Atractylenolide III Ameliorates Bile Duct Ligation-Induced Liver Fibrosis by Inhibiting the PI3K/AKT Pathway and Regulating Glutamine Metabolism
Abstract
1. Introduction
2. Results
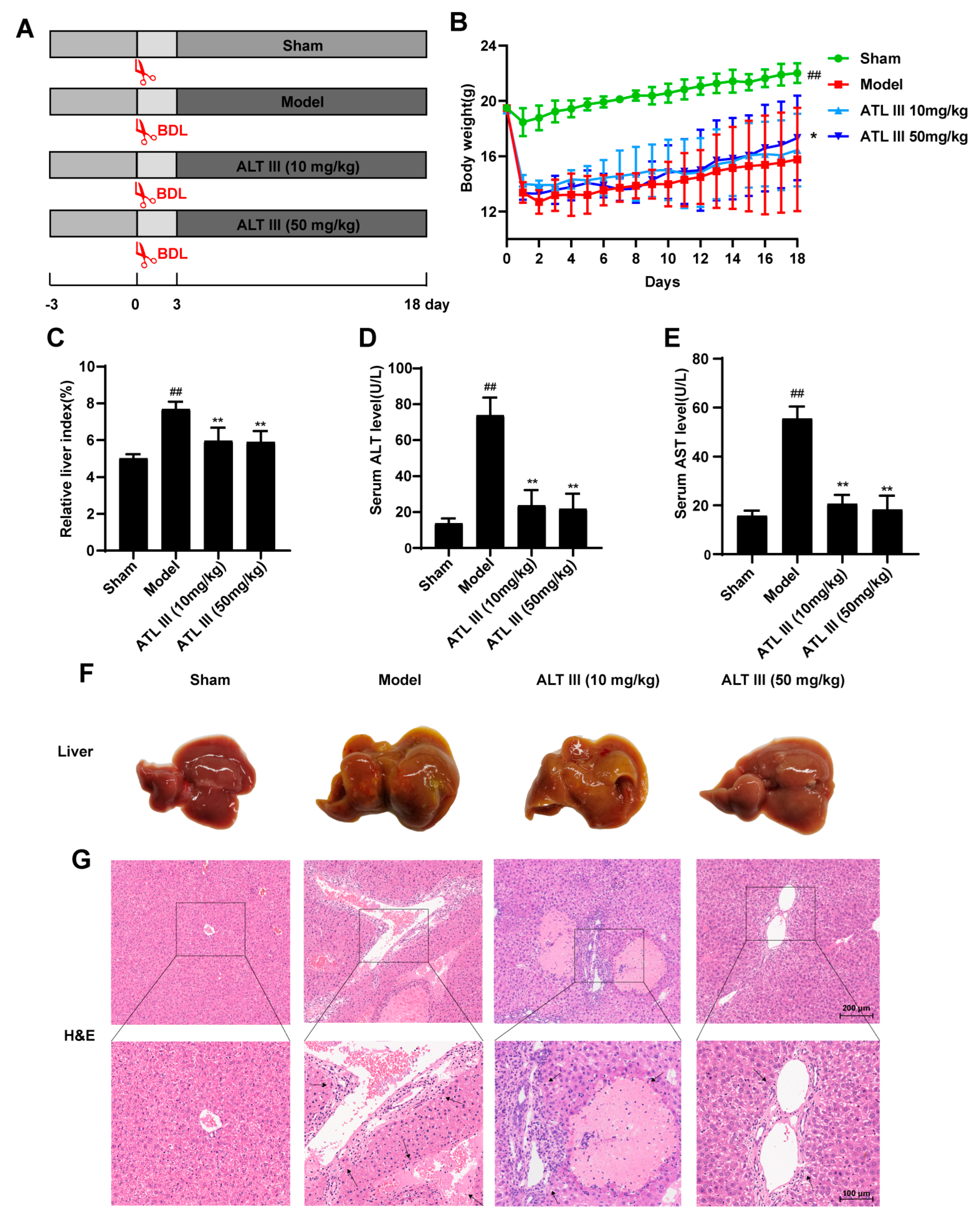
2.1. ATL III Inhibits BDL-Induced Liver Injury
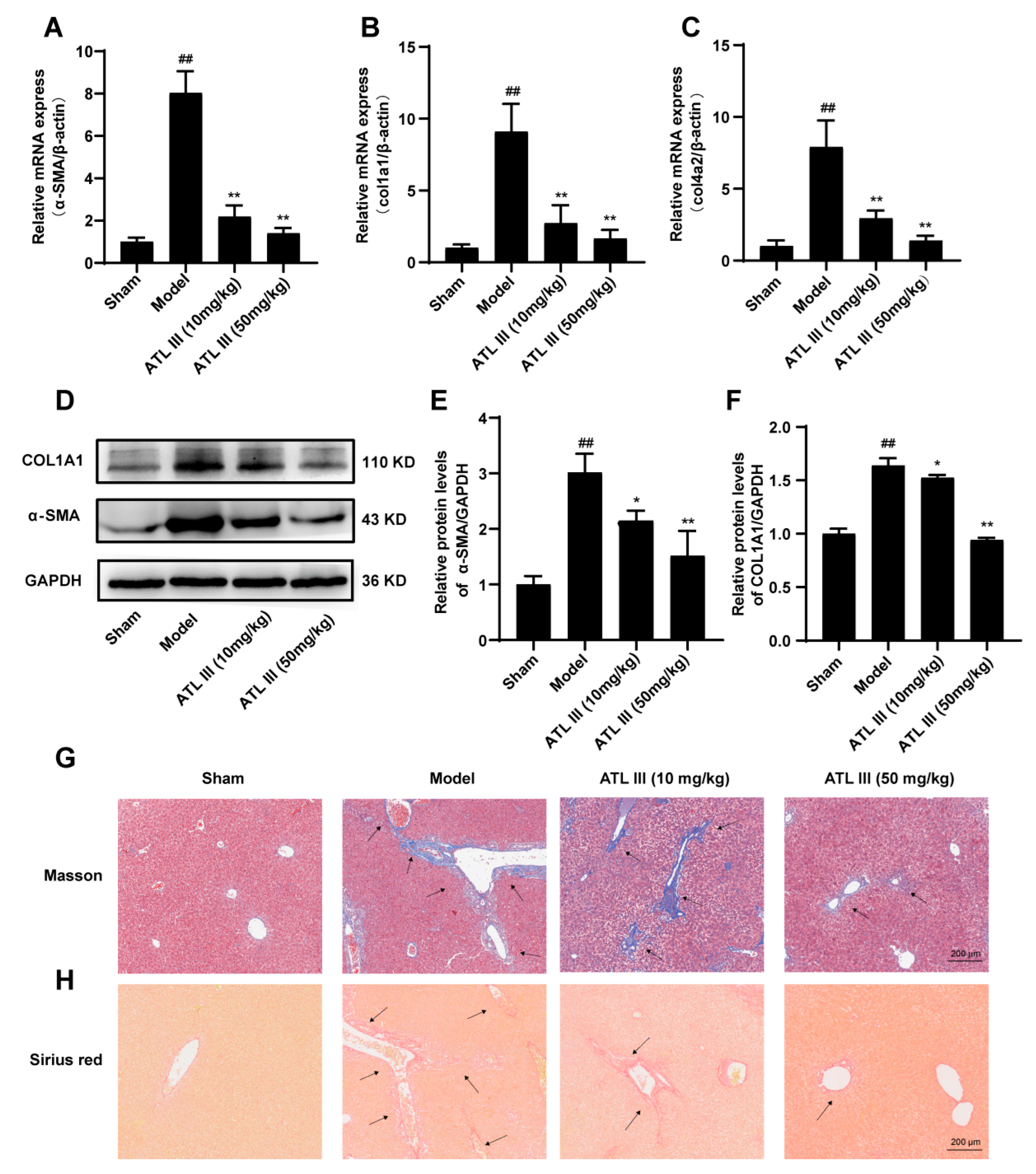
2.2. ATL III Ameliorates BDL-Induced Liver Fibrosis in Mice
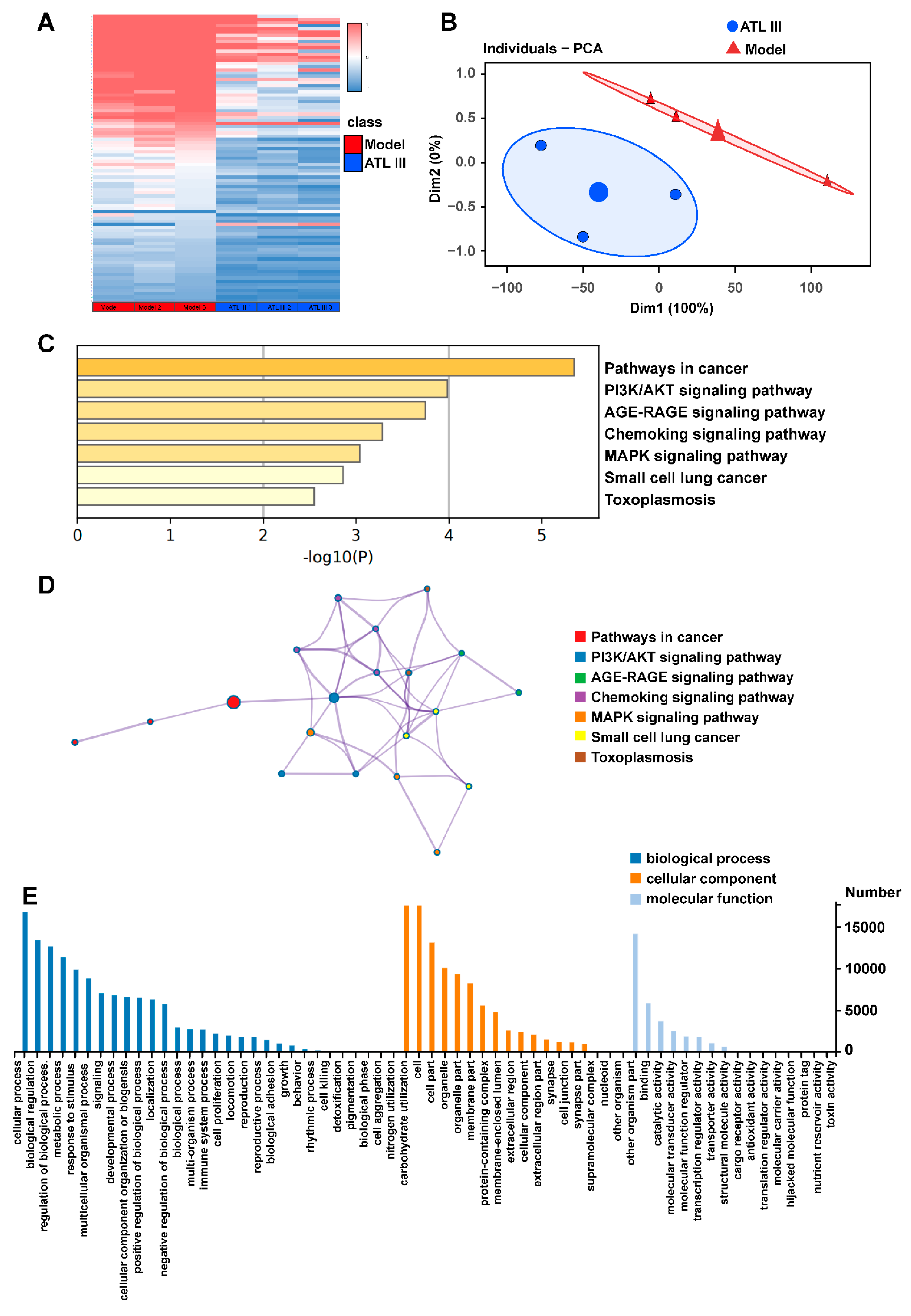
2.3. ATL III Regulates the Differentially Expressed Genes and Pathway Enrichment Analysis in the Liver of BDL Mice
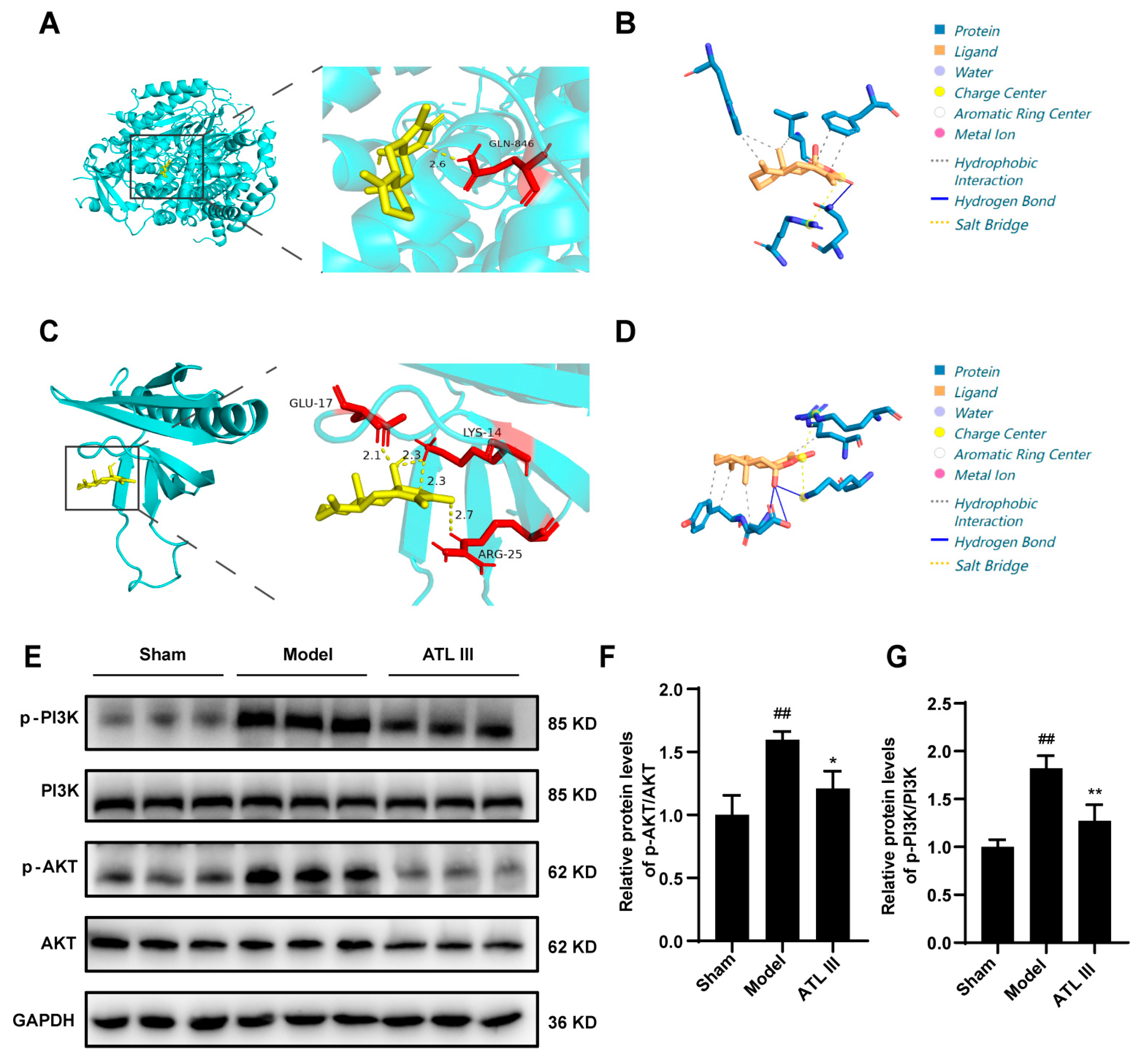
2.4. ATL III Alleviates Liver Fibrosis by Inhibiting the PI3K/AKT Pathway
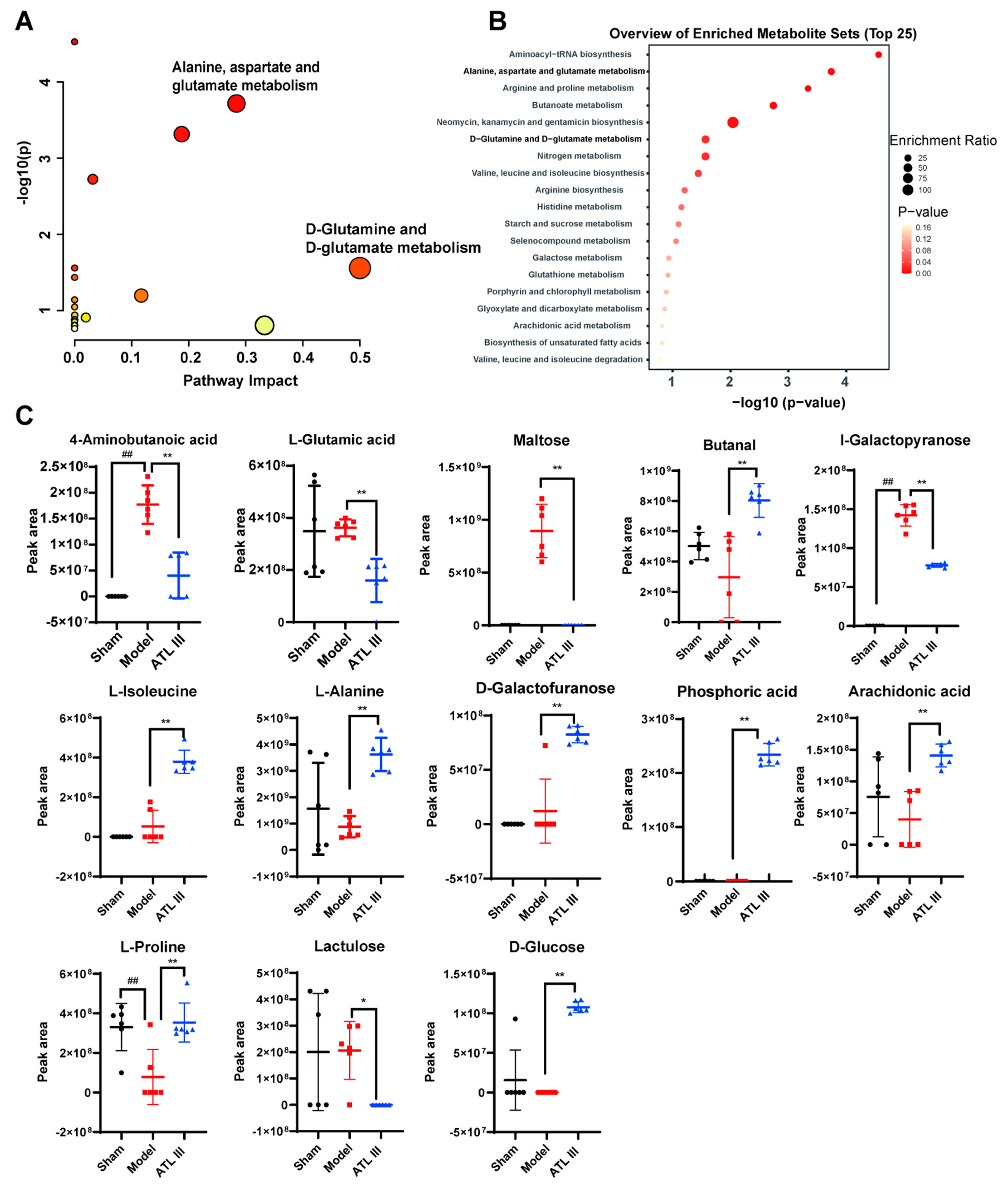
2.5. ATL III Attenuates BDL-Induced Metabolic Disorders and Regulates Glutamine Metabolism in Mice
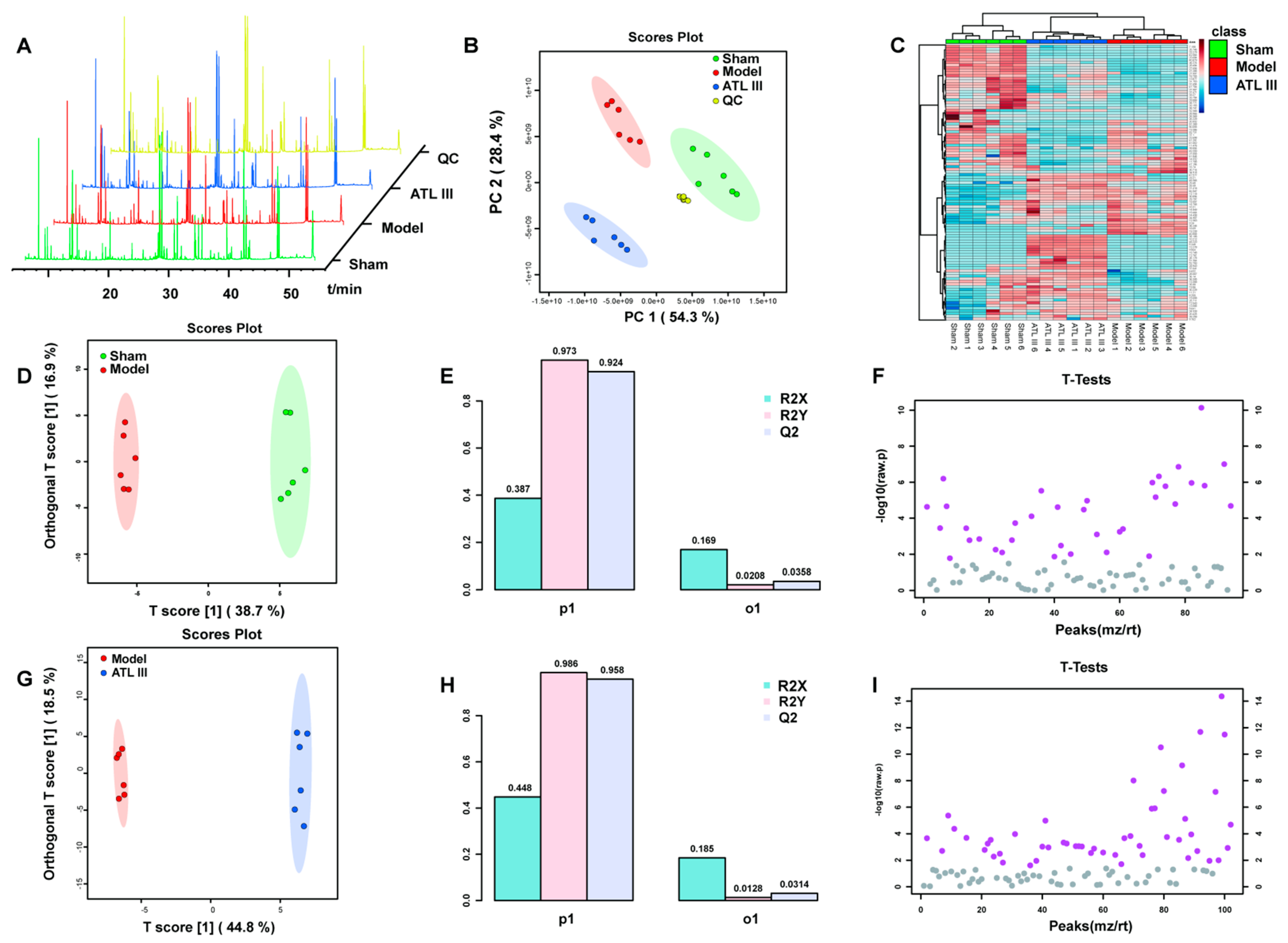
2.5.1. Multivariate Statistical Analysis
2.5.2. Multivariate Statistical Analysis
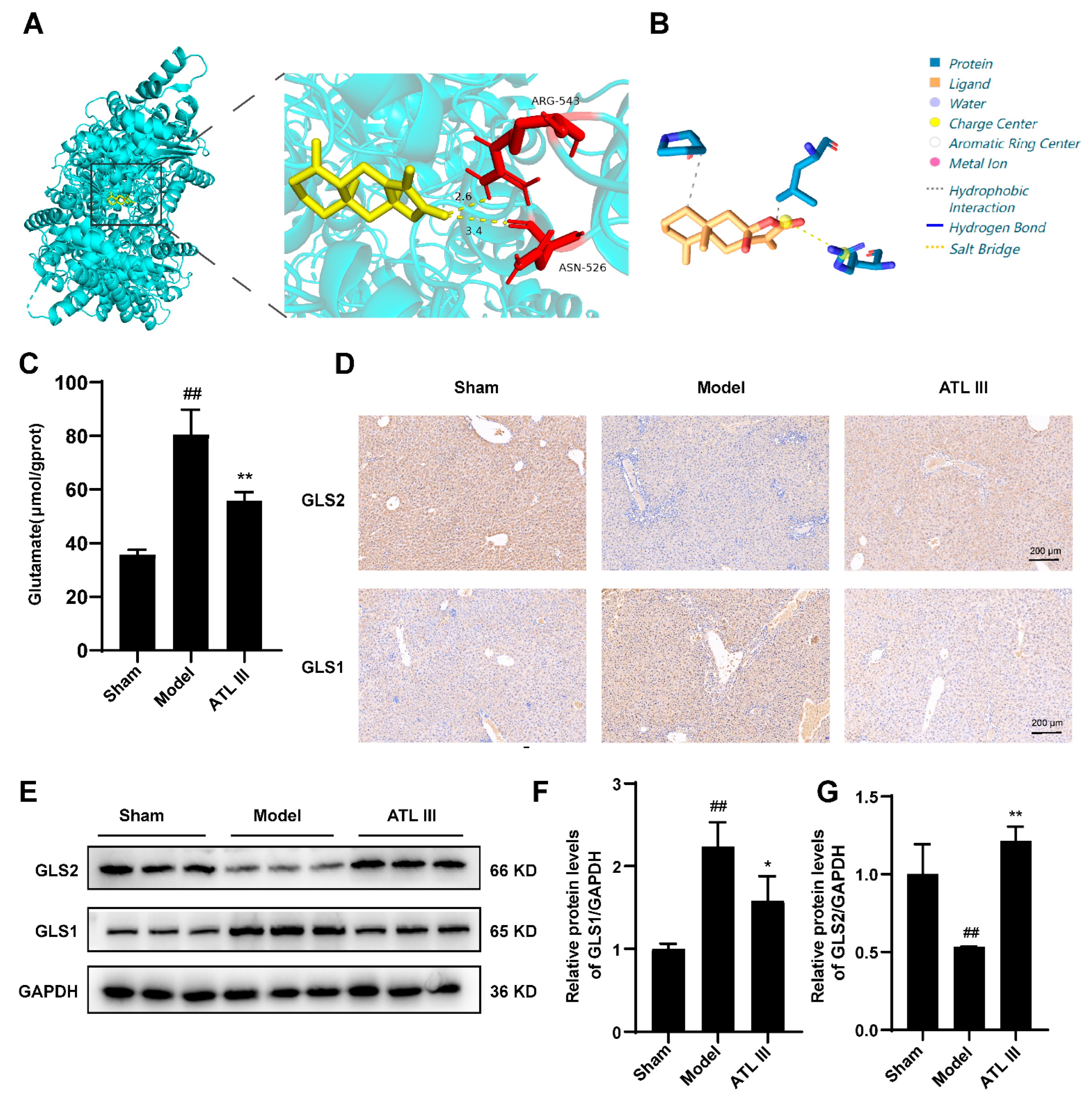
2.6. Inhibition of BDL-Induced Liver Fibrosis by ATL III through the Inhibition of Glutaminase
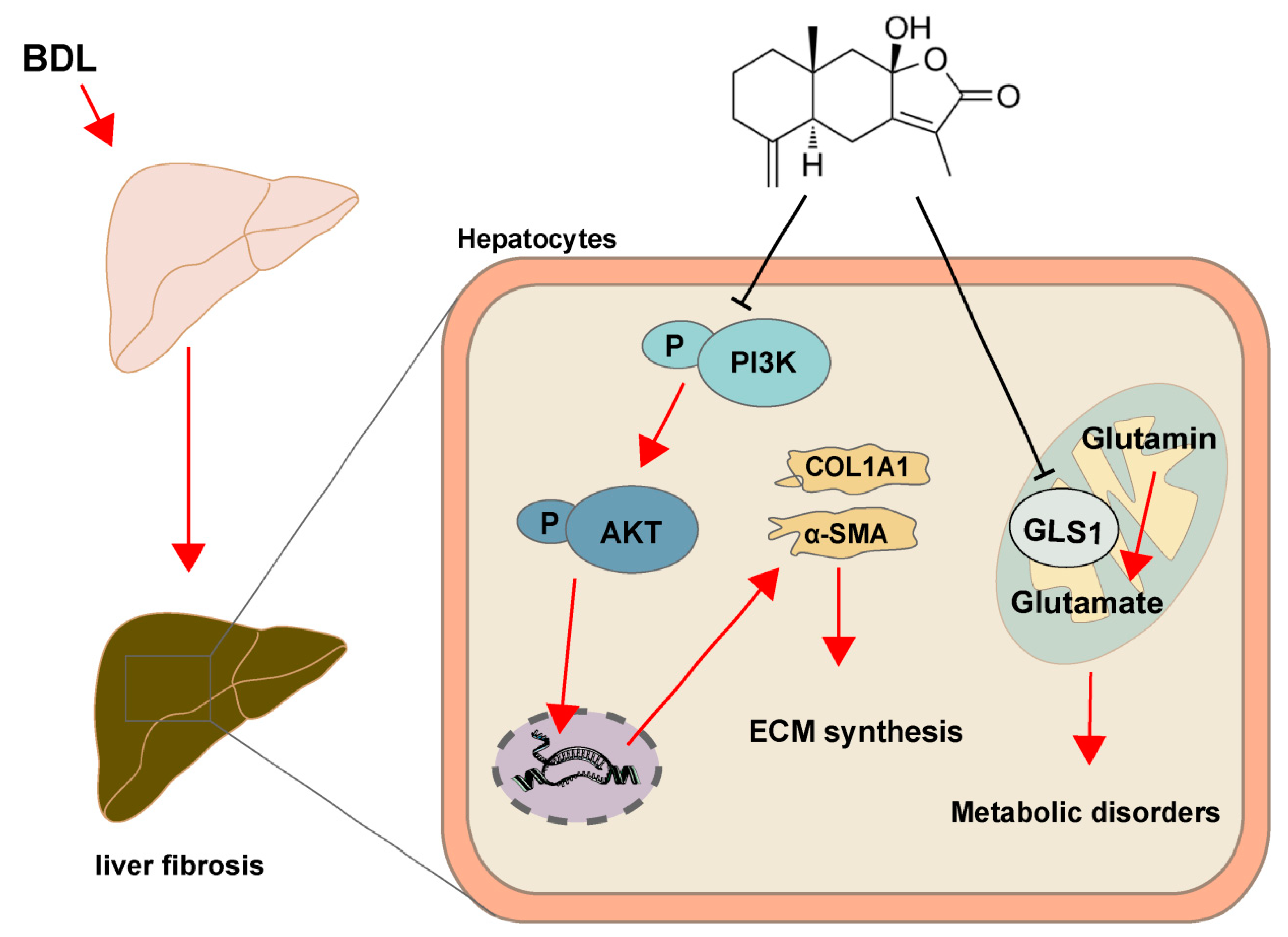
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animal Experiments
4.3. Liver Index
4.4. Histological and Immunohistological Studies
4.5. Biochemical Analyses
4.6. Western Blot
4.7. RT-qPCR
4.8. RNA Isolation and RNA-Sequencing (RNA-seq) Analysis
4.9. Metabolite Extraction and Gas Chromatography–Mass Spectrometry Analysis
4.10. Principal Component Analysis (PCA)
4.11. Partial Least Squares Discriminant Analysis (OPLS-DA)
4.12. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
4.13. Molecular Docking
4.14. Protein–Ligand Interaction Profiler (PLIP)
4.15. Cell Culture
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Cai, X.; Wang, J.; Wang, J.; Zhou, Q.; Yang, B.; He, Q.; Weng, Q. Intercellular crosstalk of hepatic stellate cells in liver fibrosis: New insights into therapy. Pharmacol. Res. 2020, 155, 104720. [Google Scholar] [PubMed]
- Friedman, S.L. Liver fibrosis—From bench to bedside. J. Hepatol. 2003, 38 (Suppl. 1), S38–S53. [Google Scholar] [PubMed]
- Elpek, G. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J. Gastroenterol. 2014, 20, 7260–7276. [Google Scholar] [CrossRef]
- Perakakis, N.; Stefanakis, K.; Mantzoros, C.S. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metab. Clin. Exp. 2020, 111, 154320. [Google Scholar]
- Govaere, O.; Cockell, S.; Tiniakos, D.; Queen, R.; Younes, R.; Vacca, M.; Alexander, L.; Ravaioli, F.; Palmer, J.; Petta, S.; et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 2020, 12, eaba4448. [Google Scholar] [PubMed]
- Zhou, Y.; Wu, R.; Cai, F.F.; Zhou, W.J.; Lu, Y.Y.; Zhang, H.; Chen, Q.L.; Sun, M.Y.; Su, S.B. Development of a novel anti-liver fibrosis formula with luteolin, licochalcone A, aloe-emodin and acacetin by network pharmacology and transcriptomics analysis. Pharm. Biol. 2021, 59, 1594–1606. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Baglieri, J.; Kisseleva, T.; Brenner, D.A. Mechanisms of liver fibrosis and its role in liver cancer. Exp. Biol. Med. 2020, 245, 96–108. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–8. [Google Scholar]
- Park, S.Y.; Le, C.T.; Sung, K.Y.; Choi, D.H.; Cho, E.H. Succinate induces hepatic fibrogenesis by promoting activation, proliferation, and migration, and inhibiting apoptosis of hepatic stellate cells. Biochem. Biophys. Res. Commun. 2018, 496, 673–678. [Google Scholar] [CrossRef]
- Song, M.Y.; Jung, H.W.; Kang, S.Y.; Park, Y.K. Atractylenolide III Enhances Energy Metabolism by Increasing the SIRT-1 and PGC1α Expression with AMPK Phosphorylation in C2C12 Mouse Skeletal Muscle Cells. Biol. Pharm. Bull. 2017, 40, 339–344. [Google Scholar]
- Shi, K.; Wang, Y.; Xiao, Y.; Tu, J.; Zhou, Z.; Cao, G.; Liu, Y. Therapeutic effects and mechanism of Atractylodis rhizoma in acute lung injury: Investigation based on an Integrated approach. Front. Pharmacol. 2023, 14, 1181951. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.H.; Han, N.R.; Kim, H.M.; Jeong, H.J. Blockade of IL-6 secretion pathway by the sesquiterpenoid atractylenolide III. J. Nat. Prod. 2011, 74, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ji, Z.H.; Liu, C.; Yu, X.Y. Neuroprotection and mechanisms of atractylenolide III in preventing learning and memory impairment induced by chronic high-dose homocysteine administration in rats. Neuroscience 2015, 290, 485–491. [Google Scholar] [CrossRef]
- Li, Q.; Tan, J.X.; He, Y.; Bai, F.; Li, S.W.; Hou, Y.W.; Ji, L.S.; Gao, Y.T.; Zhang, X.; Zhou, Z.H.; et al. Atractylenolide III ameliorates Non-Alcoholic Fatty Liver Disease by activating Hepatic Adiponectin Receptor 1-Mediated AMPK Pathway. Int. J. Biol. Sci. 2022, 18, 1594–1611. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, S.; Wu, F.; Zheng, Q.; Zhang, F.; Luo, Y.; Jian, X. Atractylenolide III reduces depressive- and anxiogenic-like behaviors in rat depression models. Neurosci. Lett. 2021, 759, 136050. [Google Scholar] [CrossRef]
- Huai, B.; Ding, J. Atractylenolide III attenuates bleomycin-induced experimental pulmonary fibrosis and oxidative stress in rat model via Nrf2/NQO1/HO-1 pathway activation. Immunopharmacol. Immunotoxicol. 2020, 42, 436–444. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, Z.; Yu, J.; Yin, L.; Zhu, A. Atractylenolide III alleviates isoflurane-induced injury in rat hippocampal neurons by activating the PI3K/Akt/mTOR pathway. J. Food Biochem. 2021, 45, e13892. [Google Scholar] [PubMed]
- Wang, F.; Li, Z.; Chen, L.; Yang, T.; Liang, B.; Zhang, Z.; Shao, J.; Xu, X.; Yin, G.; Wang, S.; et al. Inhibition of ASCT2 induces hepatic stellate cell senescence with modified proinflammatory secretome through an IL-1α/NF-κB feedback pathway to inhibit liver fibrosis. Acta Pharm. Sinica. B 2022, 12, 3618–3638. [Google Scholar]
- Lin, X.; Bai, F.; Nie, J.; Lu, S.; Lu, C.; Zhu, X.; Wei, J.; Lu, Z.; Huang, Q. Didymin Alleviates Hepatic Fibrosis Through Inhibiting ERK and PI3K/Akt Pathways via Regulation of Raf Kinase Inhibitor Protein. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016, 40, 1422–1432. [Google Scholar] [CrossRef]
- Peng, R.; Wang, S.; Wang, R.; Wang, Y.; Wu, Y.; Yuan, Y. Antifibrotic effects of tanshinol in experimental hepatic fibrosis by targeting PI3K/AKT/mTOR/p70S6K1 signaling pathways. Discov. Med. 2017, 23, 81–94. [Google Scholar]
- Wang, Q.; Wen, R.; Lin, Q.; Wang, N.; Lu, P.; Zhu, X. Wogonoside Shows Antifibrotic Effects in an Experimental Regression Model of Hepatic Fibrosis. Dig. Dis. Sci. 2015, 60, 3329–3339. [Google Scholar]
- Xu, W.; Yang, Z.; Lu, N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adhes. Migr. 2015, 9, 317–324. [Google Scholar] [CrossRef]
- Wei, L.; Chen, Q.; Guo, A.; Fan, J.; Wang, R.; Zhang, H. Asiatic acid attenuates CCl(4)-induced liver fibrosis in rats by regulating the PI3K/AKT/mTOR and Bcl-2/Bax signaling pathways. Int. Immunopharmacol. 2018, 60, 1–8. [Google Scholar] [PubMed]
- Reif, S.; Lang, A.; Lindquist, J.N.; Yata, Y.; Gabele, E.; Scanga, A.; Brenner, D.A.; Rippe, R.A. The role of focal adhesion kinase-phosphatidylinositol 3-kinase-akt signaling in hepatic stellate cell proliferation and type I collagen expression. J. Biol. Chem. 2003, 278, 8083–8090. [Google Scholar] [CrossRef] [PubMed]
- Roehlen, N.; Crouchet, E.; Baumert, T.F. Liver Fibrosis: Mechanistic Concepts and Therapeutic Perspectives. Cells 2020, 9, 875. [Google Scholar] [CrossRef] [PubMed]
- Connolly, M.K.; Bedrosian, A.S.; Mallen-St Clair, J.; Mitchell, A.P.; Ibrahim, J.; Stroud, A.; Pachter, H.L.; Bar-Sagi, D.; Frey, A.B.; Miller, G. In liver fibrosis, dendritic cells govern hepatic inflammation in mice via TNF-alpha. J. Clin. Investig. 2009, 119, 3213–3225. [Google Scholar]
- Ghallab, A.; Hofmann, U.; Sezgin, S.; Vartak, N.; Hassan, R.; Zaza, A.; Godoy, P.; Schneider, K.M.; Guenther, G.; Ahmed, Y.A.; et al. Bile Microinfarcts in Cholestasis Are Initiated by Rupture of the Apical Hepatocyte Membrane and Cause Shunting of Bile to Sinusoidal Blood. Hepatology 2019, 69, 666–683. [Google Scholar] [CrossRef]
- Abdel-Rahman, R.F.; Fayed, H.M.; Asaad, G.F.; Ogaly, H.A.; Hessin, A.F.; Salama, A.A.A.; Abd El-Rahman, S.S.; Arbid, M.S.; Mohamed, M.A.E. The involvement of TGF-β1 /FAK/α-SMA pathway in the antifibrotic impact of rice bran oil on thioacetamide-induced liver fibrosis in rats. PLoS ONE 2021, 16, e0260130. [Google Scholar] [CrossRef]
- Eisa, N.H.; Khodir, A.E.; El-Sherbiny, M.; Elsherbiny, N.M.; Said, E. Phenethyl isothiocyanate attenuates diabetic nephropathy via modulation of glycative/oxidative/inflammatory signaling in diabetic rats. Biomed. Pharmacother. 2021, 142, 111666. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.; Chen, L.; Li, J.; Zhang, H.; Guo, X. Glycine Suppresses AGE/RAGE Signaling Pathway and Subsequent Oxidative Stress by Restoring Glo1 Function in the Aorta of Diabetic Rats and in HUVECs. Oxidative Med. Cell. Longev. 2019, 2019, 4628962. [Google Scholar] [CrossRef]
- Wang, Q.; Feng, J.; Tang, L. Non-Coding RNA Related to MAPK Signaling Pathway in Liver Cancer. Int. J. Mol. Sci. 2022, 23, 11908. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Shan, L.; Cao, S.; Li, K.; Wu, Y.; Zhang, Q. Notoginsenoside R1, An Active Compound from Panax notoginseng, Inhibits Hepatic Stellate Cell Activation and Liver Fibrosis via MAPK Signaling Pathway. Am. J. Chin. Med. 2022, 50, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Pal, I.; Mandal, M. PI3K and Akt as molecular targets for cancer therapy: Current clinical outcomes. Acta Pharmacol. Sin. 2012, 33, 1441–1458. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Qian, D.W.; Jiang, S.; Shang, E.X.; Zhu, Z.H.; Duan, J.A. Scutellariae Radix and Coptidis Rhizoma Improve Glucose and Lipid Metabolism in T2DM Rats via Regulation of the Metabolic Profiling and MAPK/PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2018, 19, 3634. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, M.; Yin, X.; Guo, H.; Zhang, B.; Wang, Y.; Xiao, J.; Zou, X.; Zhang, M.; Zhuge, Y. TWEAK promotes hepatic stellate cell migration through activating EGFR/Src and PI3K/AKT pathways. Cell Biol. Int. 2020, 44, 278–285. [Google Scholar] [CrossRef]
- Du, K.; Hyun, J.; Premont, R.T.; Choi, S.S.; Michelotti, G.A.; Swiderska-Syn, M.; Dalton, G.D.; Thelen, E.; Rizi, B.S.; Jung, Y.; et al. Hedgehog-YAP Signaling Pathway Regulates Glutaminolysis to Control Activation of Hepatic Stellate Cells. Gastroenterology 2018, 154, 1465–1479.e13. [Google Scholar] [CrossRef]
- Li, J.; Ghazwani, M.; Liu, K.; Huang, Y.; Chang, N.; Fan, J.; He, F.; Li, L.; Bu, S.; Xie, W.; et al. Regulation of hepatic stellate cell proliferation and activation by glutamine metabolism. PLoS ONE 2017, 12, e0182679. [Google Scholar] [CrossRef]
- Du, K.; Chitneni, S.K.; Suzuki, A.; Wang, Y.; Henao, R.; Hyun, J.; Premont, R.T.; Naggie, S.; Moylan, C.A.; Bashir, M.R.; et al. Increased Glutaminolysis Marks Active Scarring in Nonalcoholic Steatohepatitis Progression. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 1–21. [Google Scholar] [CrossRef]
- Choi, W.M.; Kim, H.H.; Kim, M.H.; Cinar, R.; Yi, H.S.; Eun, H.S.; Kim, S.H.; Choi, Y.J.; Lee, Y.S.; Kim, S.Y.; et al. Glutamate Signaling in Hepatic Stellate Cells Drives Alcoholic Steatosis. Cell Metab. 2019, 30, 877–889.e7. [Google Scholar] [CrossRef]
- Mallat, A.; Lotersztajn, S. Glutamate Signaling in Alcohol-associated Fatty Liver: “Pas de Deux”. Hepatology 2020, 72, 350–352. [Google Scholar] [CrossRef]
- Yu, W.; Yang, X.; Zhang, Q.; Sun, L.; Yuan, S.; Xin, Y. Targeting GLS1 to cancer therapy through glutamine metabolism. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2021, 23, 2253–2268. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Liu, C.; Qin, M.; Zhang, X.; Xi, T.; Yuan, S.; Hao, H.; Xiong, J. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm. Sinica. B 2022, 12, 558–580. [Google Scholar] [CrossRef] [PubMed]
- Sufleţel, R.T.; Melincovici, C.S.; Gheban, B.A.; Toader, Z.; Mihu, C.M. Hepatic stellate cells—From past till present: Morphology, human markers, human cell lines, behavior in normal and liver pathology. Rom. J. Morphol. Embryol. = Rev. Roum. De Morphol. Et Embryol. 2020, 61, 615–642. [Google Scholar] [CrossRef]
- Dold, S.; Laschke, M.W.; Lavasani, S.; Menger, M.D.; Jeppsson, B.; Thorlacius, H. Simvastatin protects against cholestasis-induced liver injury. Br. J. Pharmacol. 2009, 156, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Chen, J.; Wu, J.; Wu, Q.; Jia, C.; Xu, Y.X.Z.; Chen, L.; Tu, W.; Yang, G.; Kong, J.; et al. Atractylenolide III ameliorates cerebral ischemic injury and neuroinflammation associated with inhibiting JAK2/STAT3/Drp1-dependent mitochondrial fission in microglia. Phytomedicine Int. J. Phytother. Phytopharm. 2019, 59, 152922. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, C.; Shi, K.; Sun, X.; Song, C.; Xu, K.; Liu, Y. Atractyloside-A ameliorates spleen deficiency diarrhea by interfering with TLR4/MyD88/NF-κB signaling activation and regulating intestinal flora homeostasis. Int. Immunopharmacol. 2022, 107, 108679. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Feng, F.; Wu, J.; Li, L.; Xu, H.; Yang, L.; Xu, K.; Wang, C. Diosmetin inhibits cell growth and proliferation by regulating the cell cycle and lipid metabolism pathway in hepatocellular carcinoma. Food Funct. 2021, 12, 12036–12046. [Google Scholar] [CrossRef] [PubMed]
- Linghang, Q.; Yiyi, X.; Guosheng, C.; Kang, X.; Jiyuan, T.; Xiong, L.; Guangzhong, W.; Shuiqing, L.; Yanju, L. Effects of Atractylodes Oil on Inflammatory Response and Serum Metabolites in Adjuvant Arthritis Rats. Biomed. Pharmacother. 2020, 127, 110130. [Google Scholar] [CrossRef]
- Feng, F.; Pan, L.; Wu, J.; Li, L.; Xu, H.; Yang, L.; Xu, K.; Wang, C. Cepharanthine inhibits hepatocellular carcinoma cell growth and proliferation by regulating amino acid metabolism and suppresses tumorigenesis in vivo. Int. J. Biol. Sci. 2021, 17, 4340–4352. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Shi, K.; Tu, J.; Ke, C.; Chen, N.; Wang, B.; Liu, Y.; Zhou, Z. Atractylenolide III Ameliorates Bile Duct Ligation-Induced Liver Fibrosis by Inhibiting the PI3K/AKT Pathway and Regulating Glutamine Metabolism. Molecules 2023, 28, 5504. https://doi.org/10.3390/molecules28145504
Wang Y, Shi K, Tu J, Ke C, Chen N, Wang B, Liu Y, Zhou Z. Atractylenolide III Ameliorates Bile Duct Ligation-Induced Liver Fibrosis by Inhibiting the PI3K/AKT Pathway and Regulating Glutamine Metabolism. Molecules. 2023; 28(14):5504. https://doi.org/10.3390/molecules28145504
Chicago/Turabian StyleWang, Yan, Kun Shi, Jiyuan Tu, Chang Ke, Niping Chen, Bo Wang, Yanju Liu, and Zhongshi Zhou. 2023. "Atractylenolide III Ameliorates Bile Duct Ligation-Induced Liver Fibrosis by Inhibiting the PI3K/AKT Pathway and Regulating Glutamine Metabolism" Molecules 28, no. 14: 5504. https://doi.org/10.3390/molecules28145504
APA StyleWang, Y., Shi, K., Tu, J., Ke, C., Chen, N., Wang, B., Liu, Y., & Zhou, Z. (2023). Atractylenolide III Ameliorates Bile Duct Ligation-Induced Liver Fibrosis by Inhibiting the PI3K/AKT Pathway and Regulating Glutamine Metabolism. Molecules, 28(14), 5504. https://doi.org/10.3390/molecules28145504





