Machine Learning Techniques Applied to the Study of Drug Transporters
Abstract
1. Introduction
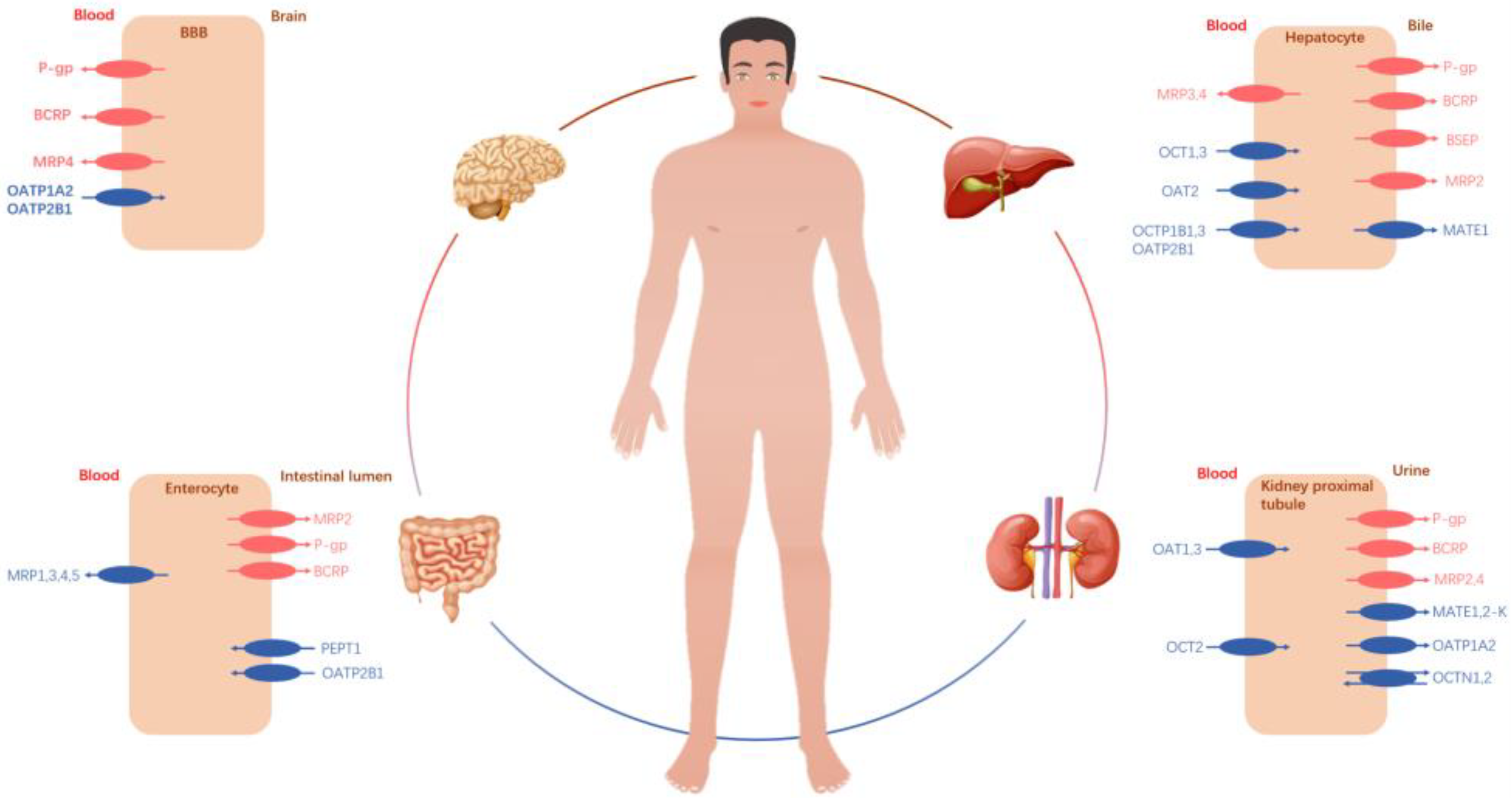
2. Drug Transporters and Important Implications
3. Machine Learning
3.1. Decision Trees and Random Forests
3.2. Neural Network
3.3. Support Vector Machine
3.4. Naïve Bayes
3.5. k-Nearest Neighbor Algorithm
4. Application of Machine Learning Methods in the Investigation of Drug Transporters
4.1. ABC Transporters
4.1.1. P-gp
4.1.2. BCRP
4.1.3. MRPs
4.1.4. BSEP
4.2. SLC Transporters
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Liang, Y.; Li, S.; Chen, L. The physiological role of drug transporters. Protein Cell 2015, 6, 334–350. [Google Scholar] [CrossRef] [PubMed]
- Wang, D. Current Research Method in Transporter Study. Adv. Exp. Med. Biol. 2019, 1141, 203–240. [Google Scholar] [PubMed]
- Shoichet, B.K.; McGovern, S.L.; Wei, B.; Irwin, J.J. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002, 6, 439–446. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Hsu, K.-C.; Chen, Y.-F.; Lin, S.-R.; Yang, J.-M. iGEMDOCK: A graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. 2011, 12, S33. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Carracedo-Reboredo, P.; Liñares-Blanco, J.; Rodríguez-Fernández, N.; Cedrón, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Comput. Struct. Biotechnol. J. 2021, 19, 4538–4558. [Google Scholar] [CrossRef]
- Cascorbi, I. Role of pharmacogenetics of ATP-binding cassette transporters in the pharmacokinetics of drugs. Pharmacol. Ther. 2006, 112, 457–473. [Google Scholar] [CrossRef]
- Meier, P.J.; Stieger, B. Bile salt transporters. Annu. Rev. Physiol. 2002, 64, 635–661. [Google Scholar] [CrossRef] [PubMed]
- Hediger, M.A.; Clémençon, B.; Burrier, R.E.; Bruford, E.A. The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol. Aspects Med. 2013, 34, 95–107. [Google Scholar] [CrossRef]
- Nigam, S.K. The SLC22 Transporter Family: A Paradigm for the Impact of Drug Transporters on Metabolic Pathways, Signaling, and Disease. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 663–687. [Google Scholar] [CrossRef]
- Gyimesi, G.; Hediger, M.A. Transporter-Mediated Drug Delivery. Molecules 2023, 28, 1151. [Google Scholar] [CrossRef]
- Zhao, J.; Zeng, Z.; Sun, J.; Zhang, Y.; Li, D.; Zhang, X.; Liu, M.; Wang, X. A Novel Model of P-Glycoprotein Inhibitor Screening Using Human Small Intestinal Organoids. Basic Clin. Pharmacol. Toxicol. 2017, 120, 250–255. [Google Scholar] [CrossRef]
- Wolf, A.; Bauer, B.; Hartz, A.M.S. ABC Transporters and the Alzheimer’s Disease Enigma. Front. Psychiatry 2012, 3, 54. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.-A.; Costales, C.; Mathialagan, S.; West, M.; Eatemadpour, S.; Lazzaro, S.; Tylaska, L.; Scialis, R.; Zhang, H.; Umland, J.; et al. Quantitative Contribution of Six Major Transporters to the Hepatic Uptake of Drugs: “SLC-Phenotyping” Using Primary Human Hepatocytes. J. Pharmacol. Exp. Ther. 2019, 370, 72–83. [Google Scholar] [CrossRef]
- Brouwer, K.L.R.; Evers, R.; Hayden, E.; Hu, S.; Li, C.Y.; Meyer Zu Schwabedissen, H.E.; Neuhoff, S.; Oswald, S.; Piquette-Miller, M.; Saran, C.; et al. Regulation of Drug Transport Proteins-From Mechanisms to Clinical Impact: A White Paper on Behalf of the International Transporter Consortium. Clin. Pharmacol. Ther. 2022, 112, 461–484. [Google Scholar] [CrossRef]
- Droździk, M.; Oswald, S.; Droździk, A. Extrahepatic Drug Transporters in Liver Failure: Focus on Kidney and Gastrointestinal Tract. Int. J. Mol. Sci. 2020, 21, 5737. [Google Scholar] [CrossRef]
- Zou, W.; Shi, B.; Zeng, T.; Zhang, Y.; Huang, B.; Ouyang, B.; Cai, Z.; Liu, M. Drug Transporters in the Kidney: Perspectives on Species Differences, Disease Status, and Molecular Docking. Front. Pharmacol. 2021, 12, 746208. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. 1943. Bull. Math. Biol. 1990, 52, 99–115. [Google Scholar] [CrossRef]
- Tkatchenko, A. Machine learning for chemical discovery. Nat. Commun. 2020, 11, 4125. [Google Scholar] [CrossRef] [PubMed]
- Klambauer, G.; Hochreiter, S.; Rarey, M. Machine Learning in Drug Discovery. J. Chem. Inf. Model. 2019, 59, 945–946. [Google Scholar] [CrossRef]
- Loftus, T.J.; Tighe, P.J.; Ozrazgat-Baslanti, T.; Davis, J.P.; Ruppert, M.M.; Ren, Y.; Shickel, B.; Kamaleswaran, R.; Hogan, W.R.; Moorman, J.R.; et al. Ideal algorithms in healthcare: Explainable, dynamic, precise, autonomous, fair, and reproducible. PLoS Digit. Health 2022, 1, e0000006. [Google Scholar] [CrossRef]
- Chen, X.; Wang, M.; Zhang, H. The use of classification trees for bioinformatics. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar] [PubMed]
- Sarker, I.H. Data Science and Analytics: An Overview from Data-Driven Smart Computing, Decision-Making and Applications Perspective. SN Comput. Sci. 2021, 2, 377. [Google Scholar] [CrossRef] [PubMed]
- Lavecchia, A. Deep learning in drug discovery: Opportunities, challenges and future prospects. Drug Discov. Today 2019, 24, 2017–2032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ai, H.; Chen, W.; Yin, Z.; Hu, H.; Zhu, J.; Zhao, J.; Zhao, Q.; Liu, H. CarcinoPred-EL: Novel models for predicting the carcinogenicity of chemicals using molecular fingerprints and ensemble learning methods. Sci. Rep. 2017, 7, 2118. [Google Scholar] [CrossRef]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Zhang, Z. Naïve Bayes classification in R. Ann. Transl. Med. 2016, 4, 241. [Google Scholar] [CrossRef]
- Uddin, S.; Haque, I.; Lu, H.; Moni, M.A.; Gide, E. Comparative performance analysis of K-nearest neighbour (KNN) algorithm and its different variants for disease prediction. Sci. Rep. 2022, 12, 6256. [Google Scholar] [CrossRef]
- Bzdok, D.; Krzywinski, M.; Altman, N. Machine learning: Supervised methods. Nat. Methods 2018, 15, 5–6. [Google Scholar] [CrossRef]
- Zhang, Z. Introduction to machine learning: K-nearest neighbors. Ann. Transl. Med. 2016, 4, 218. [Google Scholar] [CrossRef] [PubMed]
- Linton, K.J. Structure and function of ABC transporters. Physiology 2007, 22, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ecker, G.F. A Structure-Based View on ABC-Transporter Linked to Multidrug Resistance. Molecules 2023, 28, 495. [Google Scholar] [CrossRef] [PubMed]
- Kadioglu, O.; Efferth, T. A Machine Learning-Based Prediction Platform for P-Glycoprotein Modulators and Its Validation by Molecular Docking. Cells 2019, 8, 1286. [Google Scholar] [CrossRef]
- Esposito, C.; Wang, S.; Lange, U.E.W.; Oellien, F.; Riniker, S. Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates. J. Chem. Inf. Model. 2020, 60, 4730–4749. [Google Scholar] [CrossRef]
- Shaikh, N.; Sharma, M.; Garg, P. Selective Fusion of Heterogeneous Classifiers for Predicting Substrates of Membrane Transporters. J. Chem. Inf. Model. 2017, 57, 594–607. [Google Scholar] [CrossRef]
- Smajić, A.; Grandits, M.; Ecker, G.F. Using Jupyter Notebooks for re-training machine learning models. J. Cheminform. 2022, 14, 54. [Google Scholar] [CrossRef]
- Ose, A.; Toshimoto, K.; Ikeda, K.; Maeda, K.; Yoshida, S.; Yamashita, F.; Hashida, M.; Ishida, T.; Akiyama, Y.; Sugiyama, Y. Development of a Support Vector Machine-Based System to Predict Whether a Compound Is a Substrate of a Given Drug Transporter Using Its Chemical Structure. J. Pharm. Sci. 2016, 105, 2222–2230. [Google Scholar] [CrossRef]
- Ganguly, S.; Finkelstein, D.; Shaw, T.I.; Michalek, R.D.; Zorn, K.M.; Ekins, S.; Yasuda, K.; Fukuda, Y.; Schuetz, J.D.; Mukherjee, K.; et al. Metabolomic and transcriptomic analysis reveals endogenous substrates and metabolic adaptation in rats lacking Abcg2 and Abcb1a transporters. PLoS ONE 2021, 16, e0253852. [Google Scholar] [CrossRef]
- Montanari, F.; Zdrazil, B.; Digles, D.; Ecker, G.F. Selectivity profiling of BCRP versus P-gp inhibition: From automated collection of polypharmacology data to multi-label learning. J. Cheminform. 2016, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Gantner, M.E.; Peroni, R.N.; Morales, J.F.; Villalba, M.L.; Ruiz, M.E.; Talevi, A. Development and Validation of a Computational Model Ensemble for the Early Detection of BCRP/ABCG2 Substrates during the Drug Design Stage. J. Chem. Inf. Model. 2017, 57, 1868–1880. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Lei, T.; Wang, Z.; Shen, C.; Cao, D.; Hou, T. ADMET evaluation in drug discovery. 20. Prediction of breast cancer resistance protein inhibition through machine learning. J. Cheminform. 2020, 12, 16. [Google Scholar] [CrossRef]
- Hazai, E.; Hazai, I.; Ragueneau-Majlessi, I.; Chung, S.P.; Bikadi, Z.; Mao, Q. Predicting substrates of the human breast cancer resistance protein using a support vector machine method. BMC Bioinform. 2013, 14, 130. [Google Scholar] [CrossRef]
- Ding, Y.-L.; Shih, Y.-H.; Tsai, F.-Y.; Leong, M.K. In silico prediction of inhibition of promiscuous breast cancer resistance protein (BCRP/ABCG2). PLoS ONE 2014, 9, e90689. [Google Scholar] [CrossRef]
- Garg, P.; Dhakne, R.; Belekar, V. Role of breast cancer resistance protein (BCRP) as active efflux transporter on blood-brain barrier (BBB) permeability. Mol. Divers. 2015, 19, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Kharangarh, S.; Sandhu, H.; Tangadpalliwar, S.; Garg, P. Predicting Inhibitors for Multidrug Resistance Associated Protein-2 Transporter by Machine Learning Approach. Comb. Chem. High Throughput Screen. 2018, 21, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Lingineni, K.; Belekar, V.; Tangadpalliwar, S.R.; Garg, P. The role of multidrug resistance protein (MRP-1) as an active efflux transporter on blood-brain barrier (BBB) permeability. Mol. Divers. 2017, 21, 355–365. [Google Scholar] [CrossRef]
- McLoughlin, K.S.; Jeong, C.G.; Sweitzer, T.D.; Minnich, A.J.; Tse, M.J.; Bennion, B.J.; Allen, J.E.; Calad-Thomson, S.; Rush, T.S.; Brase, J.M. Machine Learning Models to Predict Inhibition of the Bile Salt Export Pump. J. Chem. Inf. Model. 2021, 61, 587–602. [Google Scholar] [CrossRef]
- Nigam, A.K.; Li, J.G.; Lall, K.; Shi, D.; Bush, K.T.; Bhatnagar, V.; Abagyan, R.; Nigam, S.K. Unique metabolite preferences of the drug transporters OAT1 and OAT3 analyzed by machine learning. J. Biol. Chem. 2020, 295, 1829–1842. [Google Scholar] [CrossRef]
- Nigam, A.K.; Ojha, A.A.; Li, J.G.; Shi, D.; Bhatnagar, V.; Nigam, K.B.; Abagyan, R.; Nigam, S.K. Molecular Properties of Drugs Handled by Kidney OATs and Liver OATPs Revealed by Chemoinformatics and Machine Learning: Implications for Kidney and Liver Disease. Pharmaceutics 2021, 13, 1720. [Google Scholar] [CrossRef] [PubMed]
- Tuerkova, A.; Bongers, B.J.; Norinder, U.; Ungvári, O.; Székely, V.; Tarnovskiy, A.; Szakács, G.; Özvegy-Laczka, C.; van Westen, G.J.P.; Zdrazil, B. Identifying Novel Inhibitors for Hepatic Organic Anion Transporting Polypeptides by Machine Learning-Based Virtual Screening. J. Chem. Inf. Model. 2022, 62, 6323–6335. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.R.; Urbina, F.; Zhang, X.; Fye, M.; Gerlach, J.; Wright, S.H.; Ekins, S. Machine Learning Models Identify New Inhibitors for Human OATP1B1. Mol. Pharm. 2022, 19, 4320–4332. [Google Scholar] [CrossRef]
- Jensen, O.; Brockmöller, J.; Dücker, C. Identification of Novel High-Affinity Substrates of OCT1 Using Machine Learning-Guided Virtual Screening and Experimental Validation. J. Med. Chem. 2021, 64, 2762–2776. [Google Scholar] [CrossRef]
- Juliano, R.L.; Ling, V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta 1976, 455, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Hyde, S.C.; Emsley, P.; Hartshorn, M.J.; Mimmack, M.M.; Gileadi, U.; Pearce, S.R.; Gallagher, M.P.; Gill, D.R.; Hubbard, R.E.; Higgins, C.F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 1990, 346, 362–365. [Google Scholar] [CrossRef]
- Dewanjee, S.; Dua, T.K.; Bhattacharjee, N.; Das, A.; Gangopadhyay, M.; Khanra, R.; Joardar, S.; Riaz, M.; Feo, V.D.; Zia-Ul-Haq, M. Natural Products as Alternative Choices for P-Glycoprotein (P-gp) Inhibition. Molecules 2017, 22, 871. [Google Scholar] [CrossRef]
- Constantinides, P.P.; Wasan, K.M. Lipid formulation strategies for enhancing intestinal transport and absorption of P-glycoprotein (P-gp) substrate drugs: In vitro/in vivo case studies. J. Pharm. Sci. 2007, 96, 235–248. [Google Scholar] [CrossRef]
- DeGorter, M.K.; Xia, C.Q.; Yang, J.J.; Kim, R.B. Drug transporters in drug efficacy and toxicity. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 249–273. [Google Scholar] [CrossRef]
- Ueda, K.; Clark, D.P.; Chen, C.J.; Roninson, I.B.; Gottesman, M.M.; Pastan, I. The human multidrug resistance (mdr1) gene. cDNA cloning and transcription initiation. J. Biol. Chem. 1987, 262, 505–508. [Google Scholar] [CrossRef]
- Doyle, L.A.; Yang, W.; Abruzzo, L.V.; Krogmann, T.; Gao, Y.; Rishi, A.K.; Ross, D.D. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. USA 1998, 95, 15665–15670. [Google Scholar] [CrossRef]
- Ni, Z.; Bikadi, Z.; Rosenberg, M.F.; Mao, Q. Structure and function of the human breast cancer resistance protein (BCRP/ABCG2). Curr. Drug Metab. 2010, 11, 603–617. [Google Scholar] [CrossRef]
- Begley, D.J. Delivery of therapeutic agents to the central nervous system: The problems and the possibilities. Pharmacol. Ther. 2004, 104, 29–45. [Google Scholar] [CrossRef]
- Sohail, M.I.; Dönmez-Cakil, Y.; Szöllősi, D.; Stockner, T.; Chiba, P. The Bile Salt Export Pump: Molecular Structure, Study Models and Small-Molecule Drugs for the Treatment of Inherited BSEP Deficiencies. Int. J. Mol. Sci. 2021, 22, 784. [Google Scholar] [CrossRef] [PubMed]
- Bongers, B.J.; Sijben, H.J.; Hartog, P.B.R.; Tarnovskiy, A.; Ijzerman, A.P.; Heitman, L.H.; van Westen, G.J.P. Proteochemometric Modeling Identifies Chemically Diverse Norepinephrine Transporter Inhibitors. J. Chem. Inf. Model. 2023, 63, 1745–1755. [Google Scholar] [CrossRef]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Mak, L.; Marcus, D.; Howlett, A.; Yarova, G.; Duchateau, G.; Klaffke, W.; Bender, A.; Glen, R.C. Metrabase: A cheminformatics and bioinformatics database for small molecule transporter data analysis and (Q)SAR modeling. J. Cheminform. 2015, 7, 31. [Google Scholar] [CrossRef]
- Montanari, F.; Knasmüller, B.; Kohlbacher, S.; Hillisch, C.; Baierová, C.; Grandits, M.; Ecker, G.F. Vienna LiverTox Workspace-A Set of Machine Learning Models for Prediction of Interactions Profiles of Small Molecules With Transporters Relevant for Regulatory Agencies. Front. Chem. 2019, 7, 899. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Yamashita, F.; Ose, A.; Maeda, K.; Sugiyama, Y.; Hashida, M. Automated extraction of information on chemical-P-glycoprotein interactions from the literature. J. Chem. Inf. Model. 2013, 53, 2506–2510. [Google Scholar] [CrossRef]
- Morgan, R.E.; van Staden, C.J.; Chen, Y.; Kalyanaraman, N.; Kalanzi, J.; Dunn, R.T.; Afshari, C.A.; Hamadeh, H.K. A multifactorial approach to hepatobiliary transporter assessment enables improved therapeutic compound development. Toxicol. Sci. 2013, 136, 216–241. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pérez, R.; Gerebtzoff, G. Identification of bile salt export pump inhibitors using machine learning: Predictive safety from an industry perspective. Artif. Intell. Life Sci. 2021, 1, 100027. [Google Scholar] [CrossRef]
- Baidya, A.T.K.; Ghosh, K.; Amin, S.A.; Adhikari, N.; Nirmal, J.; Jha, T.; Gayen, S. In silico modelling, identification of crucial molecular fingerprints, and prediction of new possible substrates of human organic cationic transporters 1 and 2. New J. Chem. 2020, 44, 4129–4143. [Google Scholar] [CrossRef]
- Malani, M.; Hiremath, M.S.; Sharma, S.; Jhunjhunwala, M.; Gayen, S.; Hota, C.; Nirmal, J. Interaction of systemic drugs causing ocular toxicity with organic cation transporter: An artificial intelligence prediction. J. Biomol. Struct. Dyn. 2023, 1–12. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Burggraaff, L.; Oranje, P.; Gouka, R.; van der Pijl, P.; Geldof, M.; van Vlijmen, H.W.T.; Ijzerman, A.P.; van Westen, G.J.P. Identification of novel small molecule inhibitors for solute carrier SGLT1 using proteochemometric modeling. J. Cheminform. 2019, 11, 15. [Google Scholar] [CrossRef]
| Transporter | ML Methods | References | |
|---|---|---|---|
| ABC | P-gp | RF | [35,36,37,38] |
| NN | [35] | ||
| SVM | [35,36,37,38,39] | ||
| k-NN | [35,37,38] | ||
| Bayes | [37,40] | ||
| Logistic regression (LR) | [37,38] | ||
| GTB | [36] | ||
| BCRP | RF | [37,38,41,42] | |
| Bayes | [37,40] | ||
| DNN | [43] | ||
| SVM | [37,38,39,41,44,45,46] | ||
| k-NN | [37,38] | ||
| XGBoost | [43] | ||
| LR | [37,38,41] | ||
| MRPs | RF | [37,38,47] | |
| SVM | [37,38,39,47,48] | ||
| Bayes | [37] | ||
| k-NN | [37,38,47] | ||
| LR | [37,38] | ||
| BSEP | RF | [38,49] | |
| SVM, LR, k-NN | [38] | ||
| SLC | OAT | RF | [50,51] |
| SVM | [39] | ||
| SNN, NB, k-NN, LR | [51] | ||
| OATP | RF | [52,53] | |
| k-NN, LR | [37,38,51,53] | ||
| XGBoost, DL | [53] | ||
| SVM | [37,38,39] | ||
| SNN | [51] | ||
| Bayes | [37,51,53] | ||
| OCT | RF, Bayes, k-NN, LR | [37] | |
| SVM | [37,39] | ||
| MATE1,2-K | SVM | [39] | |
| NET | RF | [54] | |
| Transport Protein | Data Sources | References |
|---|---|---|
| P-gp | Literature | [35] |
| P-gp | In-house dataset; ChEMBL [66] | [36] |
| P-gp, BCRP, MRPs, OATP, OCT | Metrabase [67] (http://www-metrabase.ch.cam.ac.uk, accessed on 25 July 2023); literature | [37] |
| P-gp, BCRP, MRPs, BSEP, OATP | LiverTox [68]; ChEMBL and PubChem [69] | [38] |
| P-gp, BCRP, MRPs, OAT, OATP, OCT, MATE1,2-K | Text-mining technique [70]; TP search (http://togodb.dbcls.jp/tpsearch, accessed on 25 July 2023); DIDB (http://www.druginteractioninfo.org/, accessed on 25 July 2023); PharmGKB (www.pharmgkb.org); TransPortal (http://dbts.ucsf.edu/fdatransportal, accessed on 25 July 2023); PubChem | [39] |
| P-gp, BCRP | Literature | [40] |
| BCRP | Literature; the Open PHACTS Discovery Platform | [41] |
| BCRP | Literature | [42,43,45,46] |
| BCRP | Literature; University of Washington Metabolism & Transport Drug Interaction Database (http://www.druginteractioninfo.org/, accessed on 25 July 2023); PubChem Database (http://pubchem.ncbi.nlm.nih.gov, accessed on 25 July 2023) | [44] |
| MRPs | Literature; Metrabase | [47] |
| MRPs | Literature; PubMed; TP search | [48] |
| BSEP | A proprietary BSEP assay dataset; published dataset [71] | [49] |
| BSEP | Training dataset | [72] |
| OAT | Literature; training dataset | [50] |
| OAT, OATP | PubChem | [51] |
| OATP | ChEMBL, UCSF-FDA TransPortal, DrugBank, Metrabase, IUPHAR | [52] |
| OATP | ChEMBL | [53] |
| OCT | Literature | [73] |
| OCT1 | Training dataset | [74] |
| NET | ZINC database [75]; PubChem; literature | [54] |
| SGLT1 | ChEMBL; the Spectrum Collection compound library | [76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, X.; Lin, K.; Wu, G.; Tao, X.; Zhai, X.; Lv, L.; Dong, D.; Zhu, Y.; Yang, S. Machine Learning Techniques Applied to the Study of Drug Transporters. Molecules 2023, 28, 5936. https://doi.org/10.3390/molecules28165936
Kong X, Lin K, Wu G, Tao X, Zhai X, Lv L, Dong D, Zhu Y, Yang S. Machine Learning Techniques Applied to the Study of Drug Transporters. Molecules. 2023; 28(16):5936. https://doi.org/10.3390/molecules28165936
Chicago/Turabian StyleKong, Xiaorui, Kexin Lin, Gaolei Wu, Xufeng Tao, Xiaohan Zhai, Linlin Lv, Deshi Dong, Yanna Zhu, and Shilei Yang. 2023. "Machine Learning Techniques Applied to the Study of Drug Transporters" Molecules 28, no. 16: 5936. https://doi.org/10.3390/molecules28165936
APA StyleKong, X., Lin, K., Wu, G., Tao, X., Zhai, X., Lv, L., Dong, D., Zhu, Y., & Yang, S. (2023). Machine Learning Techniques Applied to the Study of Drug Transporters. Molecules, 28(16), 5936. https://doi.org/10.3390/molecules28165936







