Bringing Nitric Oxide to the Molybdenum World—A Personal Perspective
Abstract
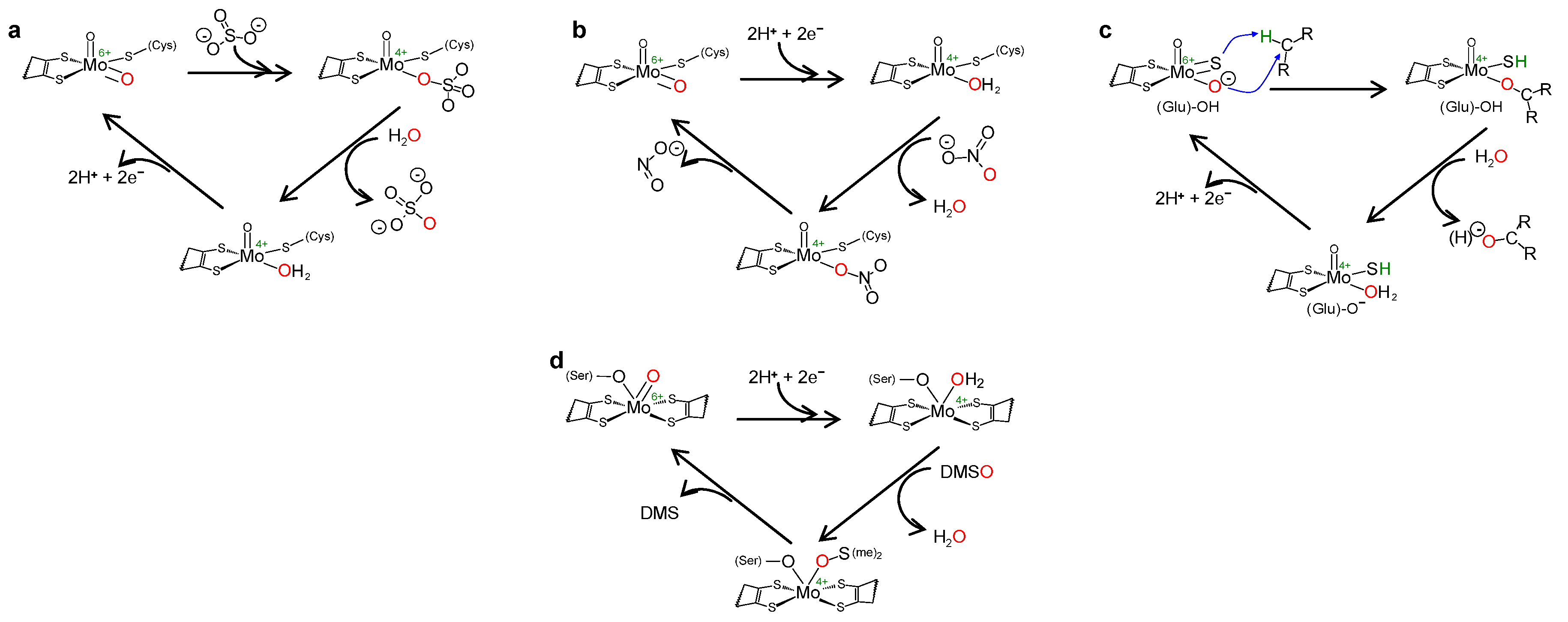
1. Context—I: The Molybdenum Side
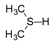 + 2e− + 2H+ →
+ 2e− + 2H+ →  + H2O
+ H2O
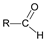 + H2O →
+ H2O → 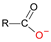 (H+) + 2e− + 2H+
(H+) + 2e− + 2H+
 + H2O →
+ H2O → 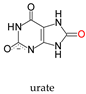 (H+) + 2e− + 2H+
(H+) + 2e− + 2H+
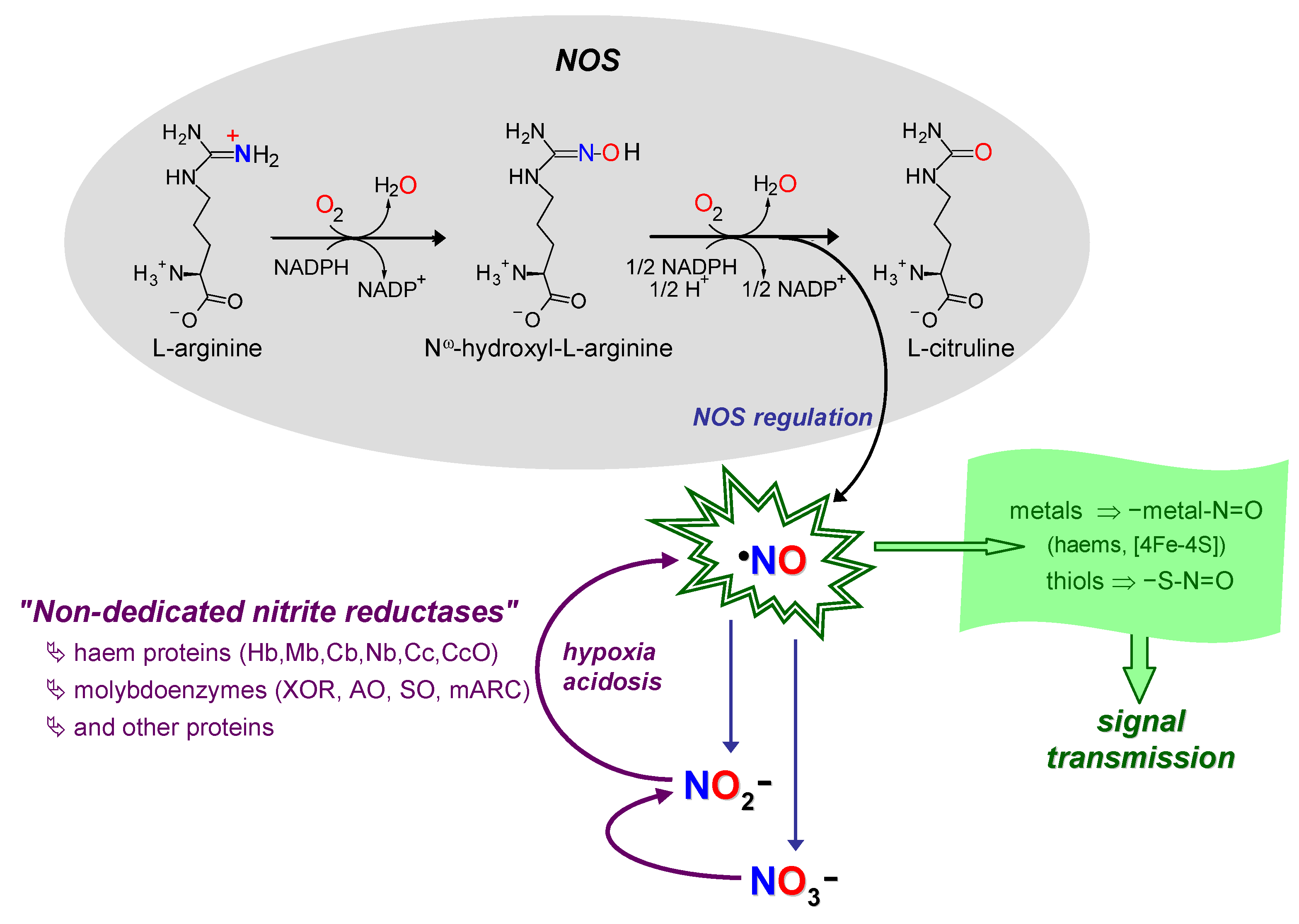
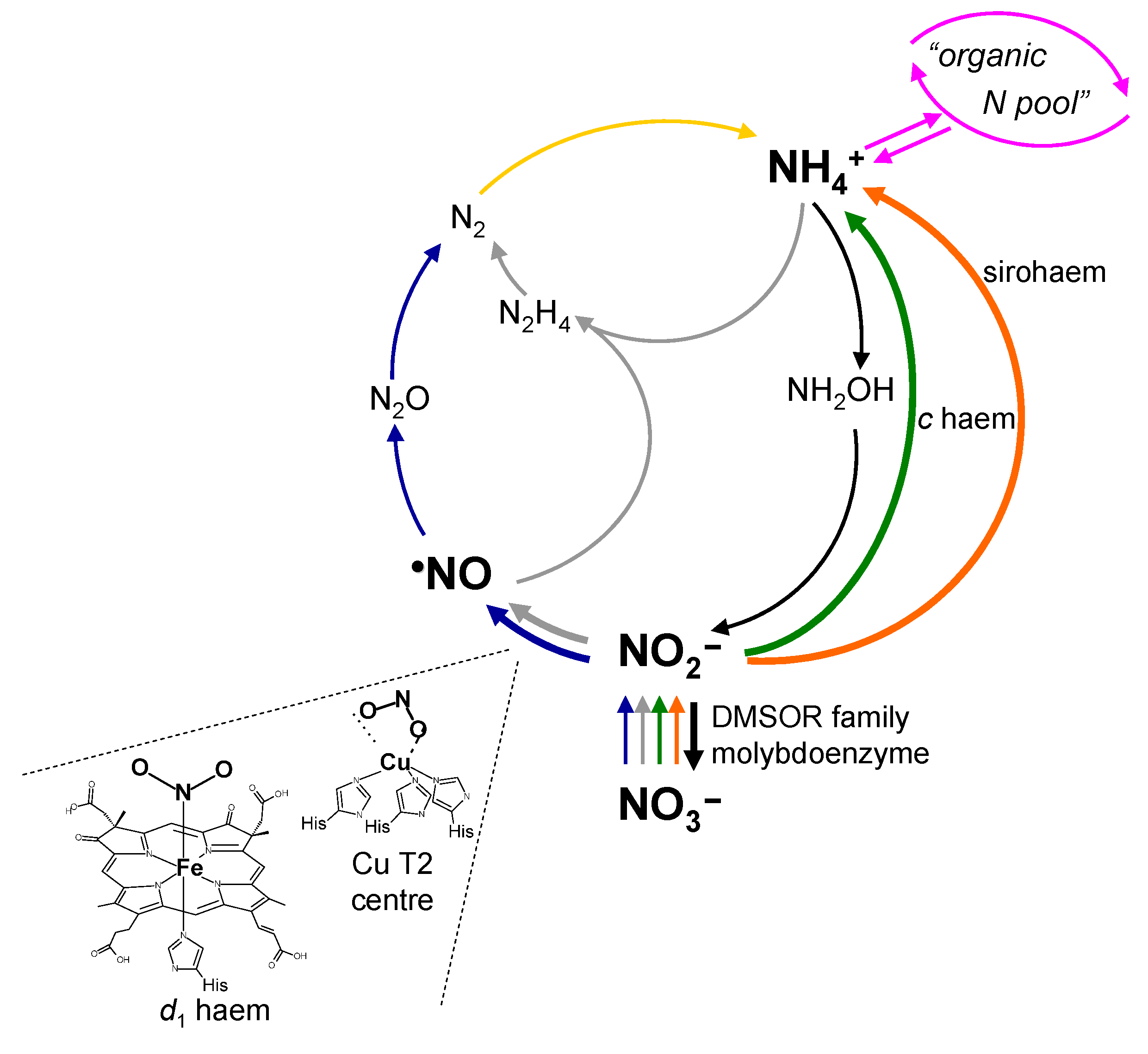
2. Context—II: The Nitric Oxide Side
3. How it Began…
- (1)
- are mammalian XO/XD and AO able to generate NO? with what magnitude and kinetics? (aiming to confirm the enzymes’ ability to reduce nitrite to NO and study the kinetics and magnitude of NO generation);
- (2)
- how is it possible for XO (and similar enzymes) to catalyse an OAT-A reaction? (aiming to establish the reaction mechanism of nitrite-derived NO formation);
- (3)
- if no other organism is known to use a “true” molybdenum-dependent nitrite reductase to reduce nitrite to NO, why would a mammalian cell be able to do so? (aiming to assess the reaction physiological feasibility and significance).
4. My Journey on the Molybdenum-Dependent, “Non-Dedicated” Nitrite Reductases
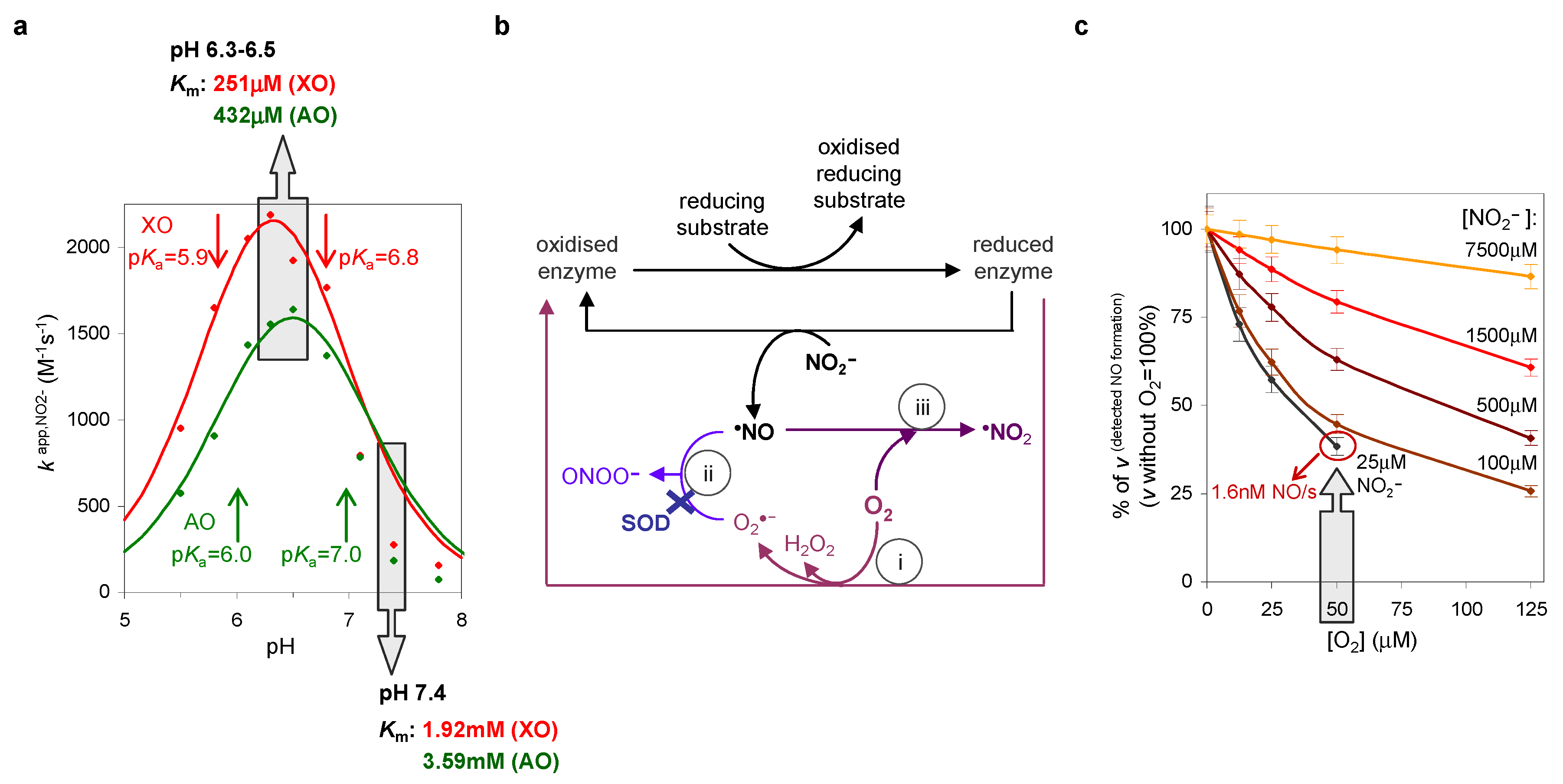
4.1. Are Mammalian XO/XD and AO Able to Generate NO?
4.2. Kinetics and Magnitude of XO/XD- and AO-Generated NO
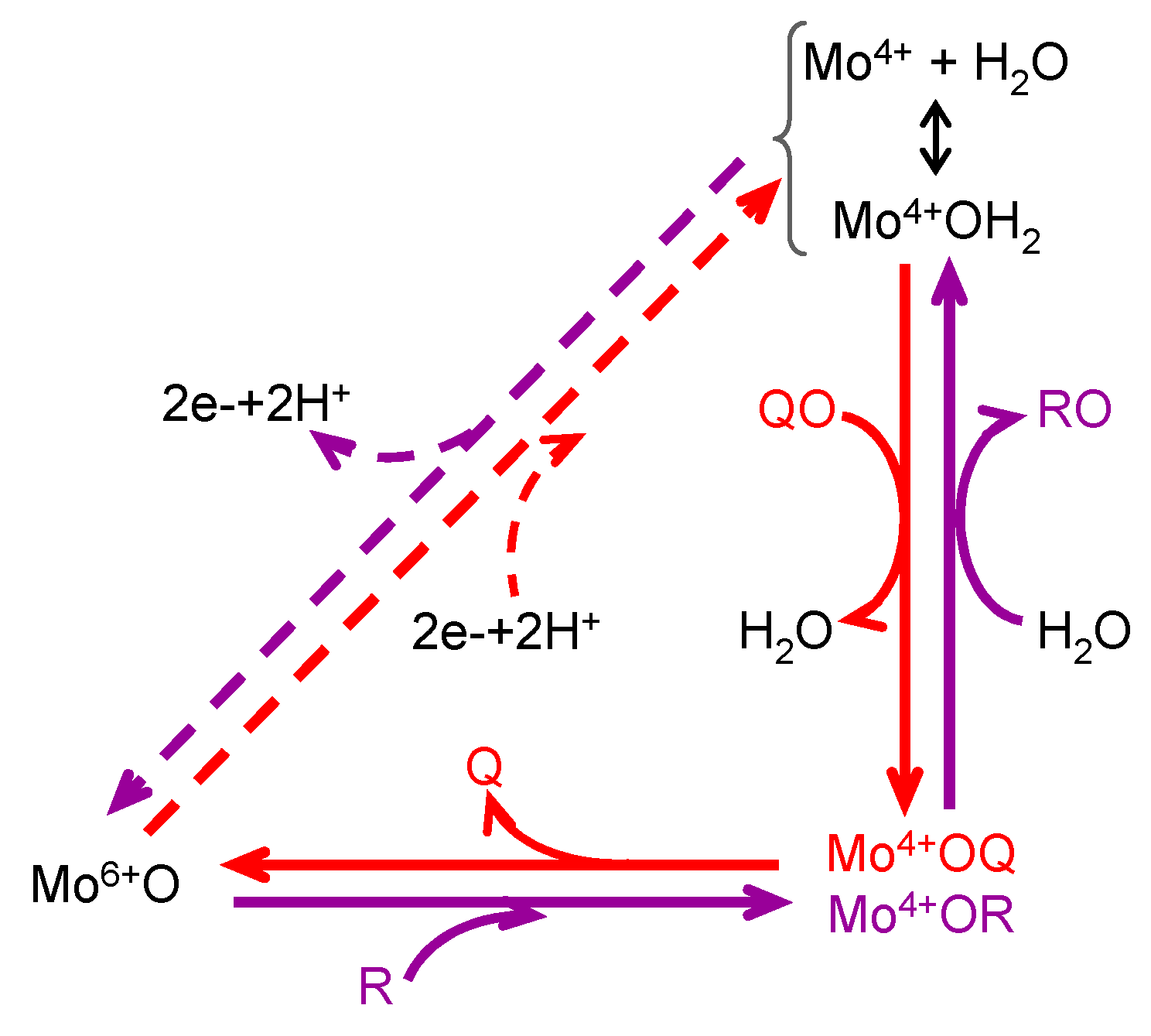
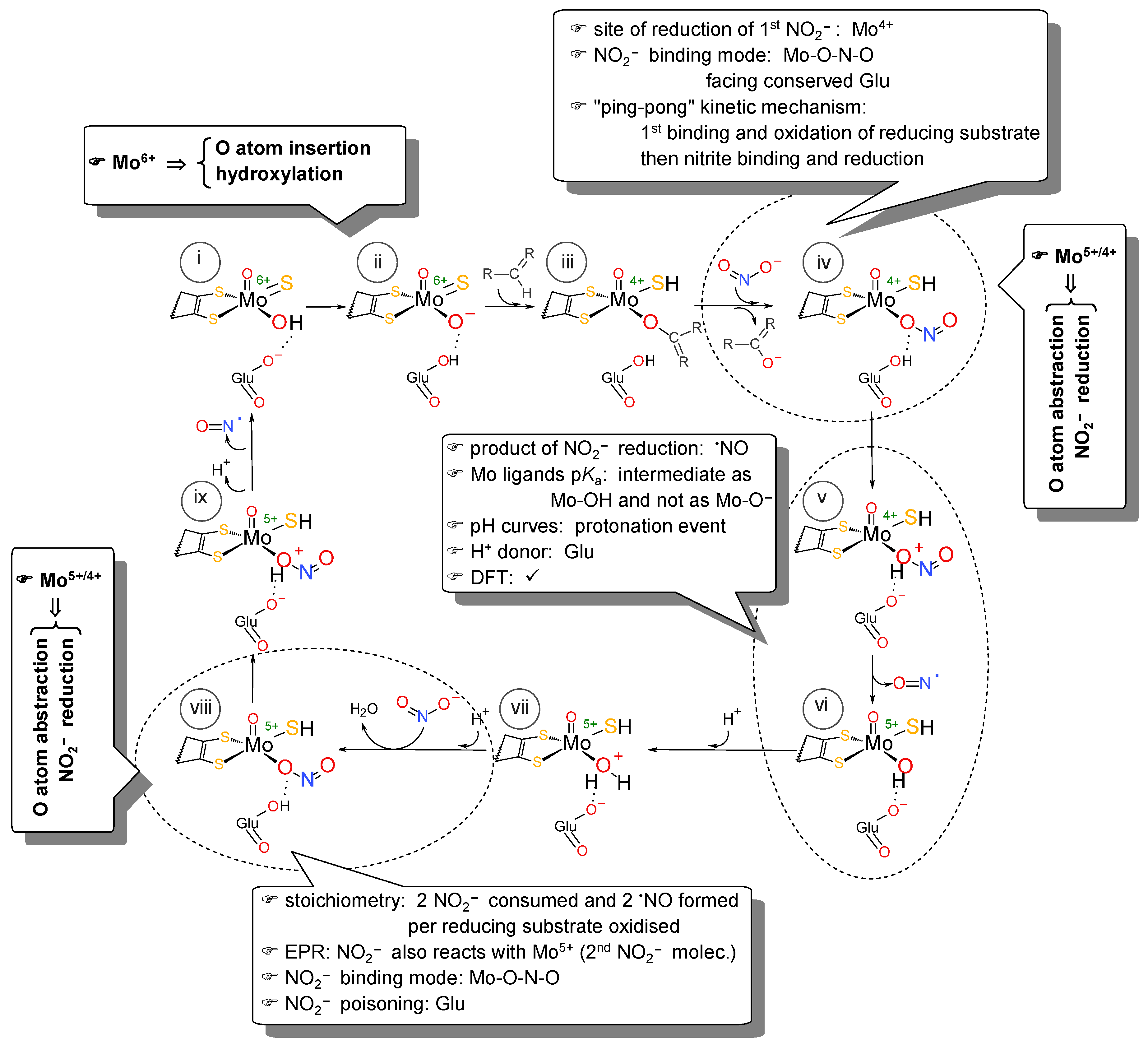
4.3. How Is It Possible for XO (and Similar Enzymes) to Catalyse an OAT-A Reaction?
 + 2e− + 2H+ →
+ 2e− + 2H+ → 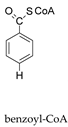 + H2O
+ H2O
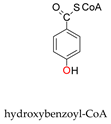
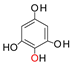 +
+  →
→  +
+ 
4.4. If No Other Organism Is Known to Use a “True” Molybdenum-Dependent Nitrite Reductase to Reduce Nitrite to NO, Why Would a Mammalian Cell Be Able to Do So?
5. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Zhang, Y.; Gladyshev, V.N. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 2008, 379, 881–899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rump, S.; Gladyshev, V.N. Comparative genomics and evolution of molybdenum utilization. Coord. Chem. Rev. 2011, 255, 1206–1217. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, V.N.; Zhang, Y. Chapter 2: Abundance, ubiquity and evolution of molybdoenzymes. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 81–99. [Google Scholar] [CrossRef]
- Wells, M.; Kanmanii, N.J.; Al Zadjali, A.M.; Janecka, J.E.; Basu, P.; Oremland, R.S.; Stolz, J.F. Methane, arsenic, selenium and the origins of the DMSO reductase family. Sci. Rep. 2020, 10, 10946. [Google Scholar]
- Hille, R. The mononuclear molybdenum enzymes. Chem. Rev. 1996, 96, 2757–2816. [Google Scholar] [CrossRef]
- Hille, R. The molybdenum oxotransferases and related enzymes. Dalton Trans. 2013, 42, 3029–3042. [Google Scholar]
- Hille, R.; Hall, J.; Basu, P. The mononuclear molybdenum enzymes. Chem. Rev. 2014, 114, 3963–4038. [Google Scholar]
- Maia, L.; Moura, J.J.G. Chapter 1: Molybdenum and tungsten-containing enzymes: An overview. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 1–80. [Google Scholar] [CrossRef]
- Seefeldt, L.C.; Dean, D.R.; Hoffman, B.M. Nitrogenase Mechanism: Electron and Proton Accumulation and N2 Reduction. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; Chapter 8; pp. 274–296. [Google Scholar] [CrossRef]
- Burén, S.; Jiménez-Vicente, E.; Echavarri-Erasun, C.; Rubio, L.M. Biosynthesis of Nitrogenase Cofactors. Chem. Rev. 2020, 120, 4921–4968. [Google Scholar] [CrossRef]
- Einsle, O.; Rees, D.C. Structural Enzymology of Nitrogenase Enzymes. Chem. Rev. 2020, 120, 4969–5004. [Google Scholar]
- Stappen, V.; Decamps, L.; Cutsail, G.E.; Bjornsson, R.; Henthorn, J.T.; Birrell, J.A.; DeBeer, S. The Spectroscopy of Nitrogenases. Chem. Rev. 2020, 120, 5005–5081. [Google Scholar] [CrossRef] [PubMed]
- Fischer, B.; Enemark, J.H.; Basu, P. A chemical approach to systematically designate the pyranopterin centers of molybdenum and tungsten enzymes and synthetic models. J. Inorg. Biochem. 1998, 72, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Basu, P.; Burgmayer, S.N.J. Pterin chemistry and its relationship to the molybdenum cofactor. Coord. Chem. Rev. 2011, 255, 1016–1038. [Google Scholar] [CrossRef]
- Williams, B.R.; Fu, Y.; Yap, G.P.A.; Burgmayer, S.J.N. Structure and Reversible Pyran Formation in Molybdenum Pyranopterin Dithiolene Models of the Molybdenum Cofactor. J. Am. Chem. Soc. 2012, 134, 1958–1987. [Google Scholar] [CrossRef][Green Version]
- Dong, C.; Yang, J.; Leimkühler, S.; Kirk, M.L. Pyranopterin Dithiolene Distortions Relevant to Electron Transfer in Xanthine Oxidase/Dehydrogenase. Inorg. Chem. 2014, 53, 7077–7079. [Google Scholar] [CrossRef]
- Rothery, R.A.; Weiner, J.H. Shifting the metallocentric molybdoenzyme paradigm: The importance of pyranopterin coordination. J. Biol. Inorg. Chem. 2015, 20, 349–372. [Google Scholar] [CrossRef]
- Maiti, B.K.; Maia, L.B.; Moro, A.J.; Lima, J.C.; Cordas, C.M.; Moura, I.; Moura, J.J.G. Unusual Reduction Mechanism of Copper in Cysteine-Rich Environment. Inorg. Chem. 2018, 57, 8078–8088. [Google Scholar] [PubMed]
- Enemark, J.H. {Moco}n, (n = 0–8): A general formalism for describing the highly covalent molybdenum cofactor of sulfite oxidase and related Mo enzymes. J. Inorg. Biochem. 2022, 231, 111801. [Google Scholar] [CrossRef]
- Yang, J.; Enemark, J.H.; Kirk, M.L. Metal-Dithiolene Bonding Contributions to Pyranopterin Molybdenum Enzyme Reactivity. Inorganics 2020, 8, 19. [Google Scholar] [PubMed]
- Kisker, C.; Schindelin, H.; Pacheco, A.; Wehbi, W.A.; Garrett, R.M.; Rajagopalan, K.V.; Enemark, J.H.; Rees, D.C. Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase. Cell 1997, 91, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Brody, M.S.; Hille, R. The kinetic behavior of chicken liver sulfite oxidase. Biochemistry 1999, 38, 6668–6677. [Google Scholar]
- Pacheco, A.; Hazzard, J.T.; Tollin, G.; Enemark, J.H. The pH dependence of intramolecular electron transfer rates in sulfite oxidase at high and low anion concentrations. J. Biol. Inorg. Chem. 1999, 4, 390–401. [Google Scholar] [CrossRef]
- Feng, C.; Kedia, R.V.; Hazzard, J.T.; Hurley, J.K.; Tollin, G.; Enemark, J.H. Effect of solution viscosity on intramolecular electron transfer in sulfite oxidase. Biochemistry 2002, 41, 5816–5821. [Google Scholar] [CrossRef]
- Peariso, K.; McNaughton, R.L.; Kirk, M.L. Active-site stereochemical control of oxygen atom transfer reactivity in sulfite oxidase. J. Am. Chem. Soc. 2002, 124, 9006–9007. [Google Scholar] [PubMed]
- Wilson, H.L.; Rajagopalan, K.V. The role of tyrosine 343 in substrate binding and catalysis by human sulfite oxidase. J. Biol. Chem. 2004, 279, 15105–15113. [Google Scholar] [PubMed]
- Peariso, K.; Helton, M.E.; Duesler, E.N.; Shadle, S.E.; Kirk, M.L. Sulfur K-edge spectroscopic investigation of second coordination sphere effects in oxomolybdenum-thiolates: Relationship to molybdenum-cysteine covalency and electron transfer in sulfite oxidase. Inorg. Chem. 2007, 46, 1259–1267. [Google Scholar]
- Bailey, S.; Rapson, T.; Johnson-Winters, K.; Astashkin, A.V.; Enemark, J.H.; Kappler, U. Molecular basis for enzymatic sulfite oxidation: How three conserved active site residues shape enzyme activity. J. Biol. Chem. 2009, 284, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Johnson-Winters, K.; Nordstrom, A.R.; Emesh, S.; Astashkin, A.V.; Rajapakshe, A.; Berry, R.E.; Tollin, G.; Enemark, J.H. Effects of interdomain tether length and flexibility on the kinetics of intramolecular electron transfer in human sulfite oxidase. Biochemistry 2010, 49, 1290–1296. [Google Scholar]
- Maiti, B.K.; Maia, L.B.; Pal, K.; Pakhira, B.; Avilés, T.; Moura, I.; Pauleta, S.R.; Nuñez, J.L.; Rizzi, A.C.; Brondino, C.D.; et al. One electron reduced square planar bis(benzene-1,2-dithiolato) copper dianionic complex and redox switch by O2/HO(-). Inorg. Chem. 2014, 53, 12799–12808. [Google Scholar] [CrossRef] [PubMed]
- Kappler, U.; Enemark, J.H. Sulfite-oxidizing enzymes. J. Biol. Inorg. Chem. 2015, 20, 253–264. [Google Scholar] [PubMed]
- Enemark, J.H. Consensus structures of the Mo(v) sites of sulfite-oxidizing enzymes derived from variable frequency pulsed EPR spectroscopy, isotopic labelling and DFT calculations. Dalton Trans. 2017, 46, 13202–13210. [Google Scholar] [CrossRef]
- Kappler, U.; Schwarz, G. The Sulfite Oxidase Family of Molybdenum Enzymes. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; Chapter 7; pp. 240–273. [Google Scholar] [CrossRef]
- Caldararu, O.; Feldt, M.; Cioloboc, D.; van Severen, M.C.; Starke, K.; Mata, R.A.; Nordlander, E.; Ryde, U. QM/MM study of the reaction mechanism of sulfite oxidase. Sci. Rep. 2018, 8, 4684. [Google Scholar]
- Gutteridge, S.; Bray, R.C.; Notton, B.A.; Fido, R.J.; Hewitt, E.J. Studies by electron-paramagnetic-resonance spectroscopy of the molybdenum centre of spinach (Spinacia oleraceaSpinacia oleracea) nitrate reductase. Biochem. J. 1983, 213, 137–142. [Google Scholar] [CrossRef]
- Solomonson, L.P.; Barber, M.J.; Howard, W.D.; Johnson, J.L.; Rajagopalan, K.V. Electron paramagnetic resonance studies on the molybdenum center of assimilatory NADH:nitrate reductase from Chlorella vulgaris. J. Biol. Chem. 1984, 259, 849–853. [Google Scholar] [PubMed]
- Campbell, W.H. Nitrate reductase structure, function and regulation: Bridging the Gap between Biochemistry and Physiology. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1999, 50, 277–303. [Google Scholar] [PubMed]
- George, G.N.; Mertens, J.A.; Campbell, W.H. Structural Changes Induced by Catalytic Turnover at the Molybdenum Site of Arabidopsis Nitrate Reductase. J. Am. Chem. Soc. 1999, 121, 9730–9731. [Google Scholar]
- Fischer, K.; Barbier, G.G.; Hecht, H.J.; Mendel, R.R.; Campbell, W.H.; Schwarz, G. Structural basis of eukaryotic nitrate reduction: Crystal structures of the nitrate reductase active site. Plant Cell 2005, 17, 1167–1179. [Google Scholar]
- Chi, J.C.; Roeper, J.; Schwarz, G.; Fischer-Schrader, K. Dual binding of 14-3-3 protein regulates Arabidopsis nitrate reductase activity. J. Biol. Inorg. Chem. 2015, 20, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Luque, E.; Chamizo-Ampudia, A.; Llamas, A.; Galvan, A.; Fernandez, E. Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 2015, 6, 899. [Google Scholar]
- Murray, K.N.; Watson, J.G.; Chaykin, S. Catalysis of the direct transfer of oxygen from nicotinamide N-oxide to xanthine by xanthine oxidase. J. Biol. Chem. 1966, 241, 4798–4801. [Google Scholar] [CrossRef]
- Hille, R.; Sprecher, H. On the mechanism of action of xanthine oxidase. Evidence in support of an oxo transfer mechanism in the molybdenum-containing hydroxylases. J. Biol. Chem. 1987, 262, 10914–10917. [Google Scholar] [CrossRef]
- Enroth, C.; Eger, B.T.; Okamoto, K.; Nishino, T.; Nishino, T.; Pai, E.F. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: Structure-based mechanism of conversion. Proc. Natl. Acad. Sci. USA 2000, 97, 10723–10728. [Google Scholar]
- Kuwabara, Y.; Nishino, T.; Okamoto, K.; Nishino, T. Unique amino acids cluster for switching from the dehydrogenase to oxidase form of xanthine oxidoreductase. Proc. Natl. Acad. Sci. USA 2003, 100, 8170–8175. [Google Scholar] [PubMed]
- Okamoto, K.; Matsumoto, K.; Hille, R.; Eger, B.T.; Pai, E.F.; Nishino, T. The crystal structure of xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition. Proc. Natl. Acad. Sci. USA 2004, 101, 7931–7936. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Okamoto, K.; Kawaguchi, Y.; Hori, H.; Matsumura, T.; Eger, B.T.; Pai, E.F.; Nishino, T. Mechanism of the conversion of xanthine dehydrogenase to xanthine oxidase: Identification of the two cysteine disulfide bonds and crystal structure of a non-convertible rat liver xanthine dehydrogenase mutant. J. Biol. Chem. 2005, 280, 24888–24894. [Google Scholar] [CrossRef]
- Nishino, T.; Okamoto, K.; Eger, B.T.; Pai, E.F.; Nishino, T. Mammalian xanthine oxidoreductase—mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J. 2008, 275, 3278–3289. [Google Scholar] [PubMed]
- Pauff, J.M.; Zhang, J.; Bell, C.E.; Hille, R. Substrate orientation in xanthine oxidase: Crystal structure of enzyme in reaction with 2-hydroxy-6-methylpurine. J. Biol. Chem. 2008, 283, 4818–4824. [Google Scholar] [PubMed]
- Sempombe, J.; Stein, B.; Kirk, M.L. Spectroscopic and Electronic Structure Studies Probing Covalency Contributions to C-H Bond Activation and Transition-State Stabilization in Xanthine Oxidase. Inorg. Chem. 2011, 50, 10919–10928. [Google Scholar] [CrossRef] [PubMed]
- Ishikita, H.; Eger, B.T.; Okamoto, K.; Nishino, T.; Pai, E.F. Protein conformational gating of enzymatic activity in xanthine oxidoreductase. J. Am. Chem. Soc. 2012, 134, 999–1009. [Google Scholar] [CrossRef]
- Okamoto, K.; Kusano, T.; Nishino, T. Chemical nature and reaction mechanisms of the molybdenum cofactor of xanthine oxidoreductase. Curr. Pharm. Des. 2013, 19, 2606–2614. [Google Scholar] [CrossRef]
- Cao, H.; Hall, J.; Hille, R. Substrate orientation and specificity in xanthine oxidase: Crystal structures of the enzyme in complex with indole-3-acetaldehyde and guanine. Biochemistry 2014, 53, 533–541. [Google Scholar]
- Nishino, T.; Okamoto, K.; Kawaguchi, Y.; Matsumura, T.; Eger, B.T.; Pai, E.F.; Nishino, T. The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase. FEBS J. 2015, 282, 3075–3090. [Google Scholar]
- Stein, B.W.; Kirk, M.L. Electronic structure contributions to reactivity in xanthine oxidase family enzymes. J. Biol. Inorg. Chem. 2015, 20, 183–194. [Google Scholar]
- Nishino, T.; Okamoto, K.; Leimkuhler, S. Enzymes of the Xanthino Oxidase Family. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; Chapter 6; pp. 192–239. [Google Scholar] [CrossRef]
- Schultz, B.E.; Hille, R.; Holm, R.H. Direct oxygen atom transfer in the mechanism of action of Rhodobacter sphaeroides dimethyl sulfoxide reductase. J. Am. Chem. Soc. 1995, 117, 827–828. [Google Scholar] [CrossRef]
- George, G.N.; Hilton, J.; Rajagopalan, K.V. X-ray Absorption Spectroscopy of Dimethyl Sulfoxide Reductase from Rhodobacter sphaeroides. J. Am. Chem. Soc. 1996, 118, 1113–1117. [Google Scholar] [CrossRef]
- Schindelin, H.; Kisker, C.; Hilton, J.; Rajagopalan, K.V.; Rees, D.C. Crystal structure of DMSO reductase: Redox-linked changes in molybdopterin coordination. Science 1996, 272, 1615–1621. [Google Scholar] [PubMed]
- Schneider, F.; Löwe, J.; Huber, R.; Schindelin, H.; Kisker, C.; Knäblein, J. Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 A resolution. J. Mol. Biol. 1996, 263, 53–69. [Google Scholar] [PubMed]
- Garton, S.D.; Hilton, J.; Oku, H.; Crouse, B.R.; Rajagopalan, K.V.; Johnson, M.K. Active Site Structures and Catalytic Mechanism of Rhodobacter sphaeroides Dimethyl Sulfoxide Reductase as Revealed by Resonance Raman Spectroscopy. J. Am. Chem. Soc. 1997, 119, 12906–12916. [Google Scholar]
- George, G.N.; Hilton, J.; Temple, C.; Prince, R.C.; Rajagopalan, K.V. Structure of the Molybdenum Site of Dimethyl Sulfoxide Reductase. J. Am. Chem. Soc. 1999, 121, 1256–1266. [Google Scholar] [CrossRef]
- Li, H.K.; Temple, C.; Rajagopalan, K.V.; Schindelin, H. The 1.3 Å Crystal Structure of Rhodobacter sphaeroides Dimethyl Sulfoxide Reductase Reveals Two Distinct Molybdenum Coordination Environments. J. Am. Chem. Soc. 2000, 122, 7673–7680. [Google Scholar]
- Magalon, A.; Ceccaldi, P.; Schoepp-Cothenet, B. The Prokaryotic Mo/W-bisPGD Family. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; Chapter 5; pp. 143–191. [Google Scholar] [CrossRef]
- Pacheco, J.; Niks, D.; Hille, R. Kinetic and spectroscopic characterization of tungsten-substituted DMSO reductase from Rhodobacter sphaeroides. J. Biol. Inorg. Chem. 2018, 23, 295–301. [Google Scholar] [CrossRef]
- Kirk, M.L.; Hille, R. Spectroscopic Studies of Mononuclear Molybdenum Enzyme Centers. Molecules 2022, 27, 4802. [Google Scholar]
- Kirk, M.L.; Lepluart, J.; Yang, J. Resonance Raman spectroscopy of pyranopterin molybdenum enzymes. J. Inorg. Biochem. 2022, 235, 111907. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.M.; Than, M.E.; Humm, A.; Huber, R.; Bourenkov, G.P.; Bartunik, H.D.; Bursakov, S.; Calvete, J.; Caldeira, J.; Carneiro, C.; et al. Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods. Structure 1999, 7, 65–79. [Google Scholar] [PubMed]
- Arnoux, P.; Sabaty, M.; Alric, J.; Frangioni, B.; Guigliarelli, B.; Adriano, J.-M.; Pignol, D. Structural and redox plasticity in the heterodimeric periplasmic nitrate reductase. Nat. Struct. Biol. 2003, 10, 928–934. [Google Scholar] [CrossRef]
- Jepson, B.J.; Anderson, L.J.; Rubio, L.M.; Taylor, C.J.; Butler, C.S.; Flores, E.; Herrero, A.; Butt, J.N.; Richardson, D.J. Tuning a nitrate reductase for function. The first spectropotentiometric characterization of a bacterial assimilatory nitrate reductase reveals novel redox properties. J. Biol. Chem. 2004, 279, 32212–32218. [Google Scholar] [CrossRef] [PubMed]
- Jormakka, M.; Richardson, D.; Byrne, B.; Iwata, S. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 2004, 12, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Bertero, M.G.; Rothery, R.A.; Boroumand, N.; Palak, M.; Blasco, F.; Ginet, N.; Weiner, J.H. Structural and biochemical characterization of a quinol binding site of Escherichia coli nitrate reductase A. J. Biol. Chem. 2005, 280, 14836–14843. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.; González, P.J.; Trincão, J.; Carvalho, A.L.; Najmudin, S.; Hettman, T.; Dieckman, S.; Moura, J.J.G.; Moura, I.; Romão, M.J. Heterodimeric nitrate reductase (NapAB) from Cupriavidus necator H16: Purification, crystallization and preliminary X-ray analysis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007, 63, 516–519. [Google Scholar]
- Jepson, B.J.N.; Mohan, S.; Clarke, T.A.; Gates, A.J.; Cole, J.A.; Butler, C.S.; Butt, J.N.; Hemmings, A.M.; Richardson, D.J. Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli. J. Biol. Chem. 2007, 282, 6425–6437. [Google Scholar] [CrossRef]
- Coelho, C.; González, P.J.; Moura, J.G.; Moura, I.; Trincão, J.; Romão, M.J. The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states. J. Mol. Biol. 2011, 408, 932–948. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.; Stolz, J.F.; Basu, P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014, 43, 676–706. [Google Scholar]
- Rendon, J.; Biaso, F.; Ceccaldi, P.; Toci, R.; Seduk, F.; Magalon, A.; Guigliarelli, B.; Grimaldi, S. Elucidating the Structures of the Low- and High-pH Mo(V) Species in Respiratory Nitrate Reductase: A Combined EPR, 14,15N HYSCORE, and DFT Study. Inorg. Chem. 2017, 56, 4423–4435. [Google Scholar] [CrossRef]
- Mintmier, B.; McGarry, J.M.; Sparacino-Watkins, C.E.; Sallmen, J.; Fischer-Schrader, K.; Magalon, A.; McCormick, J.R.; Stolz, J.F.; Schwarz, G.; Bain, D.J.; et al. Molecular cloning, expression and biochemical characterization of periplasmic nitrate reductase from Campylobacter jejuni. FEMS Microbiol. Lett. 2018, 365, fny151. [Google Scholar] [CrossRef]
- Seif Eddine, M.; Biaso, F.; Rendon, J.; Pilet, E.; Guigliarelli, B.; Magalon, A.; Grimaldi, S. 1,2H hyperfine spectroscopy and DFT modeling unveil the demethylmenasemiquinone binding mode to E. coli nitrate reductase A (NarGHI). Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148203. [Google Scholar] [CrossRef] [PubMed]
- Mintmier, B.; McGarry, J.M.; Bain, D.J.; Basu, P. Kinetic consequences of the endogenous ligand to molybdenum in the DMSO reductase family: A case study with periplasmic nitrate reductase. J. Biol. Inorg. Chem. 2021, 26, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Al-Attar, S.; Rendon, J.; Sidore, M.; Duneau, J.P.; Seduk, F.; Biaso, F.; Grimaldi, S.; Guigliarelli, B.; Magalon, A. Gating of Substrate Access and Long-Range Proton Transfer in Escherichia coli Nitrate Reductase A: The Essential Role of a Remote Glutamate Residue. ACS Catal. 2021, 11, 14303–14318. [Google Scholar] [CrossRef]
- Kumar, S.; Nicholas, D.J.D.; Williams, E.H. Definitive 15N NMR evidence that water serves as a source of ‘O’ during nitrite oxidation by Nitrobacter agilis. FEBS Lett. 1983, 152, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Sundermeyer-Klinger, H.; Meyer, W.; Warninghoff, B.; Bock, E. Membrane-bound nitrite oxidoreductase of Nitrobacter: Evidence for a nitrate reductase system. Arch. Microbiol. 1984, 140, 153–158. [Google Scholar] [CrossRef]
- DiSpirito, A.A.; Hooper, A.B. Oxygen exchange between nitrate molecules during nitrite oxidation by Nitrobacter. J. Biol. Chem. 1986, 261, 10534–10537. [Google Scholar] [CrossRef]
- Friedman, S.H.; Massefski, W.; Hollocher, T.C. Catalysis of intermolecular oxygen atom transfer by nitrite dehydrogenase of Nitrobacter agilis. J. Biol. Chem. 1986, 261, 10538–10543. [Google Scholar] [CrossRef]
- Meincke, M.; Bock, E.; Kastrau, D.; Kroneck, P.M.H. Nitrite oxidoreductase from Nitrobacter hamburgensis: Redox centers and their catalytic role. Arch. Microbiol. 1992, 158, 127–131. [Google Scholar] [CrossRef]
- Kirstein, K.; Bock, E. Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Arch. Microbiol. 1993, 160, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Spieck, E.; Muller, S.; Engel, A.; Mandelkow, E.; Patel, H.; Bock, E. Two-dimensional structure of membrane-bound nitrite oxidoreductase from Nitrobacter hamburgensis. J. Struct. Biol. 1996, 117–123. [Google Scholar] [CrossRef]
- Spieck, E.; Ehrich, S.; Aamand, J.; Bock, E. Isolation and immunocytochemical location of the nitrite-oxidizing system in nitrospira moscoviensis. Arch. Microbiol. 1998, 169, 225–230. [Google Scholar] [CrossRef]
- Martinez-Espinosa, R.M.; Dridge, E.J.; Bonete, M.J.; Butt, J.N.; Butler, C.S.; Sargent, F.; Richardson, D. Look on the positive side! The orientation, identification and bioenergetics of 'Archaeal' membrane-bound nitrate reductases. J. FEMS Microbiol. Lett. 2007, 276, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Lücker, S.; Wagner, M.; Maixner, F.; Pelletier, E.; Koch, H.; Vacherie, H.; Rattei, T.; Damstë, J.S.S.; Spieck, E.; Le Paslier, D.; et al. Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 13479–13484. [Google Scholar] [CrossRef]
- Chicano, T.M.; Dietrich, L.; de Almeida, N.M.; Akram, M.; Hartmann, E.; Leidreiter, F.; Leopoldus, D.; Mueller, M.; Sánchez, R.; Nuijten, G.H.L. Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Nat. Microbiol. 2021, 6, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Moncada, S.; Palmer, R.M.; Higgs, E.A. Nitric oxide: Physiology, pathophysiology, and pharmacology. Pharmacol Rev. 1991, 43, 109–142. [Google Scholar]
- Pfeiffer, S.; Mayer, B.; Hemmens, B. Nitric Oxide: Chemical Puzzles Posed by a Biological Messenger. Angew. Chem. Int. Edn. Engl. 1999, 38, 1714–1731. [Google Scholar] [CrossRef]
- Stuehr, D.J. Mammalian nitric oxide synthases. Biochim. Biophys. Acta 1999, 1411, 217–230. [Google Scholar] [CrossRef]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357, 593–615. [Google Scholar] [CrossRef]
- Gow, A.J.; Luchsinger, B.P.; Pawloski, J.R.; Singel, D.J.; Stamler, J.S. The oxyhemoglobin reaction of nitric oxide. Proc. Natl. Acad. Sci. USA 1999, 96, 9027–9032. [Google Scholar] [CrossRef] [PubMed]
- Brunori, M. Nitric oxide, cytochrome-c oxidase and myoglobin. Trends Biochem. Sci. 2001, 26, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Flögel, U.; Merx, M.W.; Gödecke, A.; Decking, U.K.M.; Schrader, J. Myoglobin: A scavenger of bioactive NO. Proc. Natl. Acad. Sci. USA 2001, 98, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Herold, S.; Exner, M.; Nauser, T. Kinetic and mechanistic studies of the NO*-mediated oxidation of oxymyoglobin and oxyhemoglobin. Biochemistry 2001, 40, 3385–3395. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-T.; Han, T.H.; Hyduke, D.R.; Vaughn, M.W.; Herle, H.V.; Hein, T.W.; Zhang, C.; Kuo, L.; Liao, J.C. Modulation of nitric oxide bioavailability by erythrocytes. Proc. Natl. Acad. Sci. USA 2001, 98, 11771–11776. [Google Scholar] [CrossRef]
- Witting, P.K.; Douglas, D.J.; Mauk, A.G. Reaction of human myoglobin and nitric oxide. Heme iron or protein sulfhydryl (s) nitrosation dependence on the absence or presence of oxygen. J. Biol. Chem. 2001, 276, 3991–3998. [Google Scholar] [CrossRef]
- Joshi, M.S.; Ferguson, T.B., Jr.; Han, T.H.; Hyduke, D.R.; Liao, J.C.; Rassaf, T.; Bryan, N.; Feelisch, M.; Lancaster, J.R., Jr. Nitric oxide is consumed, rather than conserved, by reaction with oxyhemoglobin under physiological conditions. Proc. Natl. Acad. Sc. USA 2002, 99, 10341–10346. [Google Scholar] [CrossRef]
- Gardner, P.R.; Gardner, A.M.; Brashear, W.T.; Suzuki, T.; Hvitved, A.N.; Setchell, K.D.R.; Olson, J.S. Hemoglobins dioxygenate nitric oxide with high fidelity. J. Inorg. Biochem. 2006, 100, 542–550. [Google Scholar] [CrossRef]
- Shiva, S.; Wang, X.; Ringwood, L.-A.; Xu, X.; Yuditskaya, S.; Annavajjhala, V.; Miyajima, H.; Hogg, N.; Harris, Z.L.; Gladwin, M.T. Ceruloplasmin is a NO oxidase and nitrite synthase that determines endocrine NO homeostasis. Nat. Chem. Biol. 2006, 2, 486–493. [Google Scholar] [CrossRef]
- Brunori, M.; Giuffre, A.; Forte, E.; Mastronicola, D.; Barone, M.C.; Sarti, P. Control of cytochrome c oxidase activity by nitric oxide. Biochim. Biophys. Acta 2004, 1655, 365–371. [Google Scholar] [CrossRef]
- Poyton, R.O.; Castello, P.R.; Ball, K.A.; Woo, D.K.; Pan, N. Mitochondria and hypoxic signaling: A new view. Ann. N. Y. Acad. Sci. 2009, 1177, 48–56. [Google Scholar] [CrossRef]
- Wink, D.A.; Darbyshire, J.F.; Nims, R.W.; Saavedra, J.E.; Ford, P.C. Reactions of the bioregulatory agent nitric oxide in oxygenated aqueous media: Determination of the kinetics for oxidation and nitrosation by intermediates generated in the NO/O2 reaction. Chem. Res. Toxicol. 1993, 6, 23–27. [Google Scholar] [CrossRef]
- Goldstein, S.; Czapski, G. Kinetics of Nitric Oxide Autoxidation in Aqueous Solution in the Absence and Presence of Various Reductants. The Nature of the Oxidizing Intermediates. J. Am. Chem. Soc. 1995, 117, 12078–12084. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Q.; Gupta, E.; Zorko, N.; Brownlee, E.; Zweier, J.L. Quantitative measurements of NO reaction kinetics with a Clark-type electrode. Nitric Oxide 2005, 13, 68–77. [Google Scholar] [CrossRef]
- Maia, L.B.; Moura, J.J.G. Putting xanthine oxidoreductase and aldehyde oxidase on the NO metabolism map: Nitrite reduction by molybdoenzymes. Redox Biol. 2018, 19, 274–289. [Google Scholar] [CrossRef]
- Johnson, G., 3rd; Tsao, P.S.; Mulloy, D.; Lefer, A.M. Cardioprotective effects of acidified sodium nitrite in myocardial ischemia with reperfusion. J. Pharmacol. Exp. Ther. 1990, 252, 35–41. [Google Scholar]
- Demoncheaux, E.A.; Higenbottam, T.W.; Foster, P.J.; Borland, C.D.; Smith, A.P.; Marriott, H.M.; Bee, D.; Akamine, S.; Davies, M.B. Circulating nitrite anions are a directly acting vasodilator and are donors for nitric oxide. Clin. Sci. 2002, 102, 77–83. [Google Scholar] [CrossRef]
- Hunter, C.J.; Dejam, A.; Blood, A.B.; Shields, H.; Kim-Shapiro, D.B.; Machado, R.F.; Tarekegn, S.; Mulla, N.; Hopper, A.O.; Schechter, A.N.; et al. Inhaled nebulized nitrite is a hypoxia-sensitive NO-dependent selective pulmonary vasodilator. Nat. Med. 2004, 10, 1122. [Google Scholar] [CrossRef]
- Webb, A.; Bond, R.; McLean, P.; Uppal, R.; Benjamin, N.; Ahluwalia, A. Reduction of nitrite to nitric oxide during ischemia protects against myocardial ischemia–reperfusion damage. Proc. Natl. Acad. Sci. USA 2004, 101, 13683–13688. [Google Scholar] [CrossRef]
- Duranski, M.R.; Greer, J.J.; Dejam, A.; Jaganmohan, S.; Hogg, N.; Langston, W.; Patel, R.P.; Yet, S.F.; Wang, X.; Kevil, C.G. Cytoprotective effects of nitrite during in vivo ischemia-reperfusion of the heart and liver. J. Clin. Investig. 2005, 115, 1232–1240. [Google Scholar] [CrossRef]
- Lu, P.; Liu, F.; Yao, Z.; Wang, C.Y.; Chen, D.D.; Tian, Y.; Zhang, J.H.; Wu, Y.H. Nitrite-derived nitric oxide by xanthine oxidoreductase protects the liver against ischemia–reperfusion injury. Hepatobiliary Pancreat Dis. Int. 2005, 4, 350. [Google Scholar]
- Lundberg, J.O.; Weitzberg, E. NO Generation From Nitrite and Its Role in Vascular Control. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 915. [Google Scholar] [CrossRef]
- Pluta, R.M.; Dejam, A.; Grimes, G.; Gladwin, M.T.; Oldfield, E.H. Nitrite infusions to prevent delayed cerebral vasospasm in a primate model of subarachnoid hemorrhage. JAMA 2005, 293, 1477–1484. [Google Scholar] [CrossRef]
- Tsuchiya, K.; Kanematsu, Y.; Yoshizumi, M.; Ohnishi, H.; Kirima, K.; Izawa, Y.; Shikishima, M.; Ishida, T.; Kondo, S.; Kagami, S.; et al. Nitrite is an alternative source of NO in vivo. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H2163–H2170. [Google Scholar] [CrossRef]
- Jung, K.-H.; Chu, K.; Ko, S.-Y.; Lee, S.-T.; Sinn, D.-I.; Park, D.-K.; Kim, J.-M.; Song, E.-C.; Kim, M.; Roh, J.K. Early Intravenous Infusion of Sodium Nitrite Protects Brain Against In Vivo Ischemia-Reperfusion Injury. Stroke 2006, 37, 2744. [Google Scholar] [CrossRef]
- Baker, J.E.; Su, J.; Fu, X.; Hsu, A.; Gross, G.J.; Tweddell, J.S.; Hogg, N. Nitrite confers protection against myocardial infarction: Role of xanthine oxidoreductase, NADPH oxidase and K(ATP) channels. J. Mol. Cell. Cardiol. 2007, 43, 437. [Google Scholar] [CrossRef]
- Bryan, N.S.; Calvert, J.W.; Elrod, J.W.; Gundewar, S.; Ji, S.Y.; Lefer, D.J. Dietary nitrite supplementation protects against myocardial ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA 2007, 104, 19144. [Google Scholar] [CrossRef]
- Dezfulian, C.; Raat, N.; Shiva, S.; Gladwin, M.T. Role of the anion nitrite in ischemia-reperfusion cytoprotection and therapeutics. Cardiovasc. Res. 2007, 75, 327–338. [Google Scholar] [CrossRef]
- Oldfield, E.H.; Cannon, R.O., 3rd; Schechter, A.N.; Gladwin, M.T. Nitrite infusion in humans and nonhuman primates: Endocrine effects, pharmacokinetics, and tolerance formation. Circulation 2007, 116, 1821–1832. [Google Scholar]
- Shiva, S.; Sack, M.N.; Greer, J.J.; Duranski, M.; Ringwood, L.A.; Burwell, L.; Wang, X.; MacArthur, P.H.; Shoja, A.; Raghavachari, N.; et al. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. J. Exp. Med. 2007, 204, 2089–2102. [Google Scholar] [CrossRef]
- Bryan, N.S.; Calvert, J.W.; Gundewar, S.; Lefer, D.J. Dietary nitrite restores NO homeostasis and is cardioprotective in endothelial nitric oxide synthase-deficient mice. Free Radic. Biol. Med. 2008, 45, 468. [Google Scholar] [CrossRef]
- Gonzalez, F.M.; Shiva, S.; Vincent, P.S.; Ringwood, L.A.; Hsu, L.Y.; Hon, Y.Y.; Aletras, A.H.; Cannon, R.O., 3rd; Gladwin, M.T.; Arai, A.E. Nitrite anion provides potent cytoprotective and antiapoptotic effects as adjunctive therapy to reperfusion for acute myocardial infarction. Circulation 2008, 117, 2986. [Google Scholar] [CrossRef]
- Maher, A.R.; Milsom, A.B.; Gunaruwan, P.; Abozguia, K.; Ahmed, I.; Weaver, R.A.; Thomas, P.; Ashrafian, H.; Born, G.V.; James, P.E.; et al. Hypoxic modulation of exogenous nitrite-induced vasodilation in humans. Circulation 2008, 117, 670–677. [Google Scholar] [CrossRef]
- Sinha, S.S.; Shiva, S.; Gladwin, M.T. Myocardial protection by nitrite: Evidence that this reperfusion therapeutic will not be lost in translation. Trends Cardiovasc. Med. 2008, 18, 163. [Google Scholar] [CrossRef]
- Raat, N.J.; Shiva, S.; Gladwin, M.T. Effects of nitrite on modulating ROS generation following ischemia and reperfusion. Adv. Drug. Deliv. Rev. 2009, 61, 339–350. [Google Scholar] [CrossRef]
- Zuckerbraun, B.S.; Shiva, S.; Ifedigbo, E.; Mathier, M.A.; Mollen, K.P.; Rao, J.; Bauer, P.M.; Choi, J.J.; Curtis, E.; Choi, A.M.; et al. Nitrite potently inhibits hypoxic and inflammatory pulmonary arterial hypertension and smooth muscle proliferation via xanthine oxidoreductase-dependent nitric oxide generation. Circulation 2010, 121, 98–109. [Google Scholar] [CrossRef]
- Alef, M.J.; Vallabhaneni, R.; Carchman, E.; Morris, S.M., Jr.; Shiva, S.; Wang, Y.; Kelley, E.E.; Tarpey, M.M.; Gladwin, M.T.; Tzeng, E.; et al. Nitrite generated NO circumvents dysregulated arginine/NOS signaling to protect against intimal hyperplasia in Sprague-Dawley rats. J. Clin. Investig. 2011, 121, 1646. [Google Scholar] [CrossRef]
- Blood, A.B.; Schroeder, H.J.; Terry, M.H.; Merrill-Henry, J.; Bragg, S.L.; Vrancken, K.; Liu, T.; Herring, J.L.; Sowers, L.C.; Wilson, S.M.; et al. Inhaled nitrite reverses hemolysis-induced pulmonary vasoconstriction in newborn lambs without blood participation. Circulation 2011, 123, 605–612. [Google Scholar] [CrossRef]
- Gilchrist, M.; Shore, A.C.; Benjamin, N. Inorganic nitrate and nitrite and control of blood pressure. Cardiovasc. Res. 2011, 89, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Kevil, C.G.; Kolluru, G.K.; Pattillo, C.B.; Giordano, T. Inorganic nitrite therapy: Historical perspective and future directions. Free Radic. Biol. Med. 2011, 51, 576–593. [Google Scholar] [CrossRef]
- Larsen, F.J.; Schiffer, T.A.; Borniquel, S.; Sahlin, K.; Ekblom, B.; Lundberg, J.O.; Weitzberg, E. Dietary inorganic nitrate improves mitochondrial efficiency in humans. Cell Metab. 2011, 13, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.O.; Carlstrom, M.; Larsen, F.J.; Weitzberg, E. Roles of dietary inorganic nitrate in cardiovascular health and disease. Cardiovasc. Res. 2011, 89, 525–532. [Google Scholar] [CrossRef]
- Murillo, D.; Kamga, C.; Mo, L.; Shiva, S. Nitrite as a mediator of ischemic preconditioning and cytoprotection. Nitric Oxide 2011, 25, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Pattillo, C.B.; Bir, S.; Rajaram, V.; Kevil, C. G Inorganic nitrite and chronic tissue ischaemia: A novel therapeutic modality for peripheral vascular diseases. Cardiovasc. Res. 2011, 89, 533–541. [Google Scholar] [CrossRef]
- Sindler, A.L.; Fleenor, B.S.; Calvert, J.W.; Marshall, K.D.; Zigler, M.L.; Lefer, D.J.; Seals, D.R. Nitrite supplementation reverses vascular endothelial dysfunction and large elastic artery stiffness with aging. Aging Cell 2011, 10, 429–437. [Google Scholar] [CrossRef]
- Baliga, R.S.; Milsom, A.B.; Ghosh, S.M.; Trinder, S.L.; Macallister, R.J.; Ahluwalia, A.; Hobbs, A.J. Dietary nitrate ameliorates pulmonary hypertension: Cytoprotective role for endothelial nitric oxide synthase and xanthine oxidoreductase. Circulation 2012, 125, 2922–2932. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.E.; Lai, Y.C.; Gladwin, M.T. Nitrate-nitrite-nitric oxide pathway in pulmonary arterial hypertension therapeutics. Ther. Circ. 2012, 125, 2824–2826. [Google Scholar] [CrossRef]
- Bueno, M.; Wang, J.; Mora, A.L.; Gladwin, M.T. Nitrite signaling in pulmonary hypertension: Mechanisms of bioactivation, signaling, and therapeutics. Antioxid. Redox Signal. 2013, 18, 1797–1809. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.M.; Kapil, V.; Fuentes-Calvo, I.; Bubb, K.J.; Pearl, V.; Milsom, A.B.; Khambata, R.; Maleki-Toyserkani, S.; Yousuf, M.; Benjamin, N.; et al. Enhanced vasodilator activity of nitrite in hypertension: Critical role for erythrocytic xanthine oxidoreductase and translational potential. Hypertension 2013, 61, 1091–1102. [Google Scholar] [CrossRef]
- Omar, S.A.; Webb, A. Nitrite reduction and cardiovascular protection. J. Mol. Cell Cardiol. 2014, 73, 57–69. [Google Scholar] [CrossRef]
- Millar, T.M.; Stevens, C.R.; Benjamin, N.; Eisenthal, R.; Harrison, R.; Blake, D.R. Xanthine oxidoreductase catalyses the reduction of nitrates and nitrite to nitric oxide under hypoxic conditions. FEBS Lett. 1998, 427, 225–228. [Google Scholar] [CrossRef]
- Zhang, Z.; Naughton, D.; Winyard, P.G.; Benjamin, N.; Blake, D.R.; Symons, M.C. Generation of nitric oxide by a nitrite reductase activity of xanthine oxidase: A potential pathway for nitric oxide formation in the absence of nitric oxide synthase activity. Biochem. Biophys. Res. Commun. 1998, 249, 767–772. [Google Scholar] [CrossRef]
- Kozlov, A.V.; Staniek, K.; Nohl, H. Nitrite reductase activity is a novel function of mammalian mitochondria. FEBS Lett. 1999, 454, 127–130. [Google Scholar] [CrossRef]
- Godber, H.L.J.; Doel, J.J.; Sapkota, G.P.; Blake, D.R.; Stevens, C.R.; Eisenthal, R.; Harrison, R. Reduction of nitrite to nitric oxide catalyzed by xanthine oxidoreductase. J. Biol. Chem. 2000, 275, 7757–7763. [Google Scholar] [CrossRef]
- Li, H.; Samouilov, A.; Liu, X.; Zweier, J.L. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrite reduction. Evaluation of its role in nitric oxide generation in anoxic tissues. J. Biol. Chem. 2001, 276, 24482–24489. [Google Scholar] [CrossRef]
- Li, H.; Samouilov, A.; Liu, X.; Zweier, J.L. Characterization of the effects of oxygen on xanthine oxidase-mediated nitric oxide formation. J. Biol. Chem. 2004, 279, 16939–16946. [Google Scholar] [CrossRef]
- Castello, P.R.; David, P.S.; McClure, T.; Crook, Z.; Poyton, R.O. Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: Implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab. 2006, 3, 277–287. [Google Scholar] [CrossRef]
- Gautier, C.; van Faassen, E.; Mikula, I.; Martasek, P.; Slama-Schwok, A. Endothelial nitric oxide synthase reduces nitrite anions to NO under anoxia. Biochem. Biophys. Res. Commun. 2006, 341, 816–821. [Google Scholar] [CrossRef]
- Li, H.; Liu, X.; Cui, H.; Chen, Y.R.; Cardounel, A.J.; Zweier, J.L. Characterization of the mechanism of cytochrome P450 reductase-cytochrome P450-mediated nitric oxide and nitrosothiol generation from organic nitrates. J. Biol. Chem. 2006, 281, 12546–12554. [Google Scholar] [CrossRef]
- Vanin, A.F.; Bevers, L.M.; Slama-Schwok, A.; van Faassen, E.E. Nitric oxide synthase reduces nitrite to NO under anoxia. Cell Mol. Life Sci. 2006, 64, 96–103. [Google Scholar] [CrossRef]
- Basu, S.; Azarova, N.A.; Font, M.D.; King, S.B.; Hogg, N.; Gladwin, M.T.; Shiva, S.; Kim-Shapiro, D.B. Nitrite reductase activity of cytochrome c. J. Biol. Chem. 2008, 283, 32590–32597. [Google Scholar] [CrossRef]
- Benamar, A.; Rolletschek, H.; Borisjuk, L.; Avelange-Macherel, M.-H.; Curien, G.; Mostefai, H.A.; Andriantsitohaina, R.; Macherel, D. Nitrite-nitric oxide control of mitochondrial respiration at the frontier of anoxia. Biochim. Biophys. Acta 2008, 1777, 1268–1275. [Google Scholar] [CrossRef] [PubMed]
- Castello, P.R.; Woo, D.K.; Ball, K.; Wojcik, J.; Liu, L.; Poyton, R.O. Oxygen-regulated isoforms of cytochrome c oxidase have differential effects on its nitric oxide production and on hypoxic signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 8203–8208. [Google Scholar] [CrossRef]
- Li, H.; Cui, H.; Kundu, T.K.; Alzawahra, W.; Zweier, J.L. Nitric oxide production from nitrite occurs primarily in tissues not in the blood: Critical role of xanthine oxidase and aldehyde oxidase. J. Biol. Chem. 2008, 283, 17855–17863. [Google Scholar] [CrossRef]
- Li, H.; Kundu, T.K.; Zweier, J.L. Characterization of the magnitude and mechanism of aldehyde oxidase-mediated nitric oxide production from nitrite. J. Biol. Chem. 2009, 284, 33850–33858. [Google Scholar] [CrossRef] [PubMed]
- Badejo, A.M., Jr.; Hodnette, C.; Dhaliwal, J.S.; Casey, D.B.; Pankey, E.; Murthy, S.N.; Nossaman, B.D.; Hyman, A.L.; Kadowitz, P. Mitochondrial aldehyde dehydrogenase mediates vasodilator responses of glyceryl trinitrate and sodium nitrite in the pulmonary vascular bed of the rat. Am. J. Physiol. Heart Circ. Physiol. 2010, 299, H819–H826. [Google Scholar] [CrossRef]
- Tiso, M.; Tejero, J.; Basu, S.; Azarov, I.; Wang, X.; Simplaceanu, V.; Frizzell, S.; Jayaraman, T.; Geary, L.; Shapiro, C.; et al. Human neuroglobin functions as a redox-regulated nitrite reductase. J. Biol. Chem. 2011, 286, 18277–18289. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hemann, C.; Abdelghany, T.M.; El-Mahdy, M.A.; Zweier, J.L. Characterization of the mechanism and magnitude of cytoglobin-mediated nitrite reduction and nitric oxide generation under anaerobic conditions. J. Biol. Chem. 2012, 278, 36623–36633. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.E.; Tejero, J.; Sun, B.; Gauthier, M.C.; Thomas, J.; Ragireddy, V.; Merchant, B.A.; Wang, J.; Azarov, I.; Basu, P.; et al. Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J. Biol. Chem. 2014, 289, 10345–10358. [Google Scholar] [CrossRef]
- Wang, J.; Krizowski, S.; Fischer-Schrader, K.; Niks, D.; Tejero, J.; Sparacino-Watkins, C.; Wang, L.; Ragireddy, V.; Frizzell, S.; Kelley, E.E.; et al. Sulfite Oxidase Catalyzes Single-Electron Transfer at Molybdenum Domain to Reduce Nitrite to Nitric Oxide. Antioxid. Redox. Signal. 2015, 23, 283–294. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, Á.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef]
- Bender, D.; Tobias Kaczmarek, A.; Niks, D.; Hille, R.; Schwarz, G. Mechanism of nitrite-dependent NO synthesis by human sulfite oxidase. Biochem. J. 2019, 476, 1805–1815. [Google Scholar] [CrossRef]
- Kaczmarek, A.T.; Strampraad, M.J.F.; Hagedoorn, P.L.; Schwarz, G. Reciprocal regulation of sulfite oxidation and nitrite reduction by mitochondrial sulfite oxidase. Nitric Oxide 2019, 89, 22–31. [Google Scholar] [CrossRef]
- Mohn, M.A.; Thaqi, B.; Fischer-Schrader, K. Isoform-Specific NO Synthesis by Arabidopsis thaliana Nitrate Reductase. Plants 2019, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Mutus, B. The catalytic mechanism for NO production by the mitochondrial enzyme, sulfite oxidase. Biochem. J. 2019, 476, 1955–1956. [Google Scholar] [CrossRef]
- Tejada-Jimenez, M.; Llamas, A.; Galván, A.; Fernández, E. Role of Nitrate Reductase in NO Production in Photosynthetic Eukaryotes. Plants 2019, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Bender, D.; Kaczmarek, A.T.; Kuester, S.; Burlina, A.B.; Schwarz, G. Oxygen and nitrite reduction by heme-deficient sulphite oxidase in a patient with mild sulphite oxidase deficiency. J. Inherit. Metab. Dis. 2020, 43, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Costa-Broseta, Á.; Castillo, M.; León, J. Post-Translational Modifications of Nitrate Reductases Autoregulates Nitric Oxide Biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 549. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kaladhar, V.C.; Fitzpatrick, T.B.; Fernie, A.R.; Møller, I.M.; Loake, G.J. Nitric oxide regulation of plant metabolism. Mol. Plant. 2022, 15, 228–242. [Google Scholar] [CrossRef]
- Timilsina, A.; Dong, W.; Hasanuzzaman, M.; Liu, B.; Hu, C. Nitrate-Nitrite-Nitric Oxide Pathway: A Mechanism of Hypoxia and Anoxia Tolerance in Plants. Int. J. Mol. Sci. 2022, 23, 11522. [Google Scholar] [CrossRef]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The evolution and future of Earth's nitrogen cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B.; Moura, J.J. How Biology handles nitrite. Chem. Rev. 2014, 114, 5273–5357. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.K.; Maia, L.B.; Silveira, C.M.; Todorovic, S.; Carreira, C.; Carepo, M.S.; Grazina, R.; Moura, I.; Pauleta, S.R.; Moura, J.J. Incorporation of molybdenum in rubredoxin: Models for mononuclear molybdenum enzymes. J. Biol. Inorg. Chem. 2015, 20, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.; Maia, L.; Pauleta, S.R.; Moura, J.J.G. A bird’s-eye view of denitrification in relation to the nitrogen cycle. In Metalloenzymes in Denitrification: Applications and Environmental Impacts; Moura, I., Moura, J.J.G., Pauleta, S.R., Maia, L., Eds.; RSC Metallobiology Series No. 9; Royal Society of Chemistry: Cambridge, UK, 2017; Chapter 1; pp. 1–10. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, N.; Dong, H.T.; Harland, J.B.; Hunt, A.P.; White, C.J. Reversing nitrogen fixation. Nat. Rev. Chem. 2018, 2, 278–289. [Google Scholar] [CrossRef]
- Lehnert, N.; Musselman, B.W.; Seefeldt, L.C. Grand challenges in the nitrogen cycle. Chem. Soc. Rev. 2021, 50, 3640–3646. [Google Scholar] [CrossRef]
- Stiefel, E.I. Proposed molecular mechanism for the action of molybdenum in enzymes: Coupled proton and electron transfer. Proc. Natl. Acad. Sci. USA 1973, 70, 988–992. [Google Scholar] [CrossRef]
- Burgmayer, S.J.N.; Stiefel, E.I. Molybdenum enzymes, cofactors, and systems: The chemical uniqueness of molybdenum. J. Chem. Educ. 1985, 62, 943–953. [Google Scholar] [CrossRef]
- Harlan, E.E.; Berg, J.M.; Holm, R.H. Thermodynamic fitness of molybdenum (IV, VI) complexes for oxygen-atom transfer reactions, including those with enzymic substrates. J. Am. Chem. Soc. 1986, 108, 6992–7000. [Google Scholar] [CrossRef]
- Holm, R.H.; Berg, J.M. Toward functional models of metalloenzyme active sites: Analog reaction systems of the molybdenum oxo transferases. Acc. Chem. Res. 1986, 19, 363–370. [Google Scholar] [CrossRef]
- Holm, R.H. Metal-centered oxygen atom transfer reactions. Chem. Rev. 1987, 87, 1401–1449. [Google Scholar] [CrossRef]
- Holm, R.H. The biologically relevant oxygen atom transfer chemistry of molybdenum: From synthetic analogue systems to enzymes Coord. Chem. Rev. 1990, 100, 183–221. [Google Scholar] [CrossRef]
- Enemark, J.H.; Young, C.G. Bioinorganic chemistry of pterin-containing molybdenum and tungsten enzymes. Adv. Inorg. Chem. 1993, 40, 1–88. [Google Scholar]
- Holm, R.H.; Donahue, J.P. A thermodynamic scale for oxygen atom transfer reactions. Polyhedron 1993, 12, 571–589. [Google Scholar] [CrossRef]
- Donahue, J.P. Thermodynamic scales for sulfur atom transfer and oxo-for-sulfido exchange reactions. Chem. Rev. 2006, 106, 4747–4783. [Google Scholar] [CrossRef] [PubMed]
- Rajapakshe, A.; Snyder, R.A.; Astashkin, A.V.; Bernardson, P.; Evans, D.J.; Young, C.G.; Evans, D.H.; Enemark, J.H. Insights into the nature of Mo (V) species in solution: Modeling catalytic cycles for molybdenum enzymes. Inorg. Chim. Acta 2009, 362, 4603–4608. [Google Scholar] [CrossRef]
- Yang, J.; Giles, L.J.; Ruppelt, C.; Mendel, R.R.; Bittner, F.; Kirk, M.L. Oxyl and hydroxyl radical transfer in mitochondrial amidoxime reducing component-catalyzed nitrite reduction. J. Am. Chem. Soc. 2015, 137, 5276–5279. [Google Scholar] [CrossRef]
- Maia, L.; Mira, L. Xanthine oxidase and aldehyde oxidase: A simple procedure for the simultaneous purification from rat liver. Arch. Biochem. Biophys. 2002, 400, 48–53. [Google Scholar] [CrossRef]
- Romão, M.J.; Archer, M.; Moura, I.; Moura, J.J.G.; LeGall, J.; Engh, R.; Schneider, M.; Hof, P.; Huber, R. Crystal structure of the xanthine oxidase-related aldehyde oxido-reductase from D. gigas. Science 1995, 270, 1170–1176. [Google Scholar] [CrossRef]
- Huber, R.; Hof, P.; Duarte, R.O.; Moura, J.J.; Moura, I.; Liu, M.Y.; LeGall, J.; Hille, R.; Archer, M.; Romão, M.J. A structure-based catalytic mechanism for the xanthine oxidase family of molybdenum enzymes. Proc. Natl. Acad. Sci. USA 1996, 93, 8846–8851. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, J.; Macieira, S.; Dias, J.M.; Huber, R.; Ascenso, C.S.; Rusnak, F.; Moura, J.J.; Moura, I.; Romão, M.J. Gene sequence and crystal structure of the aldehyde oxidoreductase from Desulfovibrio desulfuricans ATCC 27774. J. Mol. Biol. 2000, 297, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, J.M.; Dias, J.M.; Huber, R.; Moura, J.J.; Romão, M.J. Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A. J. Biol. Inorg. Chem. 2001, 6, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Krippahl, L.; Palma, N.; Moura, I.; Moura, J.J.G. Modelling the electron-transfer complex between aldehyde oxidoreductase and flavodoxin. Eur. J. Inorg. Chem. 2006, 19, 3835–3840. [Google Scholar] [CrossRef]
- Moura, J.J.G. The History of Desulfovibrio gigas Aldehyde Oxidoreductase—A Personal View. Molecules 2023, 28, 4229. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B.; Moura, J.J. Nitrite reduction by xanthine oxidase family enzymes: A new class of nitrite reductases. J. Biol. Inorg. Chem. 2011, 16, 443–460. [Google Scholar] [CrossRef]
- Maia, L.B.; Pereira, V.; Mira, L.; Moura, J.J.G. Nitrite reductase activity of rat and human xanthine oxidase, xanthine dehydrogenase, and aldehyde oxidase: Evaluation of their contribution to NO formation in vivo. Biochemistry 2015, 54, 685–710. [Google Scholar] [CrossRef]
- Maia, L.; Moura, J.J.G. Lessons from Denitrification for the Human Metabolism of Signalling Nitric Oxide. In Metalloenzymes in Denitrification: Applications and Environmental Impacts; Moura, I., Moura, J.J.G., Pauleta, S.R., Maia, L., Eds.; RSC Metallobiology Series No. 9; Royal Society of Chemistry: Cambridge, UK, 2017; Chapter 17; pp. 419–443. [Google Scholar] [CrossRef]
- Maia, L.; Moura, J.J.G. Detection of Nitric Oxide by Electron Paramagnetic Resonance Spectroscopy: Spin-Trapping with Iron-Dithiocarbamates. Methods Mol. Biol. 2016, 1424, 81–102. [Google Scholar]
- Maia, L.B.; Moura, J.J. Nitrite reduction by molybdoenzymes: A new class of nitric oxide-forming nitrite reductases. J. Biol. Inorg. Chem. 2015, 20, 403–433. [Google Scholar]
- Maia, L.; Moura, I.; Moura, J.J.G. EPR spectroscopy on mononuclear molybdenum-containing enzymes. In Future Directions in Metalloprotein and Metalloenzyme Research, Biological Magnetic Resonance; Hanson, G., Berliner, L.J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Chapter 4; Volume 33, pp. 55–101. [Google Scholar]
- Opie, L.H. Effects of regional ischemia on metabolism of glucose and fatty acids. Relative rates of aerobic and anaerobic energy production during myocardial infarction and comparison with effects of anoxia. Circ. Res. 1976, 38, 152–174. [Google Scholar]
- Cobbe, S.M.; Poole-Wilson, P.A. Tissue acidosis in myocardial hypoxia. J. Mol. Cell. Cardiol. 1980, 12, 761–770. [Google Scholar] [CrossRef]
- Momomura, S.; Ingwall, J.S.; Parker, J.A.; Sahagian, P.; Ferguson, J.J.; Grossman, W. The relationships of high energy phosphates, tissue pH, and regional blood flow to diastolic distensibility in the ischemic dog myocardium. Circ. Res. 1985, 57, 822–835. [Google Scholar] [CrossRef]
- Rodriguez, J.; Maloney, R.E.; Rassaf, T.; Bryan, N.S.; Feelisch, M. Chemical nature of nitric oxide storage forms in rat vascular tissue. Proc. Natl. Acad. Sci. USA 2003, 100, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Bryan, N.S.; Rassaf, T.; Maloney, R.E.; Rodriguez, C.M.; Saijo, F.; Rodriguez, J.R.; Feelisch, M. Cellular targets and mechanisms of nitros(yl)ation: An insight into their nature and kinetics in vivo. Proc. Natl. Acad. Sci. USA 2004, 101, 4308–4313. [Google Scholar] [CrossRef]
- Shiva, S.; Gladwin, M.T. Shining a light on tissue NO stores: Near infrared release of NO from nitrite and nitrosylated hemes. J. Mol. Cell Cardiol. 2009, 46, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Giraldez, R.R.; Panda, A.; Xia, Y.; Sanders, S.P.; Zweier, J.L. Decreased nitric-oxide synthase activity causes impaired endothelium-dependent relaxation in the postischemic heart. J. Biol. Chem. 1997, 272, 21420–21426. [Google Scholar] [CrossRef]
- Ward, J.P.T. Oxygen sensors in context. Biochim. Biophys. Acta 2008, 1777, 1–14. [Google Scholar] [CrossRef]
- Gibson, J.; Dispensa, M.; Harwood, C.S. 4-hydroxybenzoyl coenzyme A reductase (dehydroxylating) is required for anaerobic degradation of 4-hydroxybenzoate by Rhodopseudomonas palustris and shares features with molybdenum-containing hydroxylases. J. Bacteriol. 1997, 179, 634–642. [Google Scholar] [CrossRef][Green Version]
- Unciuleac, M.; Warkentin, E.; Page, C.C.; Boll, M.; Ermler, U. Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow. Structure 2004, 12, 2249–2256. [Google Scholar] [CrossRef]
- Boll, M. Key enzymes in the anaerobic aromatic metabolism catalysing Birch-like reductions. Biochim. Biophys. Acta 2005, 1707, 34–50. [Google Scholar] [CrossRef]
- Johannes, J.; Unciuleac, M.C.; Friedrich, T.; Warkentin, E.; Ermler, U.; Boll, M. Inhibitors of the molybdenum cofactor containing 4-hydroxybenzoyl-CoA reductase. Biochemistry 2008, 47, 4964–4972. [Google Scholar] [CrossRef]
- Reichenbecher, W.; Schink, B. Towards the reaction mechanism of pyrogallol-phloroglucinol transhydroxylase of Pelobacter acidigallici Biochim. Biophys. Acta 1999, 1430, 245–253. [Google Scholar]
- Benjamin, N.; O’Driscoll, F.; Dougall, H.; Duncan, C.; Smith, L.; Golden, M. Stomach NO synthesis. Nature 1994, 368, 502. [Google Scholar] [CrossRef] [PubMed]
- McKnight, G.M.; Smith, L.M.; Drummond, R.S.; Duncan, C.W.; Benjamin, N. Chemical synthesis of nitric oxide in the stomach from dietary nitrate in humans. Gut 1997, 40, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Zweier, J.L.; Samouilov, A.; Kuppusamy, P. Non-enzymatic nitric oxide synthesis in biological systems. Biochim. Biophys. Acta 1999, 1411, 250–262. [Google Scholar] [CrossRef]
- Gago, B.; Lundberg, J.O.; Barbosa, R.M.; Laranjinha, J. Red wine-dependent reduction of nitrite to nitric oxide in the stomach. Free Radical Biol. Med. 2007, 43, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Rocha, B.S.; Gago, B.; Barbosa, R.M.; Laranjinha, J. Dietary polyphenols generate nitric oxide from nitrite in the stomach and induce smooth muscle relaxation. Toxicology 2009, 265, 41–48. [Google Scholar] [CrossRef]
- Pereira, C.; Ferreira, N.R.; Rocha, B.S.; Barbosa, R.M.; Laranjinha, J. The redox interplay between nitrite and nitric oxide: From the gut to the brain. Redox Biol. 2013, 9, 276–284. [Google Scholar] [CrossRef]
- Rocha, B.S.; Nunes, C.; Pereira, C.; Barbosa, R.M.; Laranjinha, J. A shortcut to wide-ranging biological actions of dietary polyphenols: Modulation of the nitrate-nitrite-nitric oxide pathway in the gut. Food Funct. 2014, 5, 1646–1652. [Google Scholar] [CrossRef]
- Rocha, B.S.; Lundberg, J.O.; Radi, R.; Laranjinha, J. Role of nitrite, urate and pepsin in the gastroprotective effects of saliva. Redox Biol. 2016, 8, 407–414. [Google Scholar] [CrossRef]
- Hendgen-Cotta, U.B.; Merx, M.W.; Shiva, S.; Schmitz, J.; Becher, S.; Klare, J.P.; Steinhoff, H.; Goedecke, A.; Schrader, J.; Gladwin, M.T.; et al. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA 2008, 105, 10256–10261. [Google Scholar] [CrossRef]
- Shiva, S.; Huang, Z.; Grubina, R.; Sun, J.; Ringwood, L.A.; MacArthur, P.H.; Xu, X.; Murphy, E.; Darley-Usmar, V.M.; Gladwin, M.T. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ. Res. 2007, 100, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Rassaf, T.; Flögel, U.; Drexhage, C.; Hendgen-Cotta, U.; Kelm, M.; Schrader, J. Nitrite reductase function of deoxymyoglobin: Oxygen sensor and regulator of cardiac energetics and function. Circ. Res. 2007, 100, 1749–1754. [Google Scholar] [CrossRef] [PubMed]
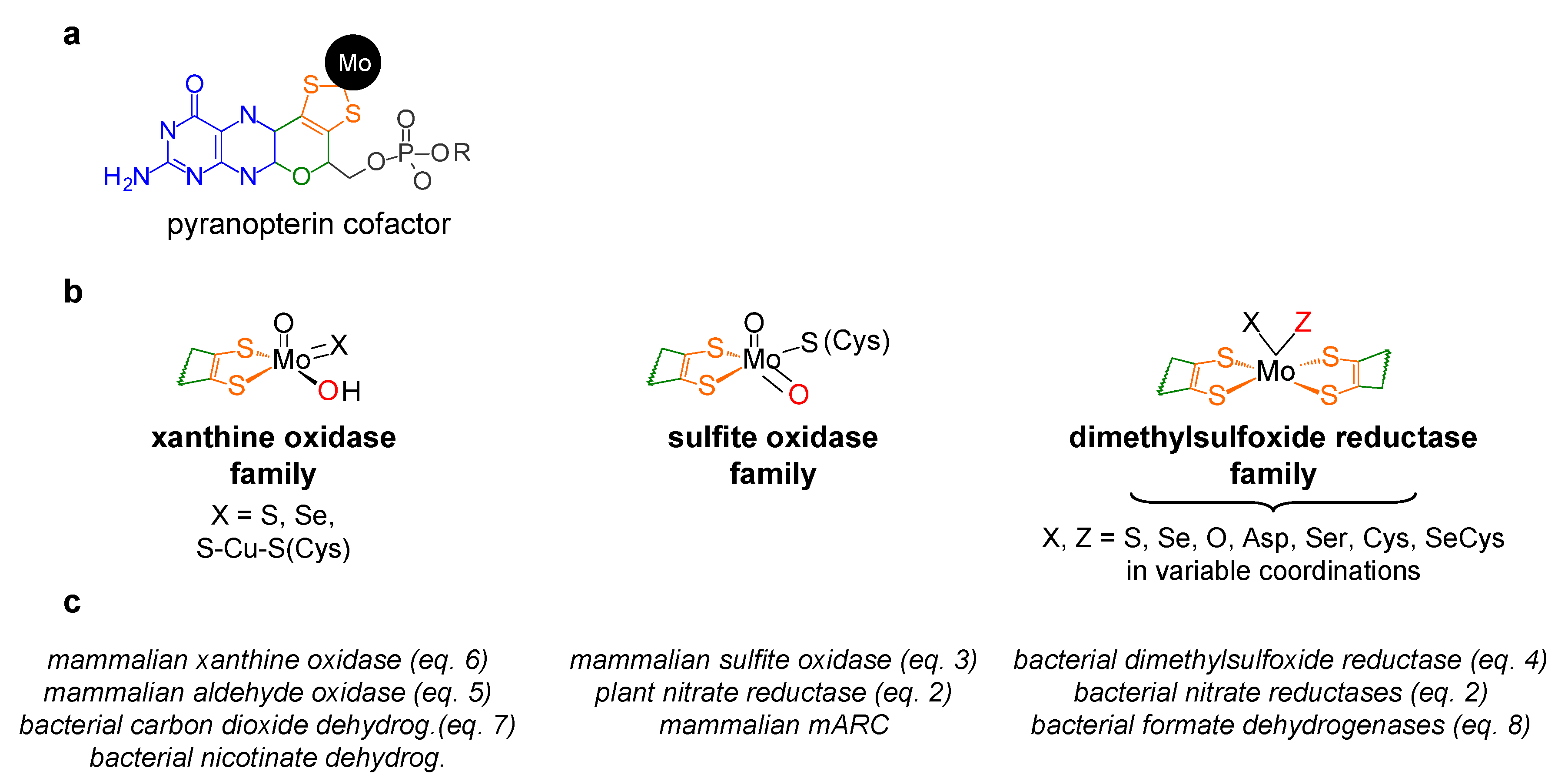

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maia, L.B. Bringing Nitric Oxide to the Molybdenum World—A Personal Perspective. Molecules 2023, 28, 5819. https://doi.org/10.3390/molecules28155819
Maia LB. Bringing Nitric Oxide to the Molybdenum World—A Personal Perspective. Molecules. 2023; 28(15):5819. https://doi.org/10.3390/molecules28155819
Chicago/Turabian StyleMaia, Luisa B. 2023. "Bringing Nitric Oxide to the Molybdenum World—A Personal Perspective" Molecules 28, no. 15: 5819. https://doi.org/10.3390/molecules28155819
APA StyleMaia, L. B. (2023). Bringing Nitric Oxide to the Molybdenum World—A Personal Perspective. Molecules, 28(15), 5819. https://doi.org/10.3390/molecules28155819






