-O-benzyl-3-O-[bis(benzyloxy)phosphoryl]-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside (11). To a cooled (0 °C) stirred solution of 9 (30 mg, 13 µmol) in dry THF (2 mL) in a Teflon tube, a solution of TBAF in THF (1 M, 15 µL, 14 µmol) was added at 0 °C and the mixture was stirred for 12 at 0 °C. The mixture was diluted with DCM (10 mL) and washed with satd. aq. NaHCO3 (3 × 5 mL), water (5 mL), and brine (5 mL). The organic phase was dried over Na2SO4, filtered over cotton, and concentrated. The residue was purified by column chromatography on silica gel (toluene/EtOAc, 2:1 → 1:1) and by gel permeation chromatography on Bio-Beads® S-X1 support (toluene-chloroform, 3:1) to give 10 (10 mg, 37%) and 11 (9 mg, 33%). 10: Rf = 0.4 (toluene-EtOAc, 1:1); = 7 (c = 1, CHCl3); 1H NMR (600 MHz, CDCl3): δ 7.31–7.10 (m, 40H, Ph), 7.06 (d, 1H, JNH′-2′ = 7.6 Hz, NH′), 6.81 (d, 1H, NH), 5.30 (m, 1H, βCHMyr), 5.07 (m, 1H, βCHMyr), 5.07–4.08 (m, 6 H, OP(O)(OCH2Ph)2), 4.95 (d, 1H, J1-2 = 3.6 Hz, H-1), 4.93–4.84 (m, 6 H, OP(O)(OCH2Ph)2), 4.49 (dd, 1H, J4′-3′ = 9.0 Hz, H-4′), 4.42 (d, 1H, J1′-2′ = 8.2 Hz, H-1′), 4.41–4.20 (m, 4H, 2×CH2-Ph), 4.37 (dd, 1H, J2′-3′ = 11.3 Hz, J3′-4′ = 9.0 Hz, H-3′), 4.18 (ddd, 1H, J4-5 = 10.2 Hz, J5-6a = 4.0 Hz, J5-6b = 1.8 Hz, H-5), 4.12 (ddd, 1H, J1-2 = 3.6 Hz, J2-3 = 10.3 Hz, J2-NH = 6.8 Hz, H-2), 3.99–3.94 (m, 1H, H-2′, H-3), 3.62 (dd, 1H, J6′a-6′b = 10.9 Hz, H-6′b), 3.57 (dd, 1H, J6a-6b = 11.0 Hz, H-6b), 3.53 (dd, 1H, H-6a′), 3.51 (dd, 1H, H-6a), 3.43 (ddd, 1H, J4′-5′ = 9.4 Hz, J5′-6a′ = 4.7 Hz, J5′-6b′ = 2.0 Hz, H-5′), 2.69–2.20 (m, 8H, 2×αCH2Myr, 2×αCH2Lau), 1.66–1.44 (m, 8H, 2×βCH2Lau, 2×γCH2Myr), 1.30–1.13 (m, 68H, CH2Myr,Lau), 0.89–0.86 (m, 12H, CH3Myr,Lau). 13C NMR (150.9 MHz, CDCl3): δ 173.62, 172.90, 172.17, 172.08 (4C, 4×C=O),138.18, 138.00, 136.15, 136.10, 135.99, 135.53, 135.13, 135.08 (8C, Cq-Ph), 128.66, 128.59, 128.55, 128.46, 128.44, 128.37, 128.33, 128.27, 128.19, 128.17, 128.06, 128.01, 127.98, 127.96, 127.60, 127.58, 127.51, 127.36 (40C, Ph), 100.53 (1C, C-1′), 98.61 (1C, C-1), 77.60 (1C, C-3′), 76.93 (1C, C-4′), 74.17 (1C, C-5′), 73.56 (1C, C-4), 73.29, 73.09 (2C, 2×CH2-Ph), 73.16 (1C, C-3), 71.33 (1C, CHMyr), 70.73 (1C, CHMyr), 70.50 (1C, C-5), 70.16, 70.17, 69.80, 69.77, 69.74, 69.66, 69.62, 69.49, 69.46 (6, 3xOP(O)(OCH2Ph)2, 68.18 (1C, C-6′), 67.91 (1C, C-6), 55.59 (1C, C-2′), 53.36 (1C, C-2), 41.91, 41.08 (4C, 2×αCMyr, 2×αCLau), 34.39, 34.56, 34.52, 34.25, 31.94, 29.72, 29.68, 29.65, 29.63, 29.58, 29.55, 29.43, 29.37, 29.27, 25.41, 25.30, 25.10, 25.01, 22.69 (CH2Myr,Lau), 14.11 (4C, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3): δ 0.68, −1.21, −1.89. HRMS (ESI-TOF) m/z: found 2140.1301, calc. for [M + Na]+ C120H171N2O24P3 + Na+: m/z = 2140.1327.
11. Rf = 0.5 (toluene-EtOAc, 1:1); = 8.0 (c = 1, CHCl3); 1H NMR (600 MHz, CDCl3): δ 7.36–7.13 (m, 40H, Ph), 7.13 (d, 1H, NH′), 6.49 (d, 1H, JNH-2 = 9.2 Hz, NH), 5.21–5.15 (m, 2H, 2×βCHMyr), 5.11–4.84 (m, 12 H, 3×OP(O)(OCH2Ph)2), 5.00 (d, 1H, H-1), 4.59 (d, 1H, J1′-2′ = 8.2 Hz, H-1′), 4.54 (m, 1H, H-3′), 4.51 (m, 1H, H-4′), 4.45–4.29 (m, 5H, 2×CH2-Ph, H-3), 4.34 (ddd, 1H, JNH-2 = 9.2 Hz, J2-3 = 9.6 Hz, J1-2 = 3.6 Hz, H-2), 4.12 (m, 1H, H-5), 3.86 (ddd, 1H, J1′-2′ = 8.2 Hz, J2′-3′ = 9.0 Hz, J2′-NH′ = 7.6 Hz, H-2′), 3.80 (dd, 1H, J3-4 = 9.0 Hz, J4-5 = 9.8 Hz, H-4), 3.70 (dd, 1H, J5-6a = 10.9 Hz, J6a-6b = 1.9 Hz, H-6a), 3.62 (dd, 1H, J5′-6′a = 10.8 Hz, J6′a-6′b = 2.3 Hz, H-6′a), 3.59 (dd, 1H, J5-6b = 4.8 Hz, H-6b), 3.58 (dd, 1H, J5′-6b′ = 4.7 Hz, H-6b′), 3.51–3.49 (m, 1H, H-5′), 2.56–2.15 (m, 8H, 2×αCH2Myr, 2×αCH2Lau), 1.66–1.48 (m, 8H, 2×βCH2Lau, 2×γCH2Myr), 1.33–1.11 (m, 68H, CH2Myr,Lau), 0.89–0.86 (m, 12H, CH3Myr,Lau). 13C NMR (150.9 MHz, CDCl3): δ 173.01, 172.98, 172.00, 169.98 (4C, 4×C=O), 138.10, 138.08, 135.64, 135.62, 135.60, 135.38, 135.34 (8C, Cq-Ph), 128.69, 128.63, 128.61, 128.58, 128.52, 128.50, 128.42, 128.39, 128.28, 128.20, 128.12, 128.05, 128.00, 127.98, 127.96, 127.60, 127.54, 127.51 (40C, Ph), 100.00 (1C, C-1′), 98.36 (1C, C-1), 81.20 (1C, C-3), 77.18 (1C, C-3′), 74.12 (1C, C-5′), 73.77 (1C, C-4′), 73.48, 73.15 (2C, 2×CH2-Ph), 71.96 (1C, C-5), 70.86, 70.65 (2C, CHMyr), 70.11, 70.07, 70.02, 69.98, 69.79, 69.76, 69.72, 69.69 (6C, 3×OP(O)(OCH2Ph)2), 69.55 (1C, C-4), 68.81 (1C, C-6′), 68.17 (1C, C-6), 55.94 (1C, C-2′), 50.92 (1C, C-2), 41.87, 40.43 (4C, 2×αCMyr, 2×αCLau), 34.67, 34.58, 34.53, 33.33, 31.93, 29.74, 29.69, 29.58, 29.53, 29.46, 29.42, 29.41, 29.39, 29.37, 29.31, 29.28 25.39, 25.28, 25.09, 25.03, 22.69 (CH2Myr,Lau), 14.12 (4C, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3): δ 0.59, 0.41, −1.87. HRMS (ESI-TOF) m/z: found 2140.1335, calc. for [M + Na]+ C120H171N2O24P3 + Na+: m/z = 2140.1327.
2-Deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-4,3-di-O-(phosphoryl)-β-d-glucopyranosyl-(1↔1)-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-3-O-(phosphoryl)-α-d-glucopyranoside (DLAM29). To a stirred solution of 11 (6 mg, 3 µmol) in toluene, MeOH (1:1, 3 mL) Pd black (10 mg) was added. The vessel was purged with Ar, the atmosphere was exchanged to hydrogen (3×), and the vessel was filled with hydrogen. The mixture was stirred for 22 h at r.t., then diluted with toluene-MeOH (1:1, 5 mL), and the solids were removed by filtration over a pad of Celite. The filtrate was concentrated and the residue was purified by gel permeation chromatography on Bio-Beads® S-X1 support (toluene–chloroform-MeOH, 1:4:2) to give DLAM29 (4 mg, 95%). 1H NMR (600 MHz, CDCl3-MeOD, 2:1): δ 5.28 (m, 1H, βCHMyr), 5.20 (m, 1H, βCHMyr), 4.94 (d, 1H, J1,2 = 3.2 Hz, H-1), 4.64 (d, 1H, J1′,2′ = 8.2 Hz, H-1′), 4.43 (m, 1H, H-3′), 4.28 (m, 1H, H-3), 4.12 (m, 1H, H-4′), 4.08 (m, 2H, H-5, H-2), 3.85–3.70 (m, 5H, H-2′, H-6a,b, H-6′a,b), 3.60 (m, 1H, H-5′), 2.63-2.52 (m, 3H, αCH2Myr), 2.42 (dd, 1H, αCH2Myr), 2.30–2.22 (m, 4H, αCH2Lau), 1.63–1.53 (m, 8H, γCH2Myr, βCH2Lau), 1.63–1.55 (m, 68H, CH2Myr,Lau), 0.84 (m, 12H, CH3Myr,Lau). 2.63–2.52 (m, (m, 8H, αCH2Myr,Lau), 1.57–1.23 (m, 72H, CH2Myr,Lau), 0.84 (m, 12H, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3/MeOD 2:1): δ 1.06, 0.79, 0.81. MALDI-TOF-MS: found m/z [M − H]− 1395.7664, calc. for C64H123N2O24P3 − H− [M − H]− 1395.7656.
2-Deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-2-deoxy-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside 3′, 4′, 4-trisphosphate (DLAM30). To a stirred solution of 10 (7 mg, 4 µmol) in dry toluene-MeOH (1:1, 3 mL), Pd black (10 mg) was added. The vessel was purged with Ar (3×) and then filled with hydrogen. The mixture was stirred for 20 h at r.t., diluted with toluene-MeOH, 1:1, the solids were removed by filtration through a syringe filter (regenerated cellulose, 0.45 µm), and the solution was concentrated. The residue was purified by gel permeation chromatography on Bio-Beads® S-X1 support (toluene-chloroform-MeOH, 1:4:2), which afforded DLAM30 (5 mg, 92%). 1H NMR (600 MHz, CDCl3-MeOD, 2:1): δ 5.26 (m, 1H, βCHMyr), 5.14 (m, 1H, βCHMyr), 4.91 (d, 1H, J1,2 = 3.4 Hz, H-1), 4.52 (d, 1H, J1′,2′ = 8.1 Hz, H-1′), 4.33 (dd, 1H, H-3′), 4.14 (dd, 1H, J3′,4′ = J4′,5′ = J4′,P′ = 9.5 Hz, H-4′), 4.10 (m, 1H, H-5), 3.98 (t, 1H, J2,3 = J3,4 = 9.4 Hz, H-4), 3.95 (dd, 1H, H-2), 3.86 (dd, 1H, H-2′), 3.80 (m, 1H, H-3), 3.78–3.65 (m, 4H, H-6a,b, H-6′a,b), 3.43 (m, 1H, H-5′), 2.61–2.50 (m, 4H, 2×αCH2Myr), 2.40 (dd, 1H, αCH2Lau), 2.25 (m, 3H, αCH2Lau), 1.56–1.49 (m, 8H, γCH2Myr, βCH2Lau), 1.57–1.23 (m, 68H, CH2Myr,Lau), and 0.84 (m, 12H, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3/MeOD 2:1): δ 1.12, 0.73, 0.43. MALDI-TOF-MS: found m/z [M − H]- 1395.6785, calc. for C64H123N2O24P3 − H− [M − H]− 1395.7656.
2-Deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside 4, 4′-bisphosphate (DLAM33). To a stirred solution of 5 (10 mg, 5 µmol) in dry toluene-MeOH (2:1, 3 mL), Pd black (10 mg) was added. The vessel was purged with Ar and then filled with hydrogen. The mixture was stirred for 20 h at r.t., diluted with toluene–MeOH, 1:1, the solids were removed by filtration through a syringe filter (regenerated cellulose, 0.45 µm), and the solvent was removed. The residue was purified by gel permeation chromatography on Bio-Beads® S-X1 support (toluene-chloroform-MeOH, 1:4:2), which afforded DLAM33 (5 mg, 80%). 1H NMR (600 MHz, CDCl3/MeOD, 3:1): δ 5.24 (m, 1H, CHMyr), 5.16 (m, 1H, CHMyr), 4.90 (m, 1H, H-1), 4.53 (m, 1H, H-1′), 4.07 (m, 1H, H-4), 3.95 (m, 3H, H-2, H-3, H-4′), 3.78–3.74 (m, 6H, H-6a,b, H-6′a,b, H-5, H-3′), 3.66 (m, 1H, H-2′), 3.35 (m, 1H, H-5′), 2.51 (m, 2H, αCH2Myr), 2.46 (m, 1H, αCH2Myr), 2.38 (m, 1H, αCH2Myr), 2.15 (m, 4H, αCH2Lau), 1.53 (m, 8H, γCH2Myr, βCH2Lau), 1.57–1.22 (m, 68H, CH2), and 0.84 (m, 12H, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3/MeOD 3:1): δ 1.43, 1.24. MALDI-TOF-MS: found m/z [M − H]− 1315.6474, calc. for C64H122N2O21P2-H- [M − H]− 1315.7994.
2-Ddeoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-β-d-glucopyranosyl-(1↔1)-2-deoxy-2-[(R)-3-(dodecanoyloxy)tetradecanoylamino]-α-d-glucopyranoside 3,3′,4,4′-tetraphosphate (DLAM36). To a stirred solution of 7 (10 mg, 4 µmol) in dry toluene-MeOH (1:1, 3 mL), Pd black (10 mg) was added. The vessel was purged with Ar and then filled with hydrogen. The mixture was stirred for 36 h at r.t., diluted with toluene-MeOH (1:2, 5 mL), the solids were removed by filtration through a syringe filter (regenerated cellulose, 0.45 µm), and the solvent was removed. The residue was purified by gel permeation chromatography on Bio-Beads® S-X1 support (toluene-chloroform-MeOH, 1:4:2), which afforded DLAM36 (5 mg, 87%). 1H NMR (600 MHz, CDCl3/MeOD 3:1): δ 5.32 (m, 1H, βCHMyr), 5.24 (m, 1H, βCHMyr), 4.98 (d, 1H, J1,2 = 3.4 Hz, H-1), 4.60 (d, 1H, J1′,2′ = 8.5 Hz, H-1′), 4.42 (q, 1H, J2,3 = J3,4 = J3,P = 9.4 Hz, H-3), 4.36 (q, 1H, J2,3 = J3,4 = J3,P = 9.8 Hz, H-3′), 4.17 (m, 4H, H-4, H-4′, H-5, H-2), 3.80 (m, 5H, H-2′, H-6a,b, H-6′a,b), 3.41 (m, 1H, H-5′), 2.58–2.53 (m, 3H, αCH2Myr), 2.45 (dd, 1H, αCH2Myr), 2.38 (m, 1H, αCH2Myr), 2.30 (t, 2H, αCH2Lau), 2.25 (t, 2H, αCH2Lau), 1.60 (m, 8H, γCH2Myr, βCH2Lau), 1.60–1.24 (m, 68H, CH2), and 0.86 (m, 12H, CH3Myr,Lau). 31P NMR (243 MHz, CDCl3/MeOD 3:1): δ 1.23, 1.01, 1.05, 0.86. MALDI-TOF-MS: found m/z 1475.5743 [M − H]−, calc. for C64H124N2O27P4 − H− 1475.7318 [M − H]−.