A First-Class Degrader Candidate Targeting Both KRAS G12D and G12V Mediated by CANDDY Technology Independent of Ubiquitination
Abstract
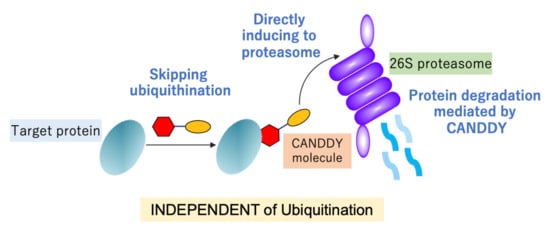
1. Introduction
2. Results
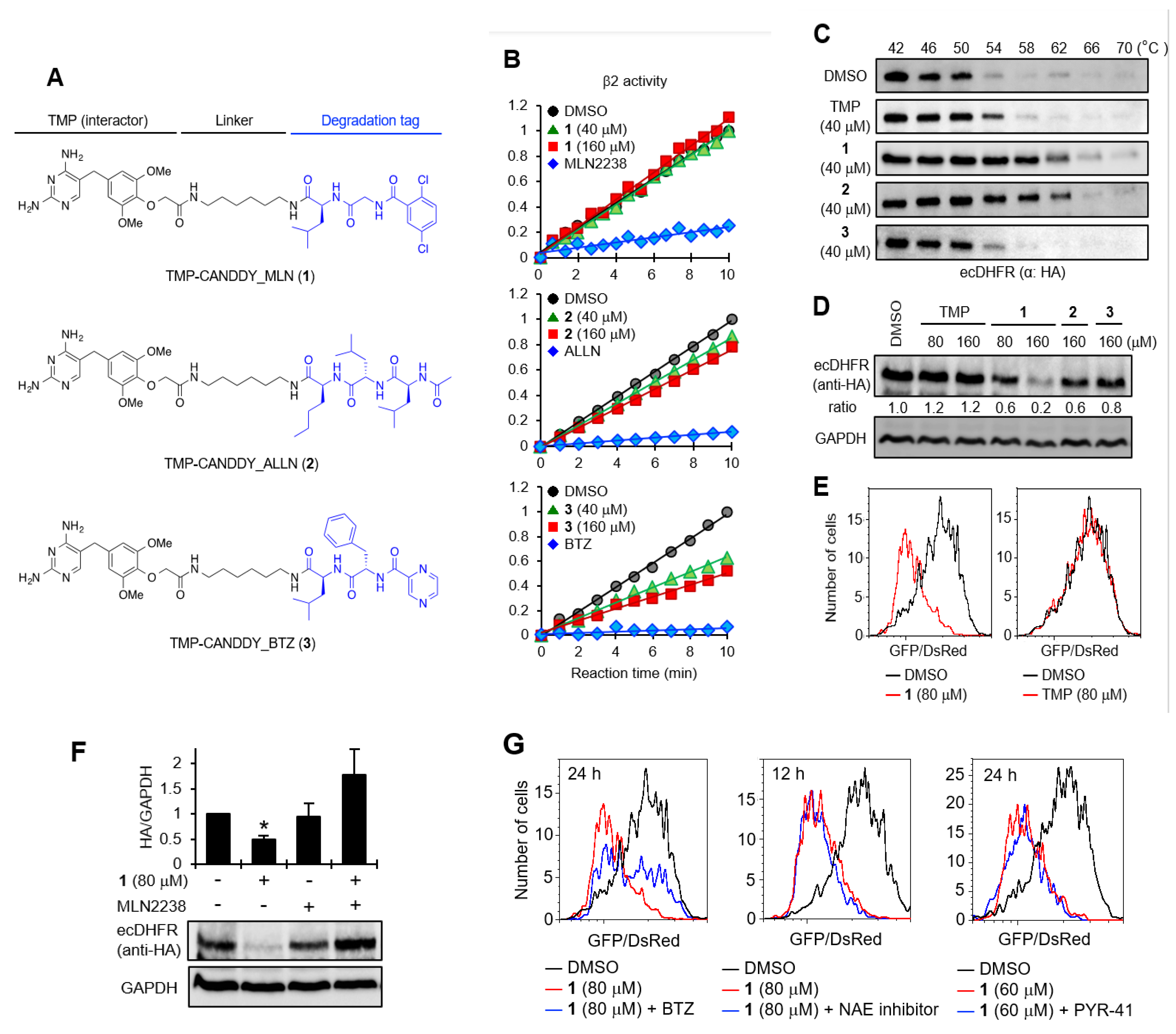
2.1. Proof of Concept for the CANDDY Strategy Mediated by Degradation Tags Derived from Proteasome Inhibitors
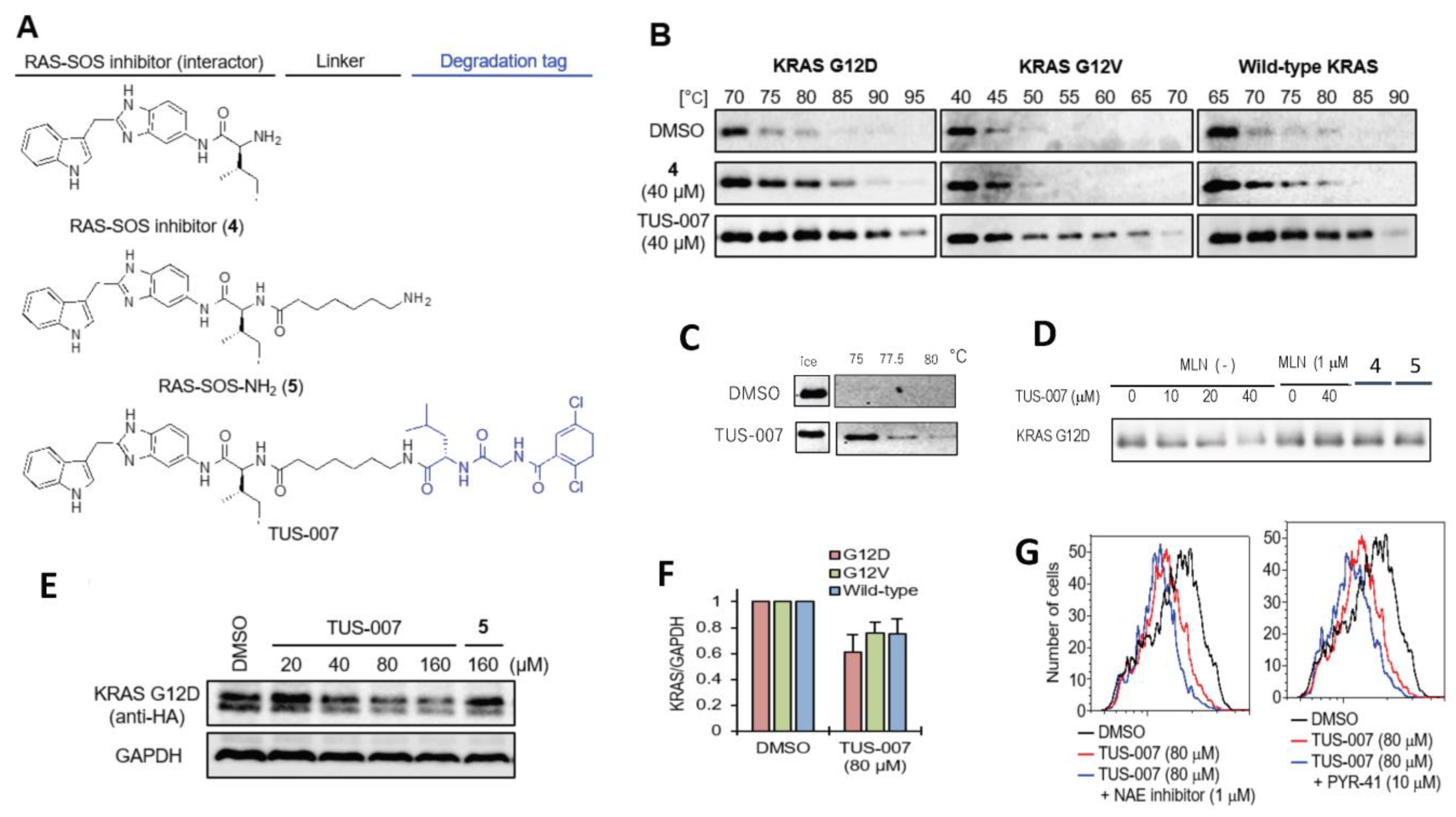
2.2. Chemical Knockdown of KRAS Mutants In Vitro by TUS-007
2.3. Apoptosis Induction and Tumor Suppression in Cetuximab-Resistant Human Colon Carcinoma Cells by TUS-007
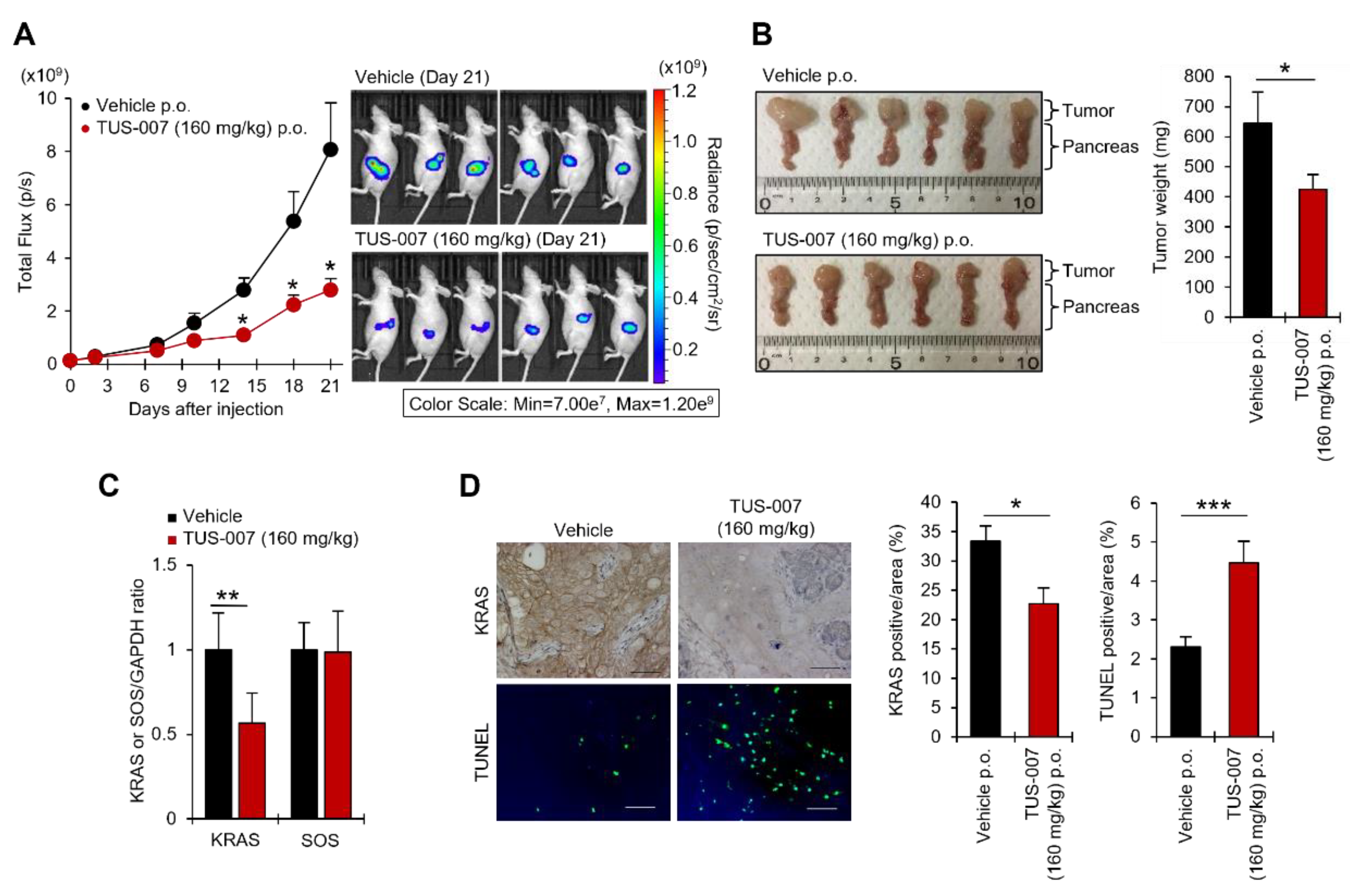
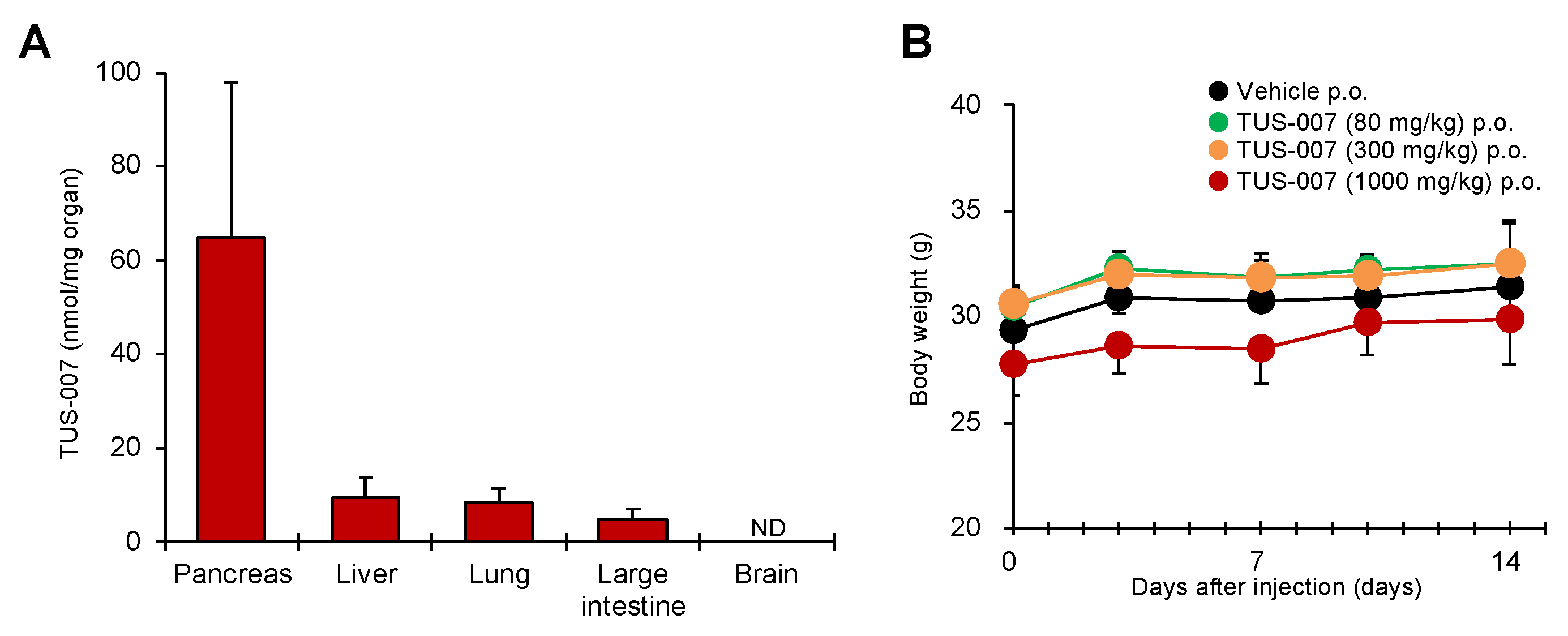
2.4. Apoptosis Induction and Tumor Suppression in Human Pancreatic Carcinoma Cells by TUS-007
3. Discussion
4. Materials and Methods
4.1. Synthesis of CANDDY Molecules
4.2. Evaluation of Affinity of TUS-007 for KRAS and Proteasome in a Thermal Shift Assay
4.3. Evaluation of KRAS Degradation Induced by TUS-007 in a Cell-Free System
4.4. Cell Proliferation Assay by WST-8
4.5. KRAS Degradation Assays of TUS-007 by Western Blotting
4.6. Analysis of Cell Cycle Distribution
4.7. KRAS Degradation Assays by Enzyme Linked Immunosorbent Assay (ELISA)
4.8. Analysis of TUS-007-Induced Apoptosis by Western Blotting
4.9. Analysis of TUS-007-Induced Apoptotic Cells by Flow Cytometry
4.10. Antitumor Evaluation of TUS-007 in a Subcutaneous Xenograft Model
4.11. Antitumor Evaluation of TUS-007 in an Orthotopic Xenograft Model
4.12. KRAS Degradation Analysis of TUS-007 in Xenograft Tumors
4.13. RAS Family Degradation Assay of TUS-007 in Normal Tissue of Mice
4.14. TUNEL Assay of Xenograft Tumors Treated with TUS-007
4.15. Pharmacokinetic Analysis
4.16. Tissue Distribution of TUS-007
4.17. In Vivo Toxicity Test of TUS-007
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Fujimori, S.; Hirai, N.; Ohashi, H.; Masuoka, K.; Nishikimi, A.; Fukui, Y.; Washio, T.; Oshikubo, T.; Yamashita, T.; Miyamoto-Sato, E. Next-generation sequencing coupled with a cell-free display technology for high-throughput production of reliable interactome data. Sci. Rep. 2012, 2, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.E.; Bayly, A.R.; Abell, C.; Skidmore, J. Small molecules, big targets: Drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discov. 2016, 15, 533–550. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Rhett, J.M.; O’Bryan, J.P. Therapeutic targeting of RAS: New Hope for drugging the “undruggable”. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118570–118586. [Google Scholar] [CrossRef] [PubMed]
- Stephen, A.G.; Esposito, D.; Bagni, R.K.; McCormick, F. Dragging Ras back in the ring. Cancer Cell 2014, 25, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Burke, J.P.; Phan, J.; Burns, M.C.; Olejniczak, E.T.; Waterson, A.G.; Lee, T.; Rossanese, O.W.; Fesik, S.W. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. 2012, 51, 6140–6143. [Google Scholar] [CrossRef]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef]
- Haigis, K.M. KRAS alleles: The devil is in the detail. Trends Cancer 2017, 3, 686–697. [Google Scholar] [CrossRef]
- Sheridan, C. Grail of RAS cancer drugs within reach. Nat. Biotechnol. 2020, 38, 6–8. [Google Scholar] [CrossRef]
- Shin, S.M.; Choi, D.-K.; Jung, W.K.; Bae, J.; Kim, J.S.; Park, S.-W.; Song, K.H.; Kim, Y.S. Antibody targeting intracellular oncogenic Ras mutants exerts anti-tumour effects after systemic administration. Nat. Commun. 2017, 8, 15090–15104. [Google Scholar] [CrossRef]
- Yin, W.; Rogge, M. Targeting RNA: A transformative therapeutic strategy. Clin. Transl. Sci. 2019, 12, 98–112. [Google Scholar] [CrossRef]
- Ross, S.J.; Revenko, A.S.; Hanson, L.L.; Ellston, R.; Staniszewska, A.; Whalley, N.; Pandey, S.K.; Revill, M.; Rooney, C.; Buckett, L.K.; et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017, 9, eaal5253–eaal5266. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature 2020, 580, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Scudellari, M. Protein-slaying drugs could be the next blockbuster therapies. Nature 2019, 567, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Cromm, P.M.; Crews, C.M. Targeted protein degradation: From chemical biology to drug discovery. Cell Chem. Biol. 2017, 24, 1181–1190. [Google Scholar] [CrossRef]
- Pettersson, M.; Crews, C.M. PROteolysis TArgeting Chimeras–(PROTACs)—Past, present and future. Drug Discov. Today Technol. 2019, 31, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Churcher, I. Protac-induced protein degradation in drug discovery: Breaking the rules or just making new ones? J. Med. Chem. 2018, 61, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Dohlman, H.G.; Campbell, S.L. Regulation of large and small G proteins by ubiquitination. J. Biol. Chem. 2019, 294, 18613–18623. [Google Scholar] [CrossRef]
- Bond, M.J.; Chu, L.; Nalawansha, D.A.; Li, K.; Crews, C.M. Targeted degradation of oncogenic KRASG12C by VHL-recruiting PROTACs. ACS Cent. Sci. 2020, 6, 1367–1375. [Google Scholar] [CrossRef]
- Miyamoto, E.; Ozawa, M. Protein Degradation Inducing Tag and Usage Thereof. U.S. Patent No. 10,976,306 B2, 13 April 2021. [Google Scholar]
- Miyamoto, E. Ras Protein Degradation Inducing Molecule and Pharmaceutical Composition. U.S. Patent No. 11052154B2, 6 July 2021. [Google Scholar]
- Shi, Y.; Long, M.J.C.; Rosenberg, M.M.; Li, S.; Kobiack, A.; Lessans, P.; Coffey, R.T.; Hedstrom, L. Boc3Arg-Linked Ligands Induce Degradation by Localizing Target Proteins to the 20S Proteasome. ACS Chem. Biol. 2016, 11, 3328–3337. [Google Scholar] [CrossRef]
- Long, M.J.C.; Gollapalli, D.R.; Hedstrom, L. Inhibitor mediated protein degradation. Chem. Biol. 2012, 19, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Berkers, C.R.; Ploegh, H.L.; Ovaa, H. Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure 2006, 14, 451–456. [Google Scholar] [CrossRef]
- Kisselev, A.F.; van der Linden, W.A.; Overkleeft, H.S. Proteasome inhibitors: An expanding army attacking a unique target. Chem. Biol. 2012, 19, 99–115. [Google Scholar] [CrossRef]
- Baccanari, D.P.; Daluge, S.; King, R.W. Inhibition of dihydrofolate reductase: Effect of reduced nicotinamide adenine dinucleotide phosphate on the selectivity and affinity of diaminobenzylpyrimidines. Biochemistry 1982, 21, 5068–5075. [Google Scholar] [CrossRef] [PubMed]
- Molina, D.M.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef]
- Lu, J.; Qian, Y.; Altieri, M.; Dong, H.; Wang, J.; Raina, K.; Hines, J.; Winkler, J.D.; Crew, A.P.; Coleman, K.; et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 2015, 22, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef]
- Raina, K.; Lu, J.; Qian, Y.; Altieri, M.; Gordon, D.; Ross, A.M.K.; Wang, J.; Chen, X.; Dong, H.; Siu, K.; et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 7124–7129. [Google Scholar] [CrossRef]
- Welsch, M.E.; Kaplan, A.; Chambers, J.M.; Stokes, M.E.; Bos, P.H.; Zask, A.; Zhang, Y.; Sanchez-Martin, M.; Badgley, M.A.; Huang, C.S.; et al. Multivalent small-molecule pan-RAS inhibitors. Cell 2017, 168, 878–889. [Google Scholar] [CrossRef]
- Pant, S.; Hubbard, J.; Martinelli, E.; Bekaii-Saab, T. Clinical update on K-Ras targeted therapy in gastrointestinal cancers. Crit. Rev. Oncol. Hematol. 2018, 130, 78–91. [Google Scholar] [CrossRef]
- Bashore, C.; Prakash, S.; Johnson, M.C.; Conrad, R.J.; Kekessie, I.A.; Scales, S.J.; Ishisoko, N.; Kleinheinz, T.; Liu, P.S.; Popovych, N.; et al. Targeted degradation via direct 26S proteasome recruitment. Nat. Chem. Bio. 2023, 19, 55–63. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyamoto-Sato, E.; Imanishi, S.; Huang, L.; Itakura, S.; Iwasaki, Y.; Ishizaka, M. A First-Class Degrader Candidate Targeting Both KRAS G12D and G12V Mediated by CANDDY Technology Independent of Ubiquitination. Molecules 2023, 28, 5600. https://doi.org/10.3390/molecules28145600
Miyamoto-Sato E, Imanishi S, Huang L, Itakura S, Iwasaki Y, Ishizaka M. A First-Class Degrader Candidate Targeting Both KRAS G12D and G12V Mediated by CANDDY Technology Independent of Ubiquitination. Molecules. 2023; 28(14):5600. https://doi.org/10.3390/molecules28145600
Chicago/Turabian StyleMiyamoto-Sato, Etsuko, Satoshi Imanishi, Lijuan Huang, Shoko Itakura, Yoichi Iwasaki, and Masamichi Ishizaka. 2023. "A First-Class Degrader Candidate Targeting Both KRAS G12D and G12V Mediated by CANDDY Technology Independent of Ubiquitination" Molecules 28, no. 14: 5600. https://doi.org/10.3390/molecules28145600
APA StyleMiyamoto-Sato, E., Imanishi, S., Huang, L., Itakura, S., Iwasaki, Y., & Ishizaka, M. (2023). A First-Class Degrader Candidate Targeting Both KRAS G12D and G12V Mediated by CANDDY Technology Independent of Ubiquitination. Molecules, 28(14), 5600. https://doi.org/10.3390/molecules28145600