Inhibitory Potential of Quercetin Derivatives Isolated from the Aerial Parts of Siegesbeckia pubescens Makino against Bacterial Neuraminidase
Abstract
1. Introduction
2. Results and Discussion
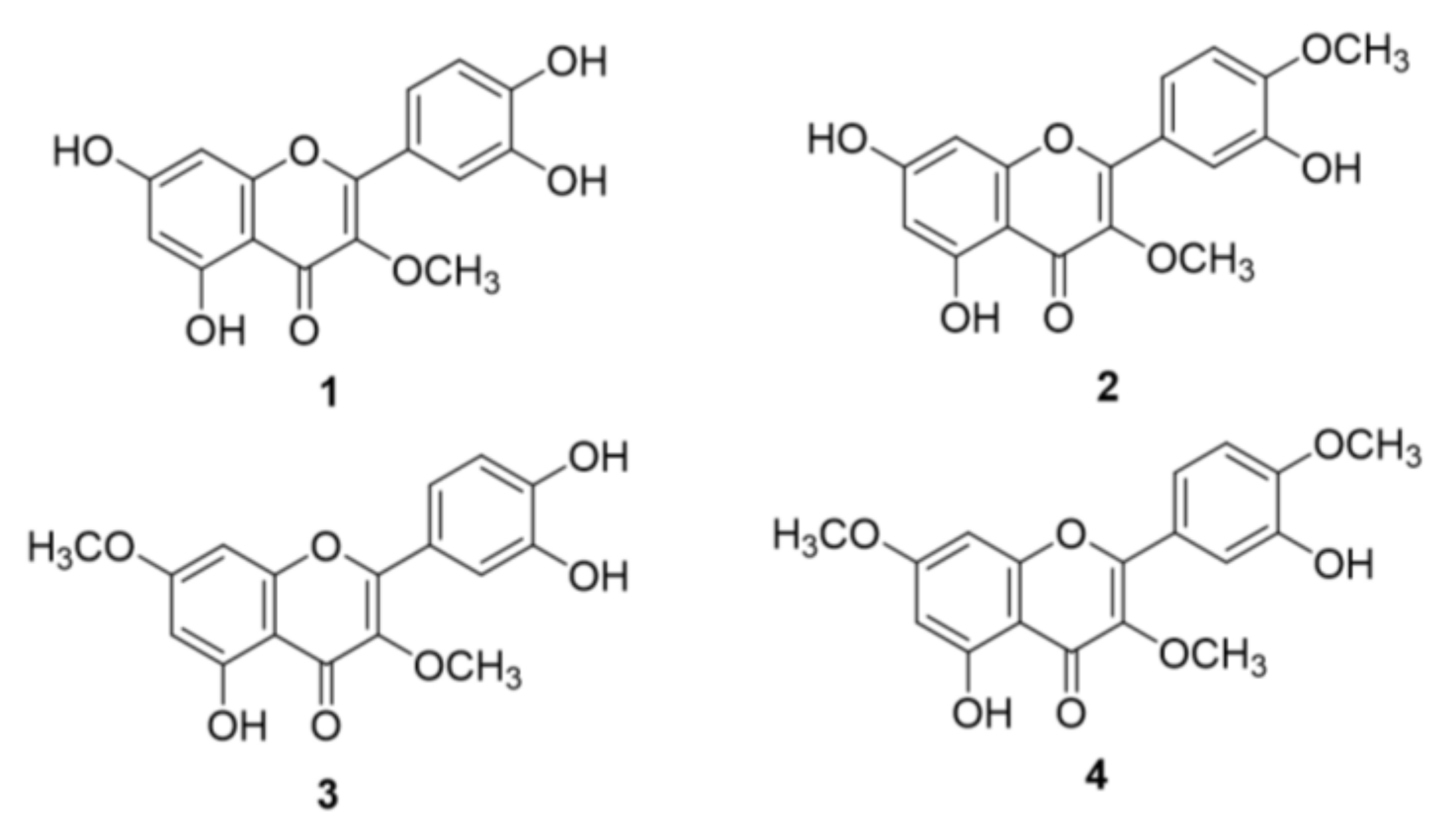
2.1. Structural Identification of O-Methylated Quercetins from S. pubescens
2.2. Inhibitory Effects of Quercetin Derivatives against Bacterial Neuraminidase
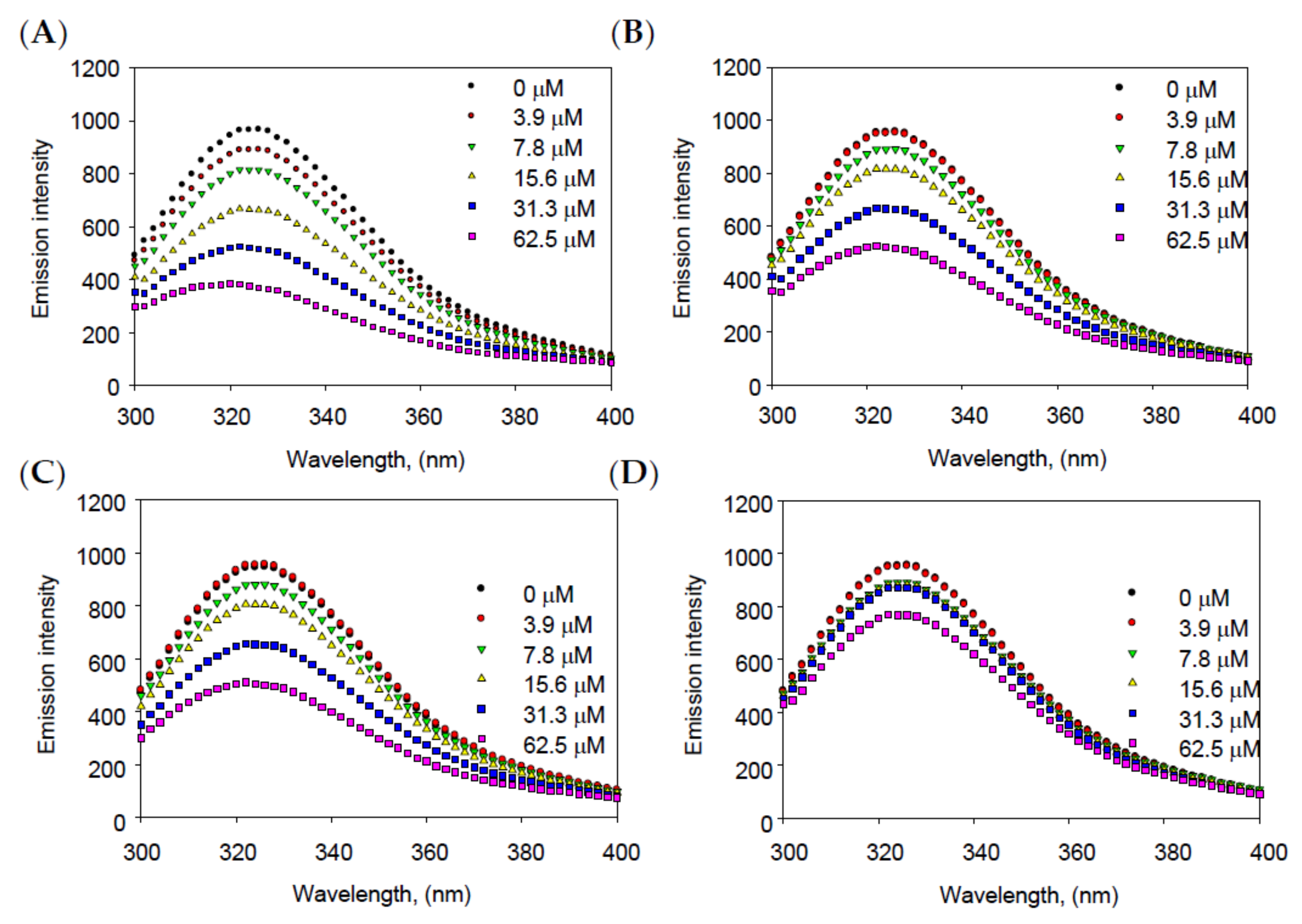
2.3. Binding Affinity between Inhibitors and Enzyme
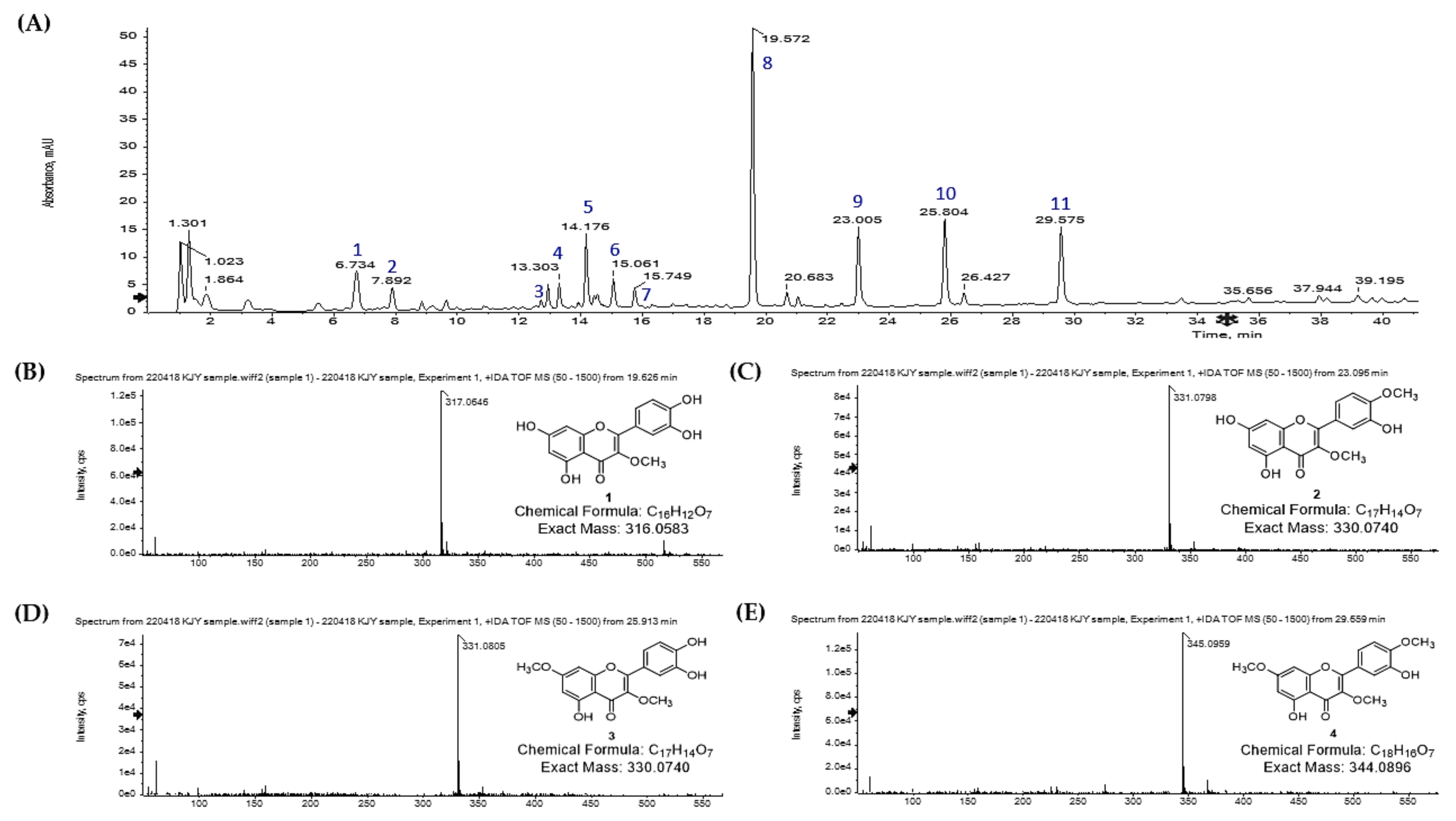
2.4. LC-Q-TOF/MS Analysis of S. pubescens Extract
3. Materials and Methods
3.1. Plant Materials and Chemicals
3.2. Instruments
3.3. Extraction, Separation, and Isolation of Quercetin Derivatives from S. pubescens
3.3.1. 3-O-Methyl Quercetin (1)
3.3.2. 3,4′-O-Dimethyl Quercetin (2)
3.3.3. 3,7-O-Dimethyl Quercetin (3)
3.3.4. 3,7,4′-O-Trimethyl Quercetin (4)
3.4. Bacterial Neuraminidase Inhibition Assay and Kinetics
3.5. Fluorescence Quenching Experiments
3.6. LC-Q-TOF/MS Analysis
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Park, M.H.; Jung, S.; Yuk, H.J.; Jang, H.J.; Kim, W.J.; Kim, D.Y.; Lim, G.T.; Lee, J.; Oh, S.R.; Lee, S.U.; et al. Rapid Identification of Isoprenylated Flavonoids Constituents with Inhibitory Activity on Bacterial Neuraminidase from Root Barks of Paper Mulberry (Broussonetia papyrifera). Int. J. Biol. Macromol. 2021, 174, 61–68. [Google Scholar] [CrossRef]
- Woo, H.S.; Shin, K.C.; Kim, J.Y.; Kim, Y.S.; Ban, Y.J.; Oh, Y.J.; Cho, H.J.; Oh, D.K.; Kim, D.W. Bakkenolides and Caffeoylquinic Acids from the Aerial Portion of Petasites japonicus and Their Bacterial Neuraminidase Inhibition Ability. Biomolecules 2020, 10, 888. [Google Scholar] [CrossRef] [PubMed]
- Baiseitova, A.; Lee, G.; Shah, A.B.; Yoon, S.; Kim, J.H.; Lee, Y.H.; Park, K.H. New Dihydrobenzoxanthone Derivatives with Bacterial Neuraminidase Inhibitory Activity Isolated from Artocarpus elasticus. Bioorg. Chem. 2022, 127, 105978. [Google Scholar] [CrossRef] [PubMed]
- Baiseitova, A.; Ban, Y.J.; Kim, J.Y.; Lee, G.; Shah, A.B.; Kim, J.H.; Lee, Y.H.; Park, K.H. Soybean Phytochemicals Responsible for Bacterial Neuraminidase Inhibition and Their Characterization by UPLC-ESI-TOF/MS. Food Funct. 2022, 13, 6923–6933. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.C.; Woo, H.S.; Kim, J.W.; Kim, Y.S.; Kim, J.Y.; Kim, J.H.; Yu, J.; Kim, Y.C.; Kim, D.W. Discovery and Characterization of Chemical Compounds That Inhibit the Function of Bacterial Neuraminidase from Codonopsis ussuriensis. Appl. Sci. 2022, 12, 6254. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, J.Y.; Son, Y.G.; Kim, K.L.; Kim, J.Y. Bacterial Neuraminidase Inhibitory Linarin from Dendranthema zawadskii. J. Appl. Biol. Chem. 2023, 66, 1–6. [Google Scholar] [CrossRef]
- Woo, S.Y.; Yang, J.Y.; Lee, H.G.; Ahn, H.J.; Lee, Y.B.; Do, S.H.; Kim, J.Y.; Seo, W.D. Changes in Metabolites with Harvest Times of Seedlings of Various Korean Oat (Avena Sativa L.) Cultivars and Their Neuraminidase Inhibitory Effects. Food Chem. 2022, 373, 131429. [Google Scholar] [CrossRef]
- Woo, H.S.; Kim, D.W.; Curtis-Long, M.J.; Lee, B.W.; Lee, J.H.; Kim, J.Y.; Kang, J.E.; Park, K.H. Potent Inhibition of Bacterial Neuraminidase Activity by Pterocarpans Isolated from the Roots of Lespedeza bicolor. Bioorg. Med. Chem. Lett. 2011, 21, 6100–6103. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and Its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Valle, J.; Solano, C.; García, B.; Toledo-Arana, A.; Lasa, I. Biofilm Switch and Immune Response Determinants at Early Stages of Infection. Trends Microbiol. 2013, 21, 364–371. [Google Scholar] [CrossRef]
- Soong, G.; Muir, A.; Gomez, M.I.; Waks, J.; Reddy, B.; Planet, P.; Singh, P.K.; Kanetko, Y.; Wolfgang, M.C.; Hsiao, Y.S.; et al. Bacterial Neuraminidase Facilitates Mucosal Infection by Participating in Biofilm Production. J. Clin. Investig. 2006, 116, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Huang, H.; Song, W.; Hu, H.; Chen, J.; Zhang, L.; Li, P.; Wu, R.; Wu, C. Preparation and Evaluation of Lipid Polymer Nanoparticles for Eradicating H. Pylori Biofilm and Impairing Antibacterial Resistance in Vitro. Int. J. Pharm. 2015, 495, 728–737. [Google Scholar] [CrossRef]
- Kim, J.H.; Ryu, Y.B.; Lee, W.S.; Kim, Y.H. Neuraminidase Inhibitory Activities of Quaternary Isoquinoline Alkaloids from Corydalis turtschaninovii Rhizome. Bioorg. Med. Chem. 2014, 22, 6047–6052. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.M.; Kim, J.Y.; Li, Z.P.; Jenis, J.; Jun Ban, Y.; Baiseitova, A.; Park, K.H. Effectiveness of Prenyl Group on Flavonoids from Epimedium koreanum Nakai on Bacterial Neuraminidase Inhibition. Molecules 2019, 24, 317. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.; Uddin, Z.; Song, Y.H.; Li, Z.P.; Kim, J.Y.; Ban, Y.J.; Park, K.H. Bacterial Neuraminidase Inhibition by Phenolic Compounds from Usnea longissima. S. Afr. J. Bot. 2019, 120, 326–330. [Google Scholar] [CrossRef]
- Wang, Y.; Curtis-Long, M.J.; Yuk, H.J.; Kim, D.W.; Tan, X.F.; Park, K.H. Bacterial Neuraminidase Inhibitory Effects of Prenylated Isoflavones from Roots of Flemingia philippinensis. Bioorg. Med. Chem. 2013, 21, 6398–6404. [Google Scholar] [CrossRef]
- Shah, A.B.; Baiseitova, A.; Kim, J.H.; Lee, Y.H.; Park, K.H. Inhibition of Bacterial Neuraminidase and Biofilm Formation by Ugonins Isolated From Helminthostachys zeylanica (L.) Hook. Front. Pharmacol. 2022, 13, 1747. [Google Scholar] [CrossRef]
- Uddin, Z.; Song, Y.H.; Curtis-Long, M.J.; Kim, J.Y.; Yuk, H.J.; Park, K.H. Potent Bacterial Neuraminidase Inhibitors, Anthraquinone Glucosides from Polygonum cuspidatum and Their Inhibitory Mechanism. J. Ethnopharmacol. 2016, 193, 283–292. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ryu, Y.B.; Curtis-Long, M.J.; Yuk, H.J.; Cho, J.K.; Kim, J.Y.; Kim, K.D.; Lee, W.S.; Park, K.H. Flavanones and Rotenoids from the Roots of Amorpha fruticosa L. That Inhibit Bacterial Neuraminidase. Food Chem. Toxicol. 2011, 49, 1849–1856. [Google Scholar] [CrossRef]
- Jang, H.; Woo Lee, J.; Gu Kim, J.; Ryoung Hong, H.; Phoung Linh Le, T.; Tae Hong, J.; Kim, Y.; Kyeong Lee, M.; Yeon Hwang, B. Nitric Oxide Inhibitory Constituents from Siegesbeckia pubescens. Bioorg. Chem. 2018, 80, 81–85. [Google Scholar] [CrossRef]
- Huh, J.E.; Baek, Y.H.; Lee, J.D.; Choi, D.Y.; Park, D.S. Therapeutic Effect of Siegesbeckia pubescens on Cartilage Protection in a Rabbit Collagenase-Induced Model of Osteoarthritis. J. Pharmacol. Sci. 2008, 107, 317–328. [Google Scholar] [CrossRef] [PubMed]
- SangHyun, L.; EunJung, N.; JungSook, K.; EunMi, S.; Xu, P.; YeongShik, K.; BakKwang, K.; BurmJong, L. Ent-Kaurane- and Ent-Pimarane-Type Diterpenoids from Siegesbeckia pubescens and Their Cytotoxicity in Caki Cells. Korean J. Crop Sci. 2005, 50, 147–150. [Google Scholar]
- Sun, Z.; Zhang, Y.; Zhou, H.; Xu, J.; Gu, Q. Diverse Diterpenoids and Sesquiterpenoids from Siegesbeckia pubescens and Their Activity against RANKL-Induced Osteoclastogenesis. Bioorg. Chem. 2021, 107, 104537. [Google Scholar] [CrossRef]
- Myagchilov, A.V.; Sokolova, L.I.; Gorovoy, P.G. New Diterpenoids of Sigesbeckia pubescens (Makino) Makino. Biointerface Res. Appl. Chem. 2021, 12, 8035–8041. [Google Scholar] [CrossRef]
- Sang, W.; Zhong, Z.; Linghu, K.; Xiong, W.; Tse, A.K.W.; Cheang, W.S.; Yu, H.; Wang, Y. Siegesbeckia pubescens Makino Inhibits Pam3CSK4-Induced Inflammation in RAW 264.7 Macrophages through Suppressing TLR1/TLR2-Mediated NF-κB Activation. Chin. Med. 2018, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Bellere, A.D.; Oh, S.; Yu, D.; Fang, M.; Yi, T.H. Antibiofilm Effect of Siegesbeckia pubescens against S. Mutans According to Environmental Factors. Appl. Sci. 2023, 13, 6179. [Google Scholar] [CrossRef]
- Wang, R.; Liu, Y.Q.; Ha, W.; Shi, Y.P.; Hwang, T.L.; Huang, G.J.; Wu, T.S.; Lee, K.H. In Vitro Anti-Inflammatory Effects of Diterpenoids and Sesquiterpenoids from Traditional Chinese Medicine Siegesbeckia pubescens. Bioorg. Med. Chem. Lett. 2014, 24, 3944–3947. [Google Scholar] [CrossRef]
- Shen, S.C.; Lee, W.R.; Lin, H.Y.; Huang, H.C.; Ko, C.H.; Yang, L.L.; Chen, Y.C. In Vitro and in Vivo Inhibitory Activities of Rutin, Wogonin, and Quercetin on Lipopolysaccharide-Induced Nitric Oxide and Prostaglandin E2 Production. Eur. J. Pharmacol. 2002, 446, 187–194. [Google Scholar] [CrossRef]
- Wang, D.; Dong, X.; Nie, Y.; Yang, W.; Li, C. A Review on Medical Plants of Genus Siegesbeckia: Phytochemical and Pharmacological Studies. Rec. Nat. Prod. 2022, 16, 537. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, J.Y.; Song, Y.H.; Li, Z.P.; Yoon, S.H.; Uddin, Z.; Jun Ban, Y.; Lee, K.W.; Park, K.H. Highly Potent Bacterial Neuraminidase Inhibitors, Chromenone Derivatives from Flemingia philippinensis. Int. J. Biol. Macromol. 2019, 128, 149–157. [Google Scholar] [CrossRef] [PubMed]
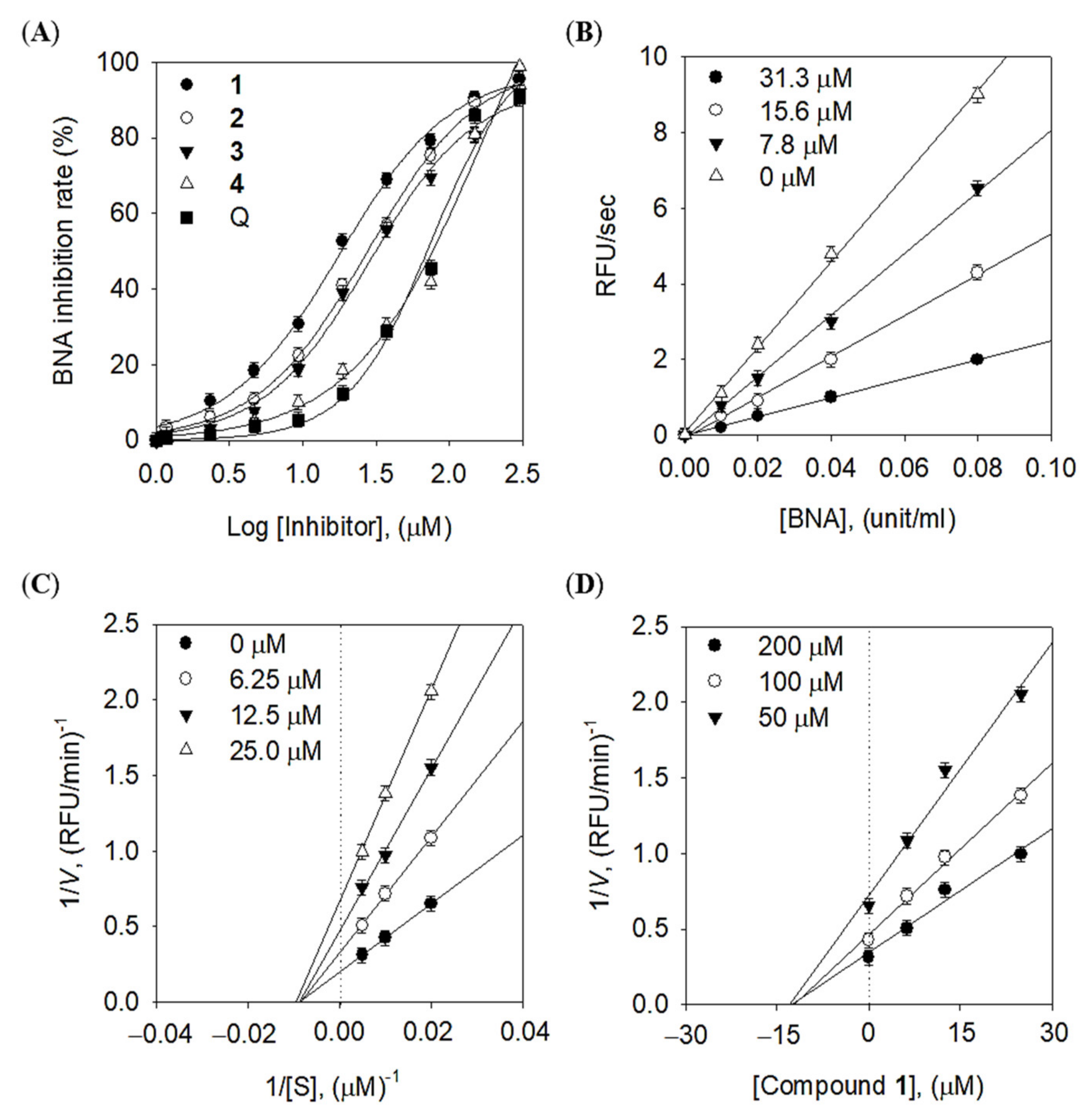
| Compounds | IC50 a, (μM) | Inhibition Mode | Ki b, (μM) |
|---|---|---|---|
| 1 | 14.0 ± 0.5 | Noncompetitive | 13.8 ± 0.2 |
| 2 | 25.8 ± 1.0 | Noncompetitive | 24.7 ± 0.5 |
| 3 | 24.3 ± 0.9 | Noncompetitive | 22.4 ± 0.9 |
| 4 | 84.1 ± 3.3 | Noncompetitive | 79.5 ± 1.8 |
| Quercetin c | 26.1 ± 0.6 | NT d | NT |
| Compounds | KSV (×105 L mol−1) | R2 | n | KA (×106 L mol−1) | R2 |
|---|---|---|---|---|---|
| 1 | 0.0252 | 0.9954 | 1.2106 | 0.05105 | 0.9938 |
| 2 | 0.0144 | 0.9968 | 1.1063 | 0.03994 | 0.9905 |
| 3 | 0.0153 | 0.9973 | 1.0959 | 0.03818 | 0.9933 |
| 4 | 0.0036 | 0.9172 | 0.7071 | 0.00021 | 0.9991 |
| Peaks | Time (min) | Observed Ion (m/z) | Calculated Ion (m/z) | Error (ppm) | Formula | Identification |
|---|---|---|---|---|---|---|
| 1 | 6.7 | 355.1012 | 355.1029 | −4.79 | C16H18O9 | chlorogenic acid |
| 2 | 7.9 | 181.0499 | 181.0500 | −0.55 | C9H8O4 | caffeic acid |
| 3 | 12.9 | 611.1612 | 611.1612 | 0 | C27H30O16 | rutin |
| 4 | 13.3 | 465.1036 | 465.1033 | +0.65 | C21H20O12 | isoquercitrin |
| 5 | 14.2 | 517.1345 | 517.1346 | −0.19 | C25H24O12 | 3,5-dicaffeoylquinic acid |
| 6 | 15.1 | 517.1346 | 517.1346 | 0 | C25H24O12 | 1,5-dicaffeoylquinic acid |
| 7 | 15.7 | 679.1665 | 679.1663 | −0.29 | C34H30O15 | 3,4,5-tricaffeoylquinic acid |
| 8 | 19.6 | 317.0642 | 317.0661 | −5.99 | C16H12O7 | 3-O-methyl quercetin |
| 9 | 23.0 | 331.0798 | 331.0818 | −6.04 | C17H14O7 | 3,4′-O-dimethyl quercetin |
| 10 | 25.8 | 331.0805 | 331.0818 | −3.93 | C17H14O7 | 3,7-O-dimethyl quercetin |
| 11 | 29.6 | 345.0961 | 345.0974 | −3.77 | C18H16O7 | 3,7,4′-O-trimethyl quercetin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, Y.G.; Kim, J.Y.; Park, J.Y.; Kim, K.D.; Park, K.H.; Kim, J.Y. Inhibitory Potential of Quercetin Derivatives Isolated from the Aerial Parts of Siegesbeckia pubescens Makino against Bacterial Neuraminidase. Molecules 2023, 28, 5365. https://doi.org/10.3390/molecules28145365
Son YG, Kim JY, Park JY, Kim KD, Park KH, Kim JY. Inhibitory Potential of Quercetin Derivatives Isolated from the Aerial Parts of Siegesbeckia pubescens Makino against Bacterial Neuraminidase. Molecules. 2023; 28(14):5365. https://doi.org/10.3390/molecules28145365
Chicago/Turabian StyleSon, Yun Gon, Ju Yeon Kim, Jae Yeon Park, Kwang Dong Kim, Ki Hun Park, and Jeong Yoon Kim. 2023. "Inhibitory Potential of Quercetin Derivatives Isolated from the Aerial Parts of Siegesbeckia pubescens Makino against Bacterial Neuraminidase" Molecules 28, no. 14: 5365. https://doi.org/10.3390/molecules28145365
APA StyleSon, Y. G., Kim, J. Y., Park, J. Y., Kim, K. D., Park, K. H., & Kim, J. Y. (2023). Inhibitory Potential of Quercetin Derivatives Isolated from the Aerial Parts of Siegesbeckia pubescens Makino against Bacterial Neuraminidase. Molecules, 28(14), 5365. https://doi.org/10.3390/molecules28145365







