The Importance of the Pyrazole Scaffold in the Design of Protein Kinases Inhibitors as Targeted Anticancer Therapies
Abstract
1. Introduction
2. The Chemical Profile of the Pyrazole Ring
3. Protein Kinases Structure and Inhibition Mechanisms
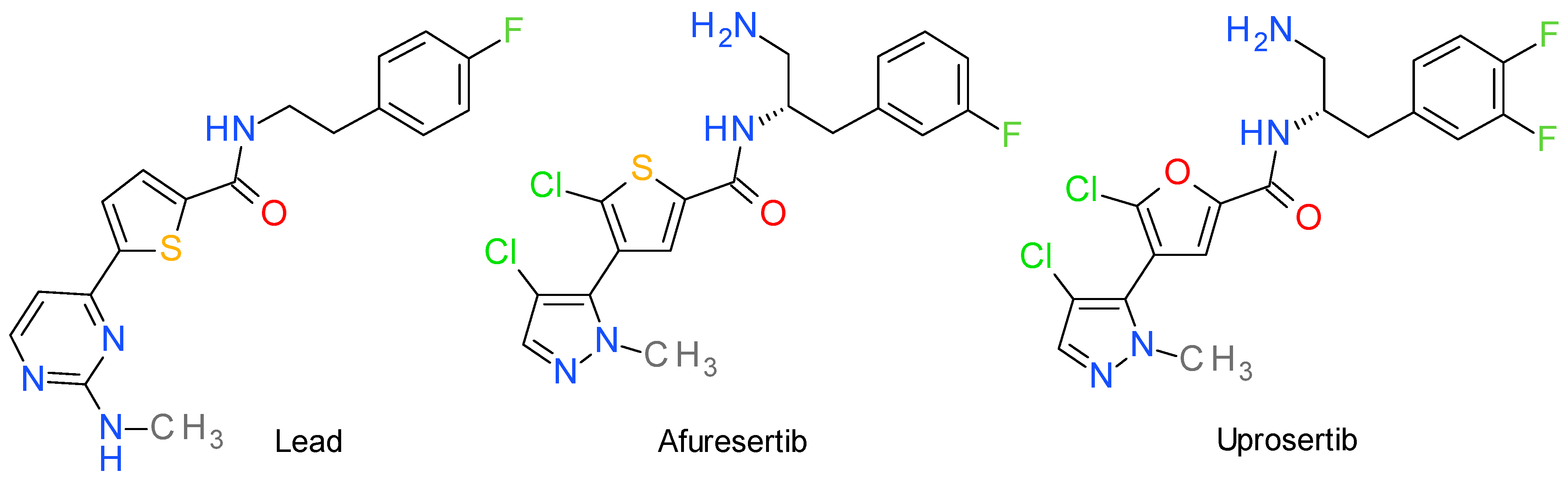
4. Akt Inhibitors
4.1. Afuresertib
4.2. Uprosertib
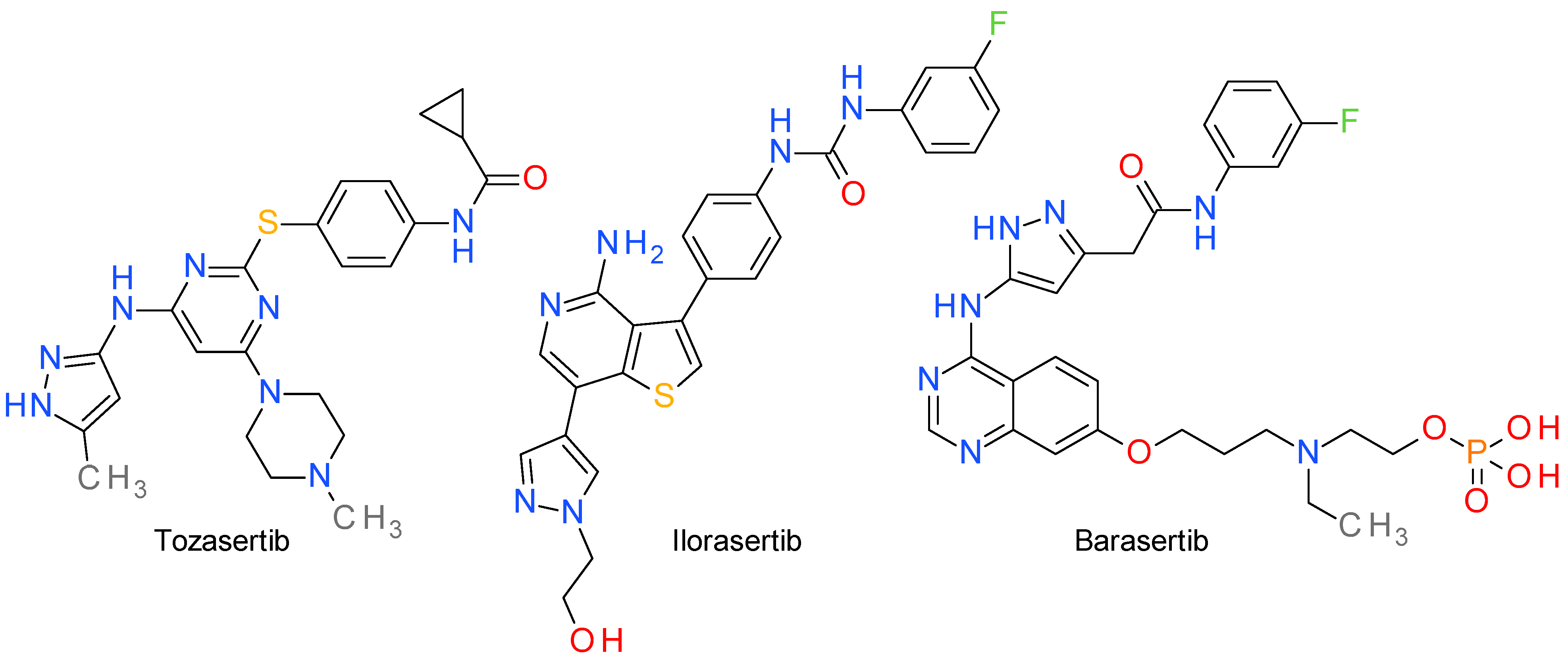
5. Aurora Kinases Inhibitors
5.1. Tozasertib
5.2. Ilorasertib
5.3. Barasertib
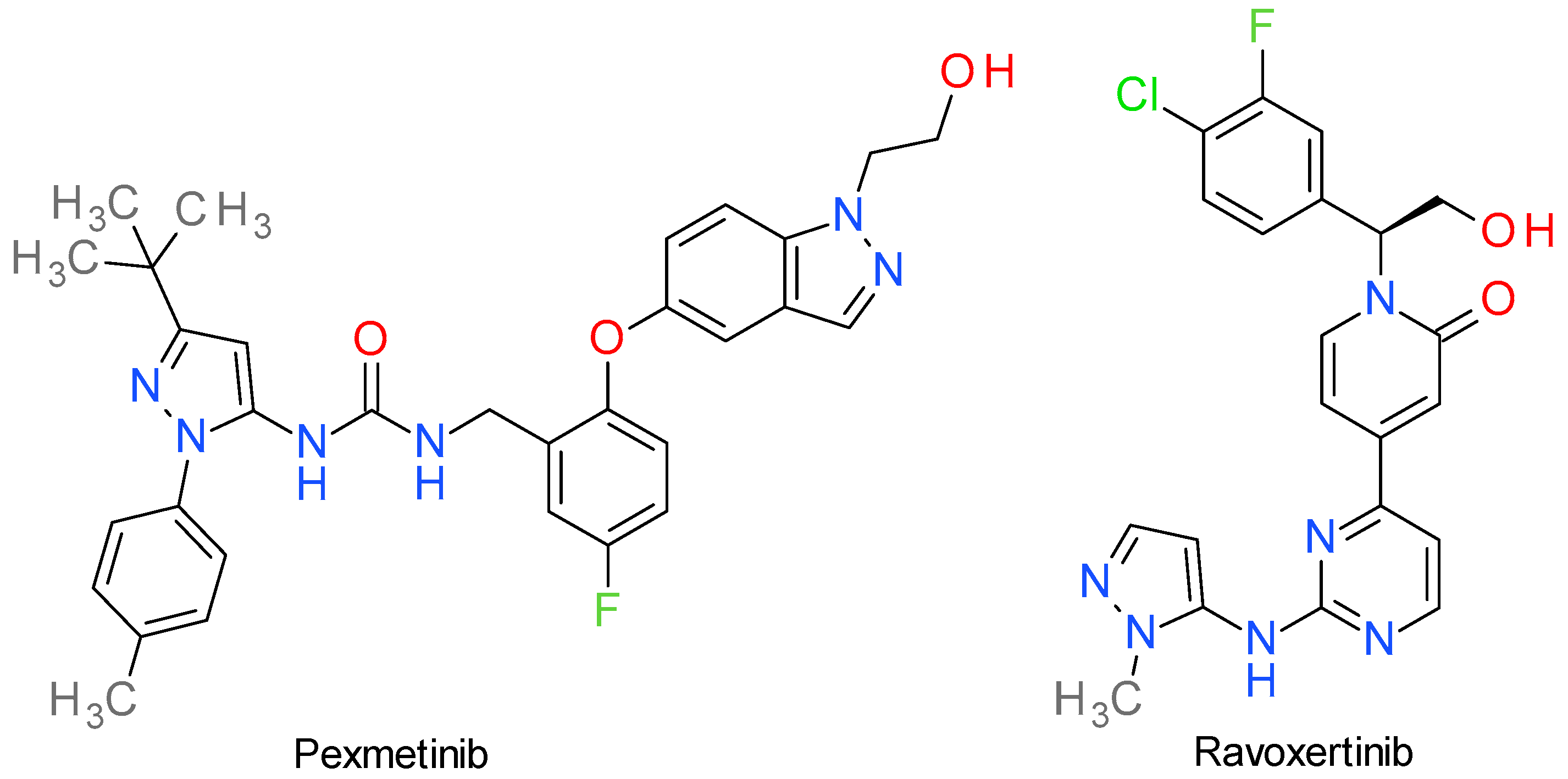
6. MAPK Inhibitors
6.1. Pexmetinib
6.2. Ravoxertinib
7. B-Raf Inhibitors
Encorafenib
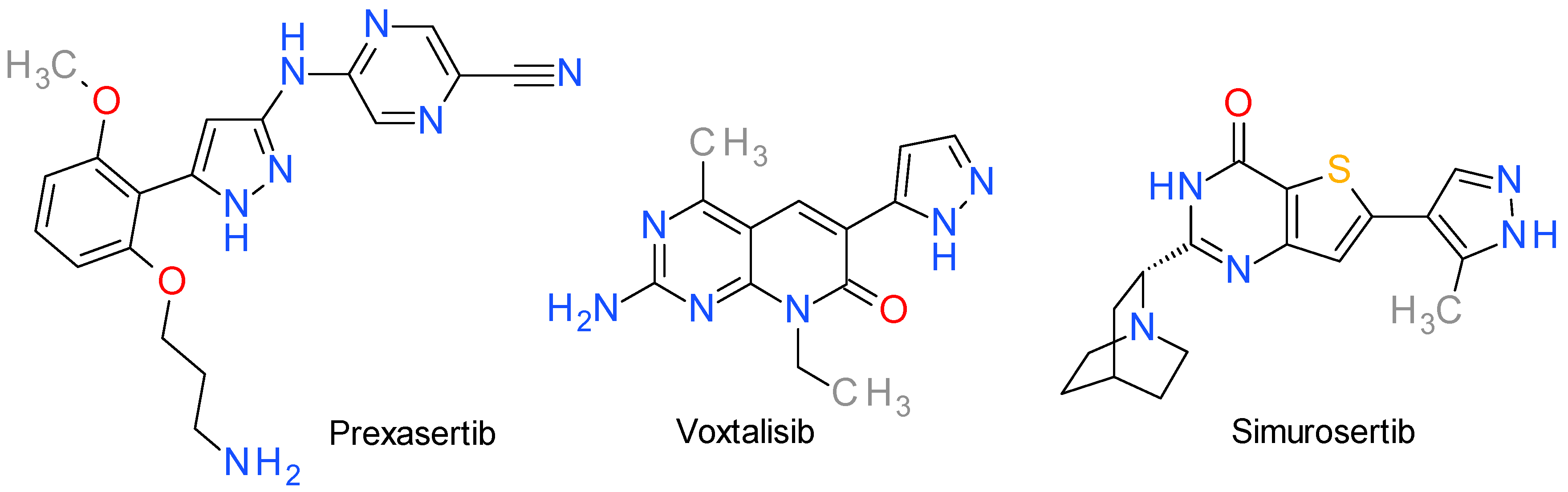
8. Inhibitors of Various Other Serine/Threonine Kinases
8.1. Prexasertib
8.2. Voxtalisib
8.3. Simurosertib
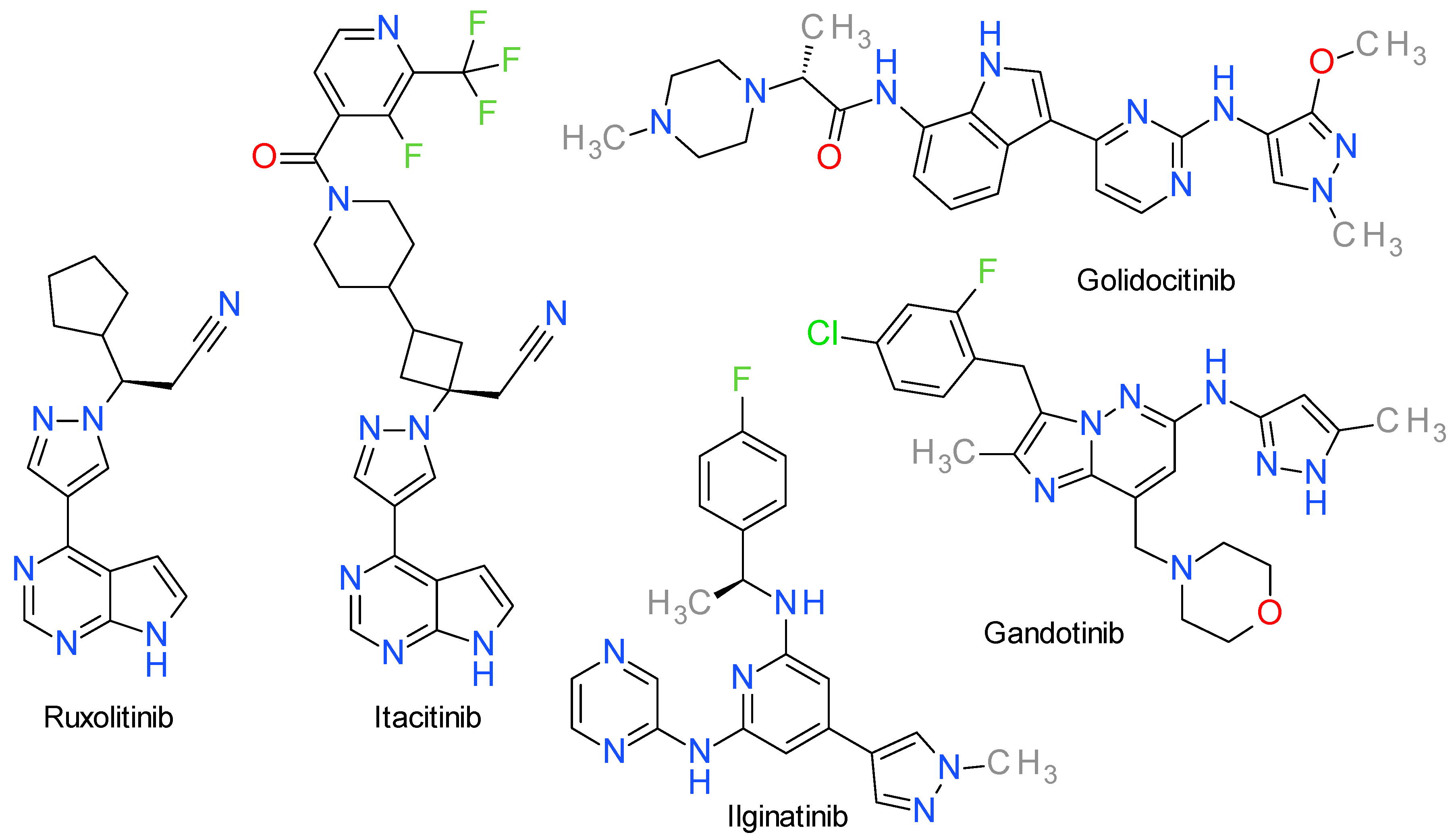
9. JAK Inhibitors
9.1. Ruxolitinib
9.2. Itacitinib
9.3. Golidocitinib
9.4. Gandotinib
9.5. Ilginatinib
10. Bcr-Abl Inhibitors
10.1. Asciminib
10.2. Rebastinib
11. c-Met Inhibitors
11.1. Crizotinib
11.2. Bozitinib
11.3. Glumetinib
11.4. Merestinib
11.5. Savolitinib
12. EGFR Inhibitors
12.1. Lazertinib
12.2. Mavelertinib
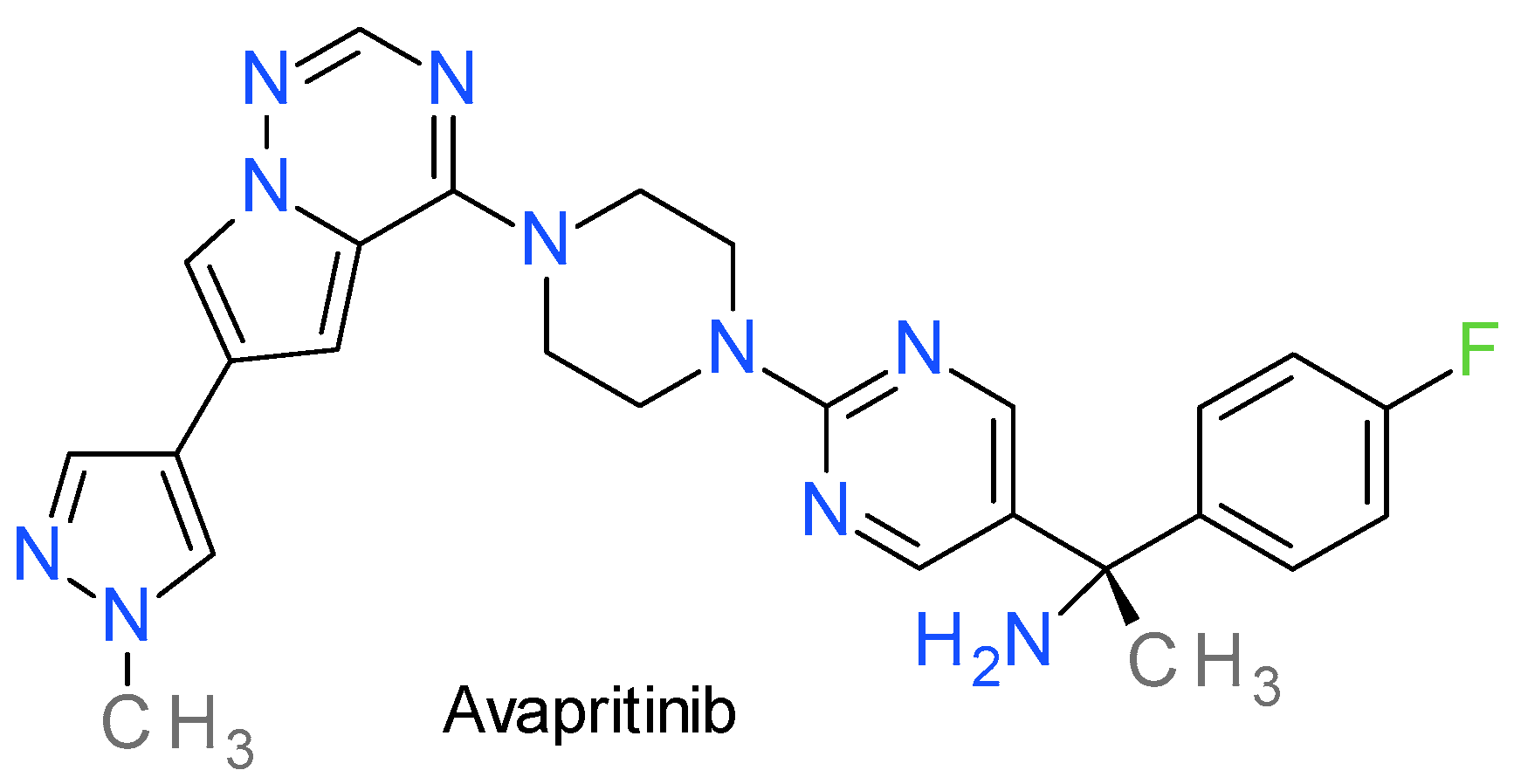
13. PDGFR Inhibitors
Avapritinib
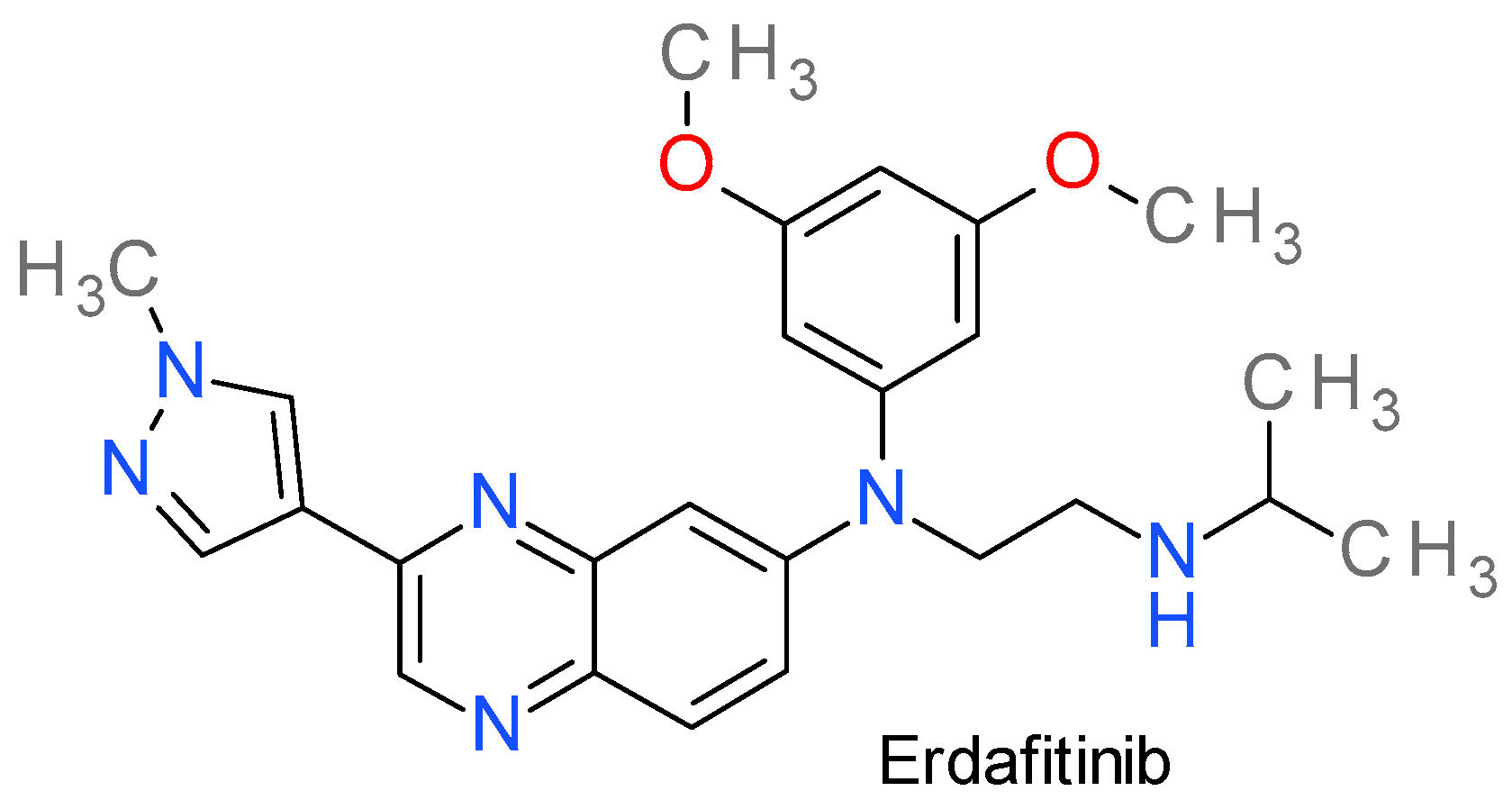
14. FGFR Inhibitors
Erdafitinib
15. RET Inhibitors
Pralsetinib
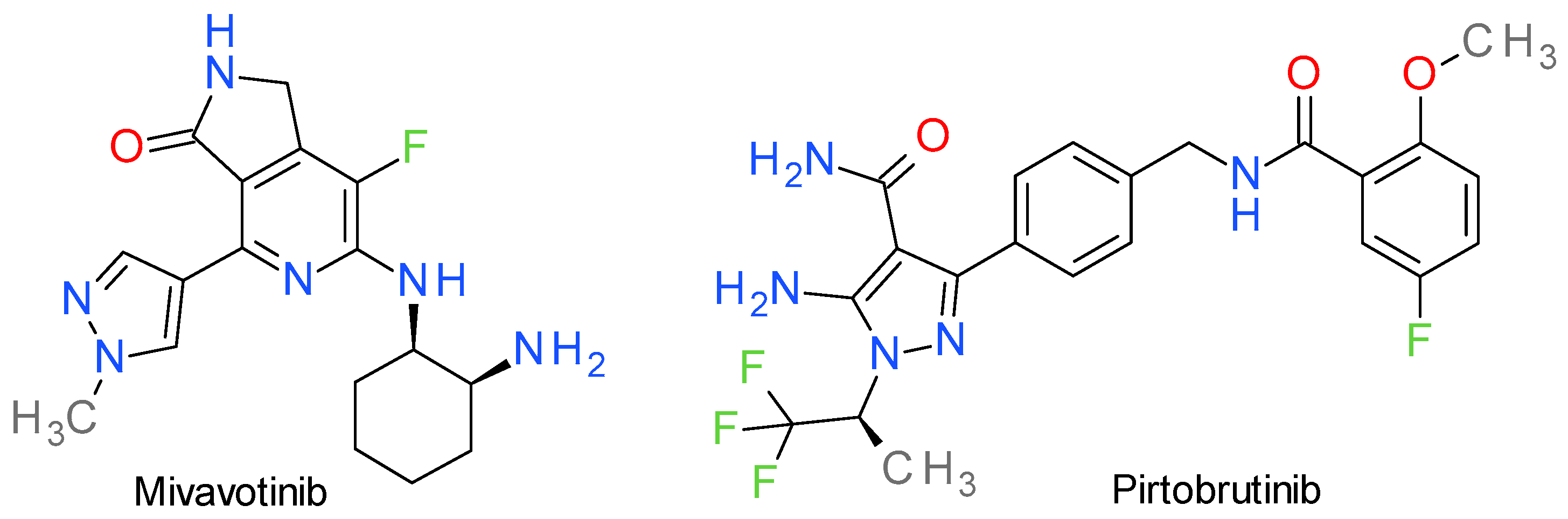
16. Inhibitors of Various Other Tyrosine Kinases
16.1. Mivavotinib
16.2. Pirtobrutinib
17. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Bournez, C.; Carles, F.; Peyrat, G.; Aci-Sèche, S.; Bourg, S.; Meyer, C.; Bonnet, P. Comparative Assessment of Protein Kinase Inhibitors in Public Databases and in PKIDB. Molecules 2020, 25, 3226. [Google Scholar] [CrossRef] [PubMed]
- Day, E.K.; Sosale, N.G.; Lazzara, M.J. Cell signaling regulation by protein phosphorylation: A multivariate, heterogeneous, and context-dependent process. Curr. Opin. Biotechnol. 2016, 40, 185–192. [Google Scholar] [CrossRef]
- Turdo, A.; D’accardo, C.; Glaviano, A.; Porcelli, G.; Colarossi, C.; Colarossi, L.; Mare, M.; Faldetta, N.; Modica, C.; Pistone, G.; et al. Targeting Phosphatases and Kinases: How to Checkmate Cancer. Front. Cell Dev. Biol. 2021, 9, 690306. [Google Scholar] [CrossRef]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Kannaiyan, R.; Mahadevan, D. A comprehensive review of protein kinase inhibitors for cancer therapy. Expert Rev. Anticancer. Ther. 2018, 18, 1249–1270. [Google Scholar] [CrossRef]
- Ion, G.N.D.; Olaru, O.T.; Nitulescu, G.; Olaru, I.I.; Tsatsakis, A.; Burykina, T.I.; Spandidos, D.A.; Nitulescu, G.M. Improving the odds of success in antitumoral drug development using scoring approaches towards heterocyclic scaffolds. Oncol. Rep. 2020, 44, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Dimova, D.; Bajorath, J. Assessing Scaffold Diversity of Kinase Inhibitors Using Alternative Scaffold Concepts and Estimating the Scaffold Hopping Potential for Different Kinases. Molecules 2017, 22, 730. [Google Scholar] [CrossRef]
- Hu, L.; Yang, Y.; Zheng, S.; Xu, J.; Ran, T.; Chen, H. Kinase Inhibitor Scaffold Hopping with Deep Learning Approaches. J. Chem. Inf. Model. 2021, 61, 4900–4912. [Google Scholar] [CrossRef] [PubMed]
- Skoreński, M.; Sieńczyk, M. The Fellowship of Privileged Scaffolds—One Structure to Inhibit Them All. Pharmaceuticals 2021, 14, 1164. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Dietrich, J. Privileged scaffolds in lead generation. Expert Opin. Drug Discov. 2015, 10, 781–790. [Google Scholar] [CrossRef]
- Schnur, D.M.; Hermsmeier, M.A.; Tebben, A.J. Are Target-Family-Privileged Substructures Truly Privileged? J. Med. Chem. 2006, 49, 2000–2009. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.S.; Ali, E.M.; Ibrahim, D.A.; Serya, R.A.; El Ella, D.A.A. Pyrazolo[3,4-d]pyrimidine based scaffold derivatives targeting kinases as anticancer agents. Futur. J. Pharm. Sci. 2016, 2, 20–30. [Google Scholar] [CrossRef]
- Xie, F.; Zhou, L.; Ge, C.; Song, X.; Yan, H. Development of pyrazolo[3,4-d]pyrimidin-4-one scaffold as novel CDK2 inhibitors: Design, synthesis, and biological evaluation. Bioorg. Med. Chem. Lett. 2022, 70, 128803. [Google Scholar] [CrossRef] [PubMed]
- Baillache, D.J.; Unciti-Broceta, A. Recent developments in anticancer kinase inhibitors based on the pyrazolo[3,4-d]pyrimidine scaffold. RSC Med. Chem. 2020, 11, 1112–1135. [Google Scholar] [CrossRef] [PubMed]
- Bendjeddou, L.Z.; Loaëc, N.; Villiers, B.; Prina, E.; Späth, G.F.; Galons, H.; Meijer, L.; Oumata, N. Exploration of the imidazo[1,2-b]pyridazine scaffold as a protein kinase inhibitor. Eur. J. Med. Chem. 2017, 125, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Tandon, N.; Luxami, V.; Kant, D.; Tandon, R.; Paul, K. Current progress, challenges and future prospects of indazoles as protein kinase inhibitors for the treatment of cancer. RSC Adv. 2021, 11, 25228–25257. [Google Scholar] [CrossRef]
- Brullo, C.; Rapetti, F.; Bruno, O. Pyrazolyl-Ureas as Interesting Scaffold in Medicinal Chemistry. Molecules 2020, 25, 3457. [Google Scholar] [CrossRef]
- Costa, R.F.; Turones, L.C.; Cavalcante, K.V.N.; Júnior, I.A.R.; Xavier, C.H.; Rosseto, L.P.; Napolitano, H.B.; Castro, P.F.D.S.; Neto, M.L.F.; Galvão, G.M.; et al. Heterocyclic Compounds: Pharmacology of Pyrazole Analogs from Rational Structural Considerations. Front. Pharmacol. 2021, 12, 666725. [Google Scholar] [CrossRef]
- Nitulescu, G.M.; Draghici, C.; Olaru, O.T. New Potential Antitumor Pyrazole Derivatives: Synthesis and Cytotoxic Evaluation. Int. J. Mol. Sci. 2013, 14, 21805–21818. [Google Scholar] [CrossRef]
- Mor, S.; Khatri, M.; Punia, R.; Sindhu, S. Recent Progress in Anticancer Agents Incorporating Pyrazole Scaffold. Mini-Rev. Med. Chem. 2022, 21, 115–163. [Google Scholar] [CrossRef]
- Bennani, F.E.; Doudach, L.; Cherrah, Y.; Ramli, Y.; Karrouchi, K.; Ansar, M.; Faouzi, M.E.A. Overview of recent developments of pyrazole derivatives as an anticancer agent in different cell line. Bioorganic Chem. 2020, 97, 103470. [Google Scholar] [CrossRef] [PubMed]
- Lusardi, M.; Spallarossa, A.; Brullo, C. Amino-Pyrazoles in Medicinal Chemistry: A Review. Int. J. Mol. Sci. 2023, 24, 7834. [Google Scholar] [CrossRef]
- Ebenezer, O.; Shapi, M.; Tuszynski, J.A. A Review of the Recent Development in the Synthesis and Biological Evaluations of Pyrazole Derivatives. Biomedicines 2022, 10, 1124. [Google Scholar] [CrossRef]
- Yet, L. 4.01—Pyrazoles. In Comprehensive Heterocyclic Chemistry III; Katritzky, A.R., Ramsden, C.A., Scriven, E.F.V., Taylor, R.J., Eds.; Elsevier: Oxford, UK, 2008; pp. 1–141. [Google Scholar]
- Faisal, M.; Saeed, A.; Hussain, S.; Dar, P.; Larik, F.A. Recent developments in synthetic chemistry and biological activities of pyrazole derivatives. J. Chem. Sci. 2019, 131, 70. [Google Scholar] [CrossRef]
- Secrieru, A.; O’neill, P.M.; Cristiano, M.L.S. Revisiting the Structure and Chemistry of 3(5)-Substituted Pyrazoles. Molecules 2020, 25, 42. [Google Scholar] [CrossRef] [PubMed]
- Karrouchi, K.; Radi, S.; Ramli, Y.; Taoufik, J.; Mabkhot, Y.N.; Al-Aizari, F.A.; Ansar, M. Synthesis and Pharmacological Activities of Pyrazole Derivatives: A Review. Molecules 2018, 23, 134. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M. Quantitative and Qualitative Analysis of the Anti-Proliferative Potential of the Pyrazole Scaffold in the Design of Anticancer Agents. Molecules 2022, 27, 3300. [Google Scholar] [CrossRef]
- Chopra, N.; Kaur, D.; Chopra, G. Nature and Hierarchy of Hydrogen-Bonding Interactions in Binary Complexes of Azoles with Water and Hydrogen Peroxide. ACS Omega 2018, 3, 12688–12702. [Google Scholar] [CrossRef]
- Zhang, K.; Ye, K.; Tang, H.; Qi, Z.; Wang, T.; Mao, J.; Zhang, X.; Jiang, S. Development and Therapeutic Implications of Tyrosine Kinase 2 Inhibitors. J. Med. Chem. 2023, 66, 4378–4416. [Google Scholar] [CrossRef]
- Fischer, P.M. Approved and Experimental Small-Molecule Oncology Kinase Inhibitor Drugs: A Mid-2016 Overview. Med. Res. Rev. 2017, 37, 314–367. [Google Scholar] [CrossRef]
- Hanks, S.K.; Hunter, T. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors: A 2023 update. Pharmacol. Res. 2023, 187, 106552. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mandiyan, V.; Suzuki, Y.; Zhang, C.; Rice, J.; Tsai, J.; Artis, D.R.; Ibrahim, P.; Bremer, R. Crystal Structures of Proto-oncogene Kinase Pim1: A Target of Aberrant Somatic Hypermutations in Diffuse Large Cell Lymphoma. J. Mol. Biol. 2005, 348, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Emrick, M.A.; Lee, T.; Starkey, P.J.; Mumby, M.C.; Resing, K.A.; Ahn, N.G. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc. Natl. Acad. Sci. USA 2006, 103, 18101–18106. [Google Scholar] [CrossRef] [PubMed]
- Mondal, J.; Tiwary, P.; Berne, B.J. How a Kinase Inhibitor Withstands Gatekeeper Residue Mutations. J. Am. Chem. Soc. 2016, 138, 4608–4615. [Google Scholar] [CrossRef]
- Eblen, S.T.; Kumar, N.V.; Shah, K.; Henderson, M.J.; Watts, C.K.W.; Shokat, K.M.; Weber, M.J. Identification of Novel ERK2 Substrates through Use of an Engineered Kinase and ATP Analogs. J. Biol. Chem. 2003, 278, 14926–14935. [Google Scholar] [CrossRef]
- Endo, S.; Satoh, Y.; Shah, K.; Takishima, K. A single amino-acid change in ERK1/2 makes the enzyme susceptible to PP1 derivatives. Biochem. Biophys. Res. Commun. 2006, 341, 261–265. [Google Scholar] [CrossRef]
- Modi, V.; Dunbrack, R.L. Defining a new nomenclature for the structures of active and inactive kinases. Proc. Natl. Acad. Sci. USA 2019, 116, 6818–6827. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacol. Res. 2019, 139, 471–488. [Google Scholar] [CrossRef]
- Roskoski, R. Hydrophobic and polar interactions of FDA-approved small molecule protein kinase inhibitors with their target enzymes. Pharmacol. Res. 2021, 169, 105660. [Google Scholar] [CrossRef] [PubMed]
- Madhusudan; Trafny, E.A.; Xuong, N.-H.; Adams, J.A.; Eyck, L.F.T.; Taylor, S.S.; Sowadski, J.M. cAMP-dependent protein kinase: Crystallographic insights into substrate recognition and phosphotransfer. Protein Sci. 1994, 3, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Kornev, A.P.; Taylor, S.S. Defining the conserved internal architecture of a protein kinase. Biochim. Biophys. Acta-Proteins Proteom. 2010, 1804, 440–444. [Google Scholar] [CrossRef]
- Dodson, C.A.; Kosmopoulou, M.; Richards, M.W.; Atrash, B.; Bavetsias, V.; Blagg, J.; Bayliss, R. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: Insights into selectivity and drug design. Biochem. J. 2010, 427, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.Y.; Yeoh, Y.; Low, T.Y. A recent update on small-molecule kinase inhibitors for targeted cancer therapy and their therapeutic insights from mass spectrometry-based proteomic analysis. FEBS J. 2022, 290, 2845–2864. [Google Scholar] [CrossRef]
- Martinez, R.; Defnet, A.; Shapiro, P. Avoiding or Co-Opting ATP Inhibition: Overview of Type III, IV, V, and VI Kinase Inhibitors. In Next Generation Kinase Inhibitors; Springer International Publishing: Cham, Switzerland, 2020; pp. 29–59. [Google Scholar]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Martorana, F.; Motta, G.; Pavone, G.; Motta, L.; Stella, S.; Vitale, S.R.; Manzella, L.; Vigneri, P. AKT Inhibitors: New Weapons in the Fight Against Breast Cancer? Front. Pharmacol. 2021, 12, 662232. [Google Scholar] [CrossRef]
- Nitulescu, G.; Van De Venter, M.; Nitulescu, G.; Ungurianu, A.; Juzenas, P.; Peng, Q.; Olaru, O.; Grădinaru, D.; Tsatsakis, A.; Tsoukalas, D.; et al. The Akt pathway in oncology therapy and beyond (Review). Int. J. Oncol. 2018, 53, 2319–2331. [Google Scholar] [CrossRef]
- Hua, H.; Zhang, H.; Chen, J.; Wang, J.; Liu, J.; Jiang, Y. Targeting Akt in cancer for precision therapy. J. Hematol. Oncol. 2021, 14, 4163–4168. [Google Scholar] [CrossRef]
- Lin, X.; Murray, J.M.; Rico, A.C.; Wang, M.X.; Chu, D.T.; Zhou, Y.; Del Rosario, M.; Kaufman, S.; Ma, S.; Fang, E.; et al. Discovery of 2-pyrimidyl-5-amidothiophenes as potent inhibitors for AKT: Synthesis and SAR studies. Bioorg. Med. Chem. Lett. 2006, 16, 4163–4168. [Google Scholar] [CrossRef]
- Dumble, M.; Crouthamel, M.-C.; Zhang, S.-Y.; Schaber, M.; Levy, D.; Robell, K.; Liu, Q.; Figueroa, D.J.; Minthorn, E.A.; Seefeld, M.A.; et al. Discovery of Novel AKT Inhibitors with Enhanced Anti-Tumor Effects in Combination with the MEK Inhibitor. PLoS ONE 2014, 9, e100880. [Google Scholar] [CrossRef]
- Huck, B.R.; Mochalkin, I. Recent progress towards clinically relevant ATP-competitive Akt inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 2838–2848. [Google Scholar] [CrossRef]
- Blagden, S.P.; Hamilton, A.L.; Mileshkin, L.; Wong, S.; Michael, A.; Hall, M.; Goh, J.C.; Lisyanskaya, A.S.; DeSilvio, M.; Frangou, E.; et al. Phase IB Dose Escalation and Expansion Study of AKT Inhibitor Afuresertib with Carboplatin and Paclitaxel in Recurrent Platinum-resistant Ovarian Cancer. Clin. Cancer Res. 2019, 25, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Arceci, R.J.; Allen, C.E.; Dunkel, I.J.; Jacobsen, E.; Whitlock, J.; Vassallo, R.; Morris, S.R.; Portnoy, A.; Reedy, B.A.; Smith, D.A.; et al. A phase IIa study of afuresertib, an oral pan-AKT inhibitor, in patients with Langerhans cell histiocytosis. Pediatr. Blood Cancer 2017, 64, e26325. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M.; Margina, D.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Saloustros, E.; Fenga, C.; Spandidos, D.A.; Libra, M.; Tsatsakis, A.M. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use (Review). Int. J. Oncol. 2015, 48, 869–885. [Google Scholar] [CrossRef]
- Kaboli, P.J.; Salimian, F.; Aghapour, S.; Xiang, S.; Zhao, Q.; Li, M.; Wu, X.; Du, F.; Zhao, Y.; Shen, J.; et al. Akt-targeted therapy as a promising strategy to overcome drug resistance in breast cancer—A comprehensive review from chemotherapy to immunotherapy. Pharmacol. Res. 2020, 156, 104806. [Google Scholar] [CrossRef] [PubMed]
- Willems, E.; Dedobbeleer, M.; Digregorio, M.; Lombard, A.; Lumapat, P.N.; Rogister, B. The functional diversity of Aurora kinases: A comprehensive review. Cell Div. 2018, 13, 7. [Google Scholar] [CrossRef]
- Tang, A.; Gao, K.; Chu, L.; Zhang, R.; Yang, J.; Zheng, J. Aurora kinases: Novel therapy targets in cancers. Oncotarget 2017, 8, 23937–23954. [Google Scholar] [CrossRef]
- Valle, A.S.-D.; Reina-Ortiz, C.; Benedi, A.; Anel, A.; Naval, J.; Marzo, I. Future prospects for mitosis-targeted antitumor therapies. Biochem. Pharmacol. 2021, 190, 114655. [Google Scholar] [CrossRef]
- Nitulescu, G.M.; Matei, L.; Aldea, I.M.; Draghici, C.; Olaru, O.T.; Bleotu, C. Ultrasound-assisted synthesis and anticancer evaluation of new pyrazole derivatives as cell cycle inhibitors. Arab. J. Chem. 2019, 12, 816–824. [Google Scholar] [CrossRef]
- Novais, P.; Silva, P.M.A.; Amorim, I.; Bousbaa, H. Second-Generation Antimitotics in Cancer Clinical Trials. Pharmaceutics 2021, 13, 1011. [Google Scholar] [CrossRef]
- Bebbington, D.; Binch, H.; Charrier, J.-D.; Everitt, S.; Fraysse, D.; Golec, J.; Kay, D.; Knegtel, R.; Mak, C.; Mazzei, F.; et al. The discovery of the potent aurora inhibitor MK-0457 (VX-680). Bioorg. Med. Chem. Lett. 2009, 19, 3586–3592. [Google Scholar] [CrossRef]
- Zhao, B.; Smallwood, A.; Yang, J.; Koretke, K.; Nurse, K.; Calamari, A.; Kirkpatrick, R.B.; Lai, Z. Modulation of kinase-inhibitor interactions by auxiliary protein binding: Crystallography studies on Aurora A interactions with VX-680 and with TPX2. Protein Sci. 2008, 17, 1791–1797. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, Y.; Wu, J.; Shen, Y.; Cheng, H.; Xiang, Y. Exploration of the selective binding mechanism of protein kinase Aurora A selectivity via a comprehensive molecular modeling study. PeerJ 2019, 7, e7832. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Mayer, R.; Venook, A.P.; O’Neill, A.F.; Beg, M.S.; LaQuaglia, M.; Kingham, P.T.; Kobos, R.; Basturk, O.; Brennan, C.; et al. Phase II Multicenter, Open-Label Study of Oral ENMD-2076 for the Treatment of Patients with Advanced Fibrolamellar Carcinoma. Oncologist 2020, 25, e1837–e1845. [Google Scholar] [CrossRef]
- Fletcher, G.C.; Brokx, R.D.; Denny, T.A.; Hembrough, T.A.; Plum, S.M.; Fogler, W.E.; Sidor, C.F.; Bray, M.R. ENMD-2076 Is an Orally Active Kinase Inhibitor with Antiangiogenic and Antiproliferative Mechanisms of Action. Mol. Cancer Ther. 2011, 10, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Choi, M.Y.; Min, K.Y.; Jo, M.G.; Kim, J.M.; Kim, H.S.; Kim, Y.M. AT9283, 1-Cyclopropyl-3-(3-(5-(Morpholinomethyl)-1H-Benzo[d] Imidazole-2-yl)-1H-Pyrazol-4-yl) Urea, Inhibits Syk to Suppress Mast Cell-Mediated Allergic Response. Biomol. Ther. 2022, 30, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.; Berdini, V.; Boulstridge, J.A.; Carr, M.G.; Cross, D.M.; Curry, J.; Devine, L.A.; Early, T.R.; Fazal, L.; Gill, A.L.; et al. Fragment-based discovery of the pyrazol-4-yl urea (AT9283), a multitargeted kinase inhibitor with potent aurora kinase activity. J. Med. Chem. 2009, 52, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Glaser, K.B.; Li, J.; Marcotte, P.A.; Magoc, T.J.; Guo, J.; Reuter, D.R.; Tapang, P.; Wei, R.-Q.; Pease, L.J.; Bui, M.H.; et al. Preclinical Characterization of ABT-348, a Kinase Inhibitor Targeting the Aurora, Vascular Endothelial Growth Factor Receptor/Platelet-Derived Growth Factor Receptor, and Src Kinase Families. Experiment 2012, 343, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Maitland, M.L.; Piha-Paul, S.; Falchook, G.; Kurzrock, R.; Nguyen, L.; Janisch, L.; Karovic, S.; McKee, M.; Hoening, E.; Wong, S.; et al. Clinical pharmacodynamic/exposure characterisation of the multikinase inhibitor ilorasertib (ABT-348) in a phase 1 dose-escalation trial. Br. J. Cancer 2018, 118, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Curtin, M.L.; Frey, R.R.; Heyman, H.R.; Soni, N.B.; Marcotte, P.A.; Pease, L.J.; Glaser, K.B.; Magoc, T.J.; Tapang, P.; Albert, D.H.; et al. Thienopyridine ureas as dual inhibitors of the VEGF and Aurora kinase families. Bioorg. Med. Chem. Lett. 2012, 22, 3208–3212. [Google Scholar] [CrossRef] [PubMed]
- Dennis, M.; Davies, M.; Oliver, S.; D’souza, R.; Pike, L.; Stockman, P. Phase I study of the Aurora B kinase inhibitor barasertib (AZD1152) to assess the pharmacokinetics, metabolism and excretion in patients with acute myeloid leukemia. Cancer Chemother. Pharmacol. 2012, 70, 461–469. [Google Scholar] [CrossRef]
- Mortlock, A.A.; Foote, K.M.; Heron, N.M.; Jung, F.H.; Pasquet, G.; Lohmann, J.-J.M.; Warin, N.; Renaud, F.; De Savi, C.; Roberts, N.J.; et al. Discovery, Synthesis, and in Vivo Activity of a New Class of Pyrazoloquinazolines as Selective Inhibitors of Aurora B Kinase. J. Med. Chem. 2007, 50, 2213–2224. [Google Scholar] [CrossRef]
- Borah, N.A.; Reddy, M.M. Aurora Kinase B Inhibition: A Potential Therapeutic Strategy for Cancer. Molecules 2021, 26, 1981. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, A.H.; Zhao, D.; Hou, J. Aurora B Inhibitors as Cancer Therapeutics. Molecules 2023, 28, 3385. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Bachegowda, L.; Morrone, K.; Winski, S.L.; Mantzaris, I.; Bartenstein, M.; Ramachandra, N.; Giricz, O.; Sukrithan, V.; Nwankwo, G.; Shahnaz, S.; et al. Pexmetinib: A Novel Dual Inhibitor of Tie2 and p38 MAPK with Efficacy in Preclinical Models of Myelodysplastic Syndromes and Acute Myeloid Leukemia. Cancer Res. 2016, 76, 4841–4849. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Y.; Xu, Y.; Sun, Q.; Liu, H.; Chen, Q.; Liu, B. BIRB796, an Inhibitor of p38 Mitogen-Activated Protein Kinase, Inhibits Proliferation and Invasion in Glioblastoma Cells. ACS Omega 2021, 6, 11466–11473. [Google Scholar] [CrossRef]
- Patterson, H.; Nibbs, R.; McInnes, I.; Siebert, S. Protein kinase inhibitors in the treatment of inflammatory and autoimmune diseases. Clin. Exp. Immunol. 2014, 176, 1–10. [Google Scholar] [CrossRef]
- Regan, J.; Capolino, A.; Cirillo, P.F.; Gilmore, T.; Graham, A.G.; Hickey, E.; Kroe, R.R.; Madwed, J.; Moriak, M.; Nelson, R.; et al. Structure−Activity Relationships of the p38α MAP Kinase Inhibitor 1-(5-tert-Butyl-2-p-tolyl-2H-pyrazol-3-yl)-3-[4-(2-morpholin-4-yl-ethoxy)naph- thalen-1-yl]urea (BIRB 796). J. Med. Chem. 2003, 46, 4676–4686. [Google Scholar] [CrossRef]
- Strâmbu, I.R.; Kobalava, Z.D.; Magnusson, B.P.; MacKinnon, A.; Parkin, J.M. Phase II Study of Single/Repeated Doses of Acumapimod (BCT197) to Treat Acute Exacerbations of COPD. COPD J. Chronic Obstr. Pulm. Dis. 2019, 16, 344–353. [Google Scholar] [CrossRef]
- Cassanello, G.; Pasquale, R.; Barcellini, W.; Fattizzo, B. Novel Therapies for Unmet Clinical Needs in Myelodysplastic Syndromes. Cancers 2022, 14, 4941. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R.J. Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol. Res. 2019, 142, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Blake, J.F.; Burkard, M.; Chan, J.; Chen, H.; Chou, K.-J.; Diaz, D.; Dudley, D.A.; Gaudino, J.J.; Gould, S.E.; Grina, J.; et al. Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development. J. Med. Chem. 2016, 59, 5650–5660. [Google Scholar] [CrossRef]
- Varga, A.; Soria, J.-C.; Hollebecque, A.; LoRusso, P.; Bendell, J.; Huang, S.-M.A.; Wagle, M.-C.; Okrah, K.; Liu, L.; Murray, E.; et al. A First-in-Human Phase I Study to Evaluate the ERK1/2 Inhibitor GDC-0994 in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2020, 26, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Schreck, K.C.; Grossman, S.A.; Pratilas, C.A. BRAF Mutations and the Utility of RAF and MEK Inhibitors in Primary Brain Tumors. Cancers 2019, 11, 1262. [Google Scholar] [CrossRef]
- Roskoski, R. RAF protein-serine/threonine kinases: Structure and regulation. Biochem. Biophys. Res. Commun. 2010, 399, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Frisone, D.; Friedlaender, A.; Malapelle, U.; Banna, G.; Addeo, A. A BRAF new world. Crit. Rev. Oncol. Hematol. 2020, 152, 103008. [Google Scholar] [CrossRef] [PubMed]
- Zaman, A.; Wu, W.; Bivona, T.G. Targeting Oncogenic BRAF: Past, Present, and Future. Cancers 2019, 11, 1197. [Google Scholar] [CrossRef]
- Subbiah, V.; Baik, C.; Kirkwood, J.M. Clinical Development of BRAF plus MEK Inhibitor Combinations. Trends Cancer 2020, 6, 797–810. [Google Scholar] [CrossRef]
- Brummer, T.; McInnes, C. RAF kinase dimerization: Implications for drug discovery and clinical outcomes. Oncogene 2020, 39, 4155–4169. [Google Scholar] [CrossRef]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef]
- Chavda, J.; Bhatt, H. Systemic review on B-RafV600E mutation as potential therapeutic target for the treatment of cancer. Eur. J. Med. Chem. 2020, 206, 112675. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Maksoud, M.S.; El-Gamal, M.I.; El-Din, M.M.G.; Oh, C.H. Design, synthesis, in vitro anticancer evaluation, kinase inhibitory effects, and pharmacokinetic profile of new 1,3,4-triarylpyrazole derivatives possessing terminal sulfonamide moiety. J. Enzym. Inhib. Med. Chem. 2019, 34, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Khan, P.S.; Rajesh, P.; Rajendra, P.; Chaskar, M.G.; Rohidas, A.; Jaiprakash, S. Recent advances in B-RAF inhibitors as anticancer agents. Bioorg. Chem. 2022, 120, 105597. [Google Scholar] [CrossRef] [PubMed]
- El-Din, M.M.G.; El-Gamal, M.I.; Abdel-Maksoud, M.S.; Yoo, K.H.; Oh, C.-H. Design, synthesis, broad-spectrum antiproliferative activity, and kinase inhibitory effect of triarylpyrazole derivatives possessing arylamides or arylureas moieties. Eur. J. Med. Chem. 2016, 119, 122–131. [Google Scholar] [CrossRef]
- Gamal El-Din, M.M.; El-Gamal, M.I.; Abdel-Maksoud, M.S.; Yoo, K.H.; Baek, D.; Choi, J.; Lee, H.; Oh, C.H. Design, synthesis, and in vitro antiproliferative and kinase inhibitory effects of pyrimidinylpyrazole derivatives terminating with arylsulfonamido or cyclic sulfamide substituents. J. Enzym. Inhib. Med. Chem. 2016, 31, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Ammar, U.M.; Abdel-Maksoud, M.S.; Ali, E.M.; Mersal, K.I.; Yoo, K.H.; Oh, C.-H. Structural optimization of imidazothiazole derivatives affords a new promising series as B-Raf V600E inhibitors; synthesis, in vitro assay and in silico screening. Bioorg. Chem. 2020, 100, 103967. [Google Scholar] [CrossRef] [PubMed]
- Ros, J.; Saoudi, N.; Baraibar, I.; Salva, F.; Tabernero, J.; Elez, E. Encorafenib plus cetuximab for the treatment of BRAF-V600E-mutated metastatic colorectal cancer. Ther. Adv. Gastroenterol. 2022, 15, 17562848221110644. [Google Scholar] [CrossRef]
- Ayala-Aguilera, C.C.; Valero, T.; Lorente-Macías, Á.; Baillache, D.J.; Croke, S.; Unciti-Broceta, A. Small Molecule Kinase Inhibitor Drugs (1995–2021): Medical Indication, Pharmacology, and Synthesis. J. Med. Chem. 2022, 65, 1047–1131. [Google Scholar] [CrossRef]
- Angius, G.; Tomao, S.; Stati, V.; Vici, P.; Bianco, V.; Tomao, F. Prexasertib, a checkpoint kinase inhibitor: From preclinical data to clinical development. Cancer Chemother. Pharmacol. 2020, 85, 9–20. [Google Scholar] [CrossRef]
- King, C.; Diaz, H.B.; McNeely, S.; Barnard, D.; Dempsey, J.; Blosser, W.; Beckmann, R.; Barda, D.; Marshall, M.S. LY2606368 Causes Replication Catastrophe and Antitumor Effects through CHK1-Dependent Mechanisms. Mol. Cancer Ther. 2015, 14, 2004–2013. [Google Scholar] [CrossRef]
- Belkadi, A.; Kenouche, S.; Melkemi, N.; Daoud, I.; Djebaili, R. Molecular docking/dynamic simulations, MEP, ADME-TOX-based analysis of xanthone derivatives as CHK1 inhibitors. Struct. Chem. 2022, 33, 833–858. [Google Scholar] [CrossRef]
- Yu, P.; Laird, A.D.; Du, X.; Wu, J.; Won, K.-A.; Yamaguchi, K.; Hsu, P.P.; Qian, F.; Jaeger, C.T.; Zhang, W.; et al. Characterization of the Activity of the PI3K/mTOR Inhibitor XL765 (SAR245409) in Tumor Models with Diverse Genetic Alterations Affecting the PI3K Pathway. Mol. Cancer Ther. 2014, 13, 1078–1091. [Google Scholar] [CrossRef]
- Marone, R.; Cmiljanovic, V.; Giese, B.; Wymann, M.P. Targeting phosphoinositide 3-kinase—Moving towards therapy. Biochim. Biophys. Acta-Proteins Proteom. 2008, 1784, 159–185. [Google Scholar] [CrossRef] [PubMed]
- Montagnoli, A.; Moll, J.; Colotta, F. Targeting Cell Division Cycle 7 Kinase: A New Approach for Cancer Therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 4503–4508. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Diderichsen, P.M.; Gupta, N. Assessment of Effects of Investigational TAK-931, an Oral Cell Division Cycle 7 Kinase Inhibitor on the QTc Intervals in Patients With Advanced Solid Tumors. Clin. Pharmacol. Drug Dev. 2022, 11, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Kurasawa, O.; Homma, M.; Oguro, Y.; Miyazaki, T.; Mori, K.; Uchiyama, N.; Iwai, K.; Ohashi, A.; Hara, H.; Yoshida, S.; et al. 2-Aminomethylthieno[3,2-d]pyrimidin-4(3H)-ones bearing 3-methylpyrazole hinge binding moiety: Highly potent, selective, and time-dependent inhibitors of Cdc7 kinase. Bioorg. Med. Chem. 2017, 25, 3658–3670. [Google Scholar] [CrossRef]
- Kurasawa, O.; Miyazaki, T.; Homma, M.; Oguro, Y.; Imada, T.; Uchiyama, N.; Iwai, K.; Yamamoto, Y.; Ohori, M.; Hara, H.; et al. Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 Inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Antitumor Agent. J. Med. Chem. 2020, 63, 1084–1104. [Google Scholar] [CrossRef] [PubMed]
- Kuboki, Y.; Shimizu, T.; Yonemori, K.; Kojima, T.; Kondo, S.; Koganemaru, S.; Iwasa, S.; Harano, K.; Koyama, T.; Lu, V.; et al. Safety, Tolerability, and Pharmacokinetics of TAK-931, a Cell Division Cycle 7 Inhibitor, in Patients with Advanced Solid Tumors: A Phase I First-in-Human Study. Cancer Res. Commun. 2022, 2, 1426–1435. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Shawky, A.M.; Almalki, F.A.; Abdalla, A.N.; Abdelazeem, A.H.; Gouda, A.M. A Comprehensive Overview of Globally Approved JAK Inhibitors. Pharmaceutics 2022, 14, 1001. [Google Scholar] [CrossRef]
- Klein, K.; Stoiber, D.; Sexl, V.; Witalisz-Siepracka, A. Untwining Anti-Tumor and Immunosuppressive Effects of JAK Inhibitors—A Strategy for Hematological Malignancies? Cancers 2021, 13, 2611. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Vaddi, K.; Liu, P.; Manshouri, T.; Li, J.; Scherle, P.A.; Caulder, E.; Wen, X.; Li, Y.; Waeltz, P.; et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: Therapeutic implications for the treatment of myeloproliferative neoplasms. Blood 2010, 115, 3109–3117. [Google Scholar] [CrossRef]
- Davis, R.R.; Li, B.; Yun, S.Y.; Chan, A.; Nareddy, P.; Gunawan, S.; Ayaz, M.; Lawrence, H.R.; Reuther, G.W.; Lawrence, N.J.; et al. Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof. J. Med. Chem. 2021, 64, 2228–2241. [Google Scholar] [CrossRef]
- Hoy, S.M. Ruxolitinib Cream 1.5%: A Review in Mild to Moderate Atopic Dermatitis. Am. J. Clin. Dermatol. 2023, 24, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Assadiasl, S.; Fatahi, Y.; Mosharmovahed, B.; Mohebbi, B.; Nicknam, M.H. Baricitinib: From Rheumatoid Arthritis to COVID-19. J. Clin. Pharmacol. 2021, 61, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Alavi, A.; Hamzavi, I.; Brown, K.; Santos, L.L.; Zhu, Z.; Liu, H.; Howell, M.D.; Kirby, J.S. Janus kinase 1 inhibitor INCB054707 for patients with moderate-to-severe hidradenitis suppurativa: Results from two phase II studies. Br. J. Dermatol. 2022, 186, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Dell’Avalle, C.; D’Amico, F.; Gabbiadini, R.; Dal Buono, A.; Pugliese, N.; Zilli, A.; Furfaro, F.; Fiorino, G.; Allocca, M.; Peyrin-Biroulet, L.; et al. JAK inhibitors in crohn’s disease: Ready to go? Expert Opin. Investig. Drugs 2022, 31, 145–161. [Google Scholar] [CrossRef]
- Page, K.M.; Suarez-Farinas, M.; Suprun, M.; Zhang, W.; Garcet, S.; Fuentes-Duculan, J.; Li, X.; Scaramozza, M.; Kieras, E.; Banfield, C.; et al. Molecular and Cellular Responses to the TYK2/JAK1 Inhibitor PF-06700841 Reveal Reduction of Skin Inflammation in Plaque Psoriasis. J. Invest. Dermatol. 2020, 140, 1546–1555. [Google Scholar] [CrossRef]
- Garmezy, B.; Schaefer, J.K.; Mercer, J.; Talpaz, M. A provider’s guide to primary myelofibrosis: Pathophysiology, diagnosis, and management. Blood Rev. 2021, 45, 100691. [Google Scholar] [CrossRef]
- Chifotides, H.T.; Bose, P.; Verstovsek, S. Momelotinib: An emerging treatment for myelofibrosis patients with anemia. J. Hematol. Oncol. 2022, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Beinhoff, P.; Sabharwal, L.; Udhane, V.; Maranto, C.; LaViolette, P.S.; Jacobsohn, K.M.; Tsai, S.; Iczkowski, K.A.; Wang, L.; Hall, W.A.; et al. Second-Generation Jak2 Inhibitors for Advanced Prostate Cancer: Are We Ready for Clinical Development? Cancers 2021, 13, 5204. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Clayton, J.R.; Walgren, R.; Zhao, B.; Evans, R.J.; Smith, M.C.; Heinz-Taheny, K.M.; Kreklau, E.L.; Bloem, L.; Pitou, C.; et al. Discovery and characterization of LY2784544, a small-molecule tyrosine kinase inhibitor of JAK2V617F. Blood Cancer J. 2013, 3, e109. [Google Scholar] [CrossRef] [PubMed]
- Verstovsek, S.; Mesa, R.A.; Salama, M.E.; Li, L.; Pitou, C.; Nunes, F.P.; Price, G.L.; Giles, J.L.; D’Souza, D.N.; Walgren, R.A.; et al. A phase 1 study of the Janus kinase 2 (JAK2)(V617F) inhibitor, gandotinib (LY2784544), in patients with primary myelofibrosis, polycythemia vera, and essential thrombocythemia. Leuk. Res. 2017, 61, 89–95. [Google Scholar] [CrossRef]
- Xu, P.; Shen, P.; Yu, B.; Xu, X.; Ge, R.; Cheng, X.; Chen, Q.; Bian, J.; Li, Z.; Wang, J. Janus kinases (JAKs): The efficient therapeutic targets for autoimmune diseases and myeloproliferative disorders. Eur. J. Med. Chem. 2020, 192, 112155. [Google Scholar] [CrossRef]
- Nakaya, Y.; Shide, K.; Naito, H.; Niwa, T.; Horio, T.; Miyake, J.; Shimoda, K. Effect of NS-018, a selective JAK2V617F inhibitor, in a murine model of myelofibrosis. Blood Cancer J. 2014, 4, e174. [Google Scholar] [CrossRef]
- Ren, R. Mechanisms of BCR–ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer 2005, 5, 172–183. [Google Scholar] [CrossRef]
- Loscocco, F.; Visani, G.; Galimberti, S.; Curti, A.; Isidori, A. BCR-ABL Independent Mechanisms of Resistance in Chronic Myeloid Leukemia. Front. Oncol. 2019, 9, 939. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Huang, H.; Lei, X.; Tang, G.; Cao, X.; Peng, J. Recent advances in Bcr-Abl tyrosine kinase inhibitors for overriding T315I mutation. Chem. Biol. Drug Des. 2021, 97, 649–664. [Google Scholar] [CrossRef]
- Roskoski, R. Targeting BCR-Abl in the treatment of Philadelphia-chromosome positive chronic myelogenous leukemia. Pharmacol. Res. 2022, 178, 106156. [Google Scholar] [CrossRef]
- Amarante-Mendes, G.P.; Rana, A.; Datoguia, T.S.; Hamerschlak, N.; Brumatti, G. BCR-ABL1 Tyrosine Kinase Complex Signaling Transduction: Challenges to Overcome Resistance in Chronic Myeloid Leukemia. Pharmaceutics 2022, 14, 215. [Google Scholar] [CrossRef] [PubMed]
- Carofiglio, F.; Lopalco, A.; Lopedota, A.; Cutrignelli, A.; Nicolotti, O.; Denora, N.; Stefanachi, A.; Leonetti, F. Bcr-Abl Tyrosine Kinase Inhibitors in the Treatment of Pediatric CML. Int. J. Mol. Sci. 2020, 21, 4469. [Google Scholar] [CrossRef] [PubMed]
- Réa, D.; Hughes, T.P. Development of asciminib, a novel allosteric inhibitor of BCR-ABL1. Crit. Rev. Oncol. Hematol. 2022, 171, 103580. [Google Scholar] [CrossRef] [PubMed]
- El-Gamal, M.I.; Zaraei, S.-O.; Madkour, M.M.; Anbar, H.S. Evaluation of Substituted Pyrazole-Based Kinase Inhibitors in One Decade (2011–2020): Current Status and Future Prospects. Molecules 2022, 27, 330. [Google Scholar] [CrossRef]
- Wang, Q.; Han, J.; Sorochinsky, A.; Landa, A.; Butler, G.; Soloshonok, V.A. The Latest FDA-Approved Pharmaceuticals Containing Fragments of Tailor-Made Amino Acids and Fluorine. Pharmaceuticals 2022, 15, 999. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhuang, Y.; Chen, Q. Current scenario of pyrazole hybrids with in vivo therapeutic potential against cancers. Eur. J. Med. Chem. 2023, 257, 115495. [Google Scholar] [CrossRef]
- Teng, M.; Luskin, M.R.; Cowan-Jacob, S.W.; Ding, Q.; Fabbro, D.; Gray, N.S. The Dawn of Allosteric BCR-ABL1 Drugs: From a Phenotypic Screening Hit to an Approved Drug. J. Med. Chem. 2022, 65, 7581–7594. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Z.; Feng, J.; Pan, G.; He, X.; Lv, M.; Chen, H.; Jiang, W.; Ji, J.; Yang, M. Synthesis and biological evaluation of novel aromatic amide derivatives as potential BCR-ABL inhibitors. Bioorg. Med. Chem. Lett. 2023, 81, 129144. [Google Scholar] [CrossRef]
- He, J.; Li, Z.; Dhawan, G.; Zhang, W.; Sorochinsky, A.E.; Butler, G.; Soloshonok, V.A.; Han, J. Fluorine-containing drugs approved by the FDA in 2021. Chin. Chem. Lett. 2022, 34, 107578. [Google Scholar] [CrossRef]
- Manley, P.W.; Huth, F.; Moussaoui, S.; Schoepfer, J. A kinase inhibitor which specifically targets the ABL myristate pocket (STAMP), but unlike asciminib crosses the blood–brain barrier. Bioorg. Med. Chem. Lett. 2023, 59, 128577. [Google Scholar] [CrossRef]
- Tanaka, M.; Siemann, D.W. Therapeutic Targeting of the Gas6/Axl Signaling Pathway in Cancer. Int. J. Mol. Sci. 2021, 22, 9953. [Google Scholar] [CrossRef]
- Chan, W.W.; Wise, S.C.; Kaufman, M.D.; Ahn, Y.M.; Ensinger, C.L.; Haack, T.; Hood, M.M.; Jones, J.; Lord, J.W.; Lu, W.P.; et al. Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036. Cancer Cell 2011, 19, 556–568. [Google Scholar] [CrossRef]
- Phadke, S.; Lopez-Barcons, L.; Vandecan, N.; Wu, Z.; Johnson, T.K.; Lachacz, E.J.; Merajver, S.D.; Soellner, M.B. Insights into the modular design of kinase inhibitors and application to Abl and Axl. RSC Med. Chem. 2022, 13, 64–71. [Google Scholar] [CrossRef]
- Pan, Y.-L.; Zeng, S.-X.; Hao, R.-R.; Liang, M.-H.; Shen, Z.-R.; Huang, W.-H. The progress of small-molecules and degraders against BCR-ABL for the treatment of CML. Eur. J. Med. Chem. 2022, 238, 114442. [Google Scholar] [CrossRef]
- Fang, L.; Vilas-Boas, J.; Chakraborty, S.; Potter, Z.E.; Register, A.C.; Seeliger, M.A.; Maly, D.J. How ATP-Competitive Inhibitors Allosterically Modulate Tyrosine Kinases That Contain a Src-like Regulatory Architecture. ACS Chem. Biol. 2020, 15, 2005–2016. [Google Scholar] [CrossRef]
- van Linden, O.P.J.; Kooistra, A.J.; Leurs, R.; de Esch, I.J.P.; de Graaf, C. KLIFS: A Knowledge-Based Structural Database To Navigate Kinase–Ligand Interaction Space. J. Med. Chem. 2014, 57, 249–277. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Talpaz, M.; Smith, H.P.; Snyder, D.S.; Khoury, J.; Bhalla, K.N.; Pinilla-Ibarz, J.; Larson, R.; Mitchell, D.; Wise, S.C.; et al. Phase 1 dose-finding study of rebastinib (DCC-2036) in patients with relapsed chronic myeloid leukemia and acute myeloid leukemia. Haematologica 2017, 102, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.-P.; Tong, X.-M.; Wang, M.-H. Oncogenic mechanism-based pharmaceutical validation of therapeutics targeting MET receptor tyrosine kinase. Ther. Adv. Med. Oncol. 2021, 13, 17588359211006957. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Rao, B.; Lou, J.; Li, J.; Liu, Z.; Li, A.; Cui, G.; Ren, Z.; Yu, Z. The Function of the HGF/c-Met Axis in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2020, 8, 55. [Google Scholar] [CrossRef]
- Rehman, S.; Dy, G.K. MET Inhibition in Non-Small Cell Lung Cancer. Eur. Med. J. 2019, 4, 100–111. [Google Scholar] [CrossRef]
- Gavine, P.R.; Ren, Y.; Han, L.; Lv, J.; Fan, S.; Zhang, W.; Xu, W.; Liu, Y.J.; Zhang, T.; Fu, H.; et al. Volitinib, a potent and highly selective c-Met inhibitor, effectively blocks c-Met signaling and growth in c-MET amplified gastric cancer patient-derived tumor xenograft models. Mol. Oncol. 2015, 9, 323–333. [Google Scholar] [CrossRef]
- Parikh, P.K.; Ghate, M.D. Recent advances in the discovery of small molecule c-Met Kinase inhibitors. Eur. J. Med. Chem. 2018, 143, 1103–1138. [Google Scholar] [CrossRef]
- Van Der Steen, N.; Giovannetti, E.; Pauwels, P.; Peters, G.J.; Hong, D.S.; Cappuzzo, F.; Hirsch, F.R.; Rolfo, C. cMET Exon 14 Skipping: From the Structure to the Clinic. J. Thorac. Oncol. 2016, 11, 1423–1432. [Google Scholar] [CrossRef]
- Liang, H.; Wang, M. MET Oncogene in Non-Small Cell Lung Cancer: Mechanism of MET Dysregulation and Agents Targeting the HGF/c-Met Axis. OncoTargets Ther. 2020, 13, 2491–2510. [Google Scholar] [CrossRef]
- Giunta, E.F.; Signori, A.; West, H.J.; Metro, G.; Friedlaender, A.; Parikh, K.; Banna, G.L.; Addeo, A. Beyond Crizotinib: A Systematic Review and Meta-Analysis of the Next-Generation ALK Inhibitors as First-Line Treatment for ALK-Translocated Lung Cancer. Front. Oncol. 2022, 12, 921854. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, Q.; Zhang, L.; Hu, S.; Chen, T.; Li, H.; Chen, Y.; Xu, Y.; Lu, T. Discovery, optimization and biological evaluation for novel c-Met kinase inhibitors. Eur. J. Med. Chem. 2018, 143, 491–502. [Google Scholar] [CrossRef]
- Collie, G.W.; Koh, C.M.; O’neill, D.J.; Stubbs, C.J.; Khurana, P.; Eddershaw, A.; Snijder, A.; Mauritzson, F.; Barlind, L.; Dale, I.L.; et al. Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors. ACS Med. Chem. Lett. 2019, 10, 1322–1327. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.J.; Tran-Dubé, M.; Shen, H.; Nambu, M.; Kung, P.-P.; Pairish, M.; Jia, L.; Meng, J.; Funk, L.; Botrous, I.; et al. Structure Based Drug Design of Crizotinib (PF-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal–Epithelial Transition Factor (c-MET) Kinase and Anaplastic Lymphoma Kinase (ALK). J. Med. Chem. 2011, 54, 6342–6363. [Google Scholar] [CrossRef] [PubMed]
- Heigener, D.F.; Reck, M. Crizotinib. In Small Molecules in Oncology; Recent Results in Cancer Research; Martens, U.M., Ed.; Springer International Publishing: Cham, Switzerland, 2018; Volume 211, pp. 57–65. [Google Scholar]
- Drilon, A.; Clark, J.W.; Weiss, J.; Ou, S.-H.I.; Camidge, D.R.; Solomon, B.J.; Otterson, G.A.; Villaruz, L.C.; Riely, G.J.; Heist, R.S.; et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat. Med. 2020, 26, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Bolcaen, J.; Nair, S.; Driver, C.H.S.; Boshomane, T.M.G.; Ebenhan, T.; Vandevoorde, C. Novel Receptor Tyrosine Kinase Pathway Inhibitors for Targeted Radionuclide Therapy of Glioblastoma. Pharmaceuticals 2021, 14, 626. [Google Scholar] [CrossRef]
- Noonan, A.; Pawlik, T.M. Hepatocellular carcinoma: An update on investigational drugs in phase I and II clinical trials. Expert Opin. Investig. Drugs 2019, 28, 941–949. [Google Scholar] [CrossRef]
- Cao, Y.; Luo, C.; Yang, P.; Li, P.; Wu, C. Indazole scaffold: A generalist for marketed and clinical drugs. Med. Chem. Res. 2021, 30, 501–518. [Google Scholar] [CrossRef]
- Ai, J.; Chen, Y.; Peng, X.; Ji, Y.; Xi, Y.; Shen, Y.; Yang, X.; Su, Y.; Sun, Y.-M.; Gao, Y.; et al. Preclinical Evaluation of SCC244 (Glumetinib), a Novel, Potent, and Highly Selective Inhibitor of c-Met in MET-dependent Cancer Models. Mol. Cancer Ther. 2018, 17, 751–762. [Google Scholar] [CrossRef]
- Lu, S.; Yu, Y.; Zhou, J.; Goto, K.; Li, X.; Sakakibara-Konishi, J.; Nishino, K.; Kentaro, T.; Wu, L.; Min, X.; et al. Abstract CT034: Phase II study of SCC244 in NSCLC patients harboring MET exon 14 skipping (METex14) mutations (GLORY study). Cancer Res. 2022, 82, CT034. [Google Scholar] [CrossRef]
- Brazel, D.; Zhang, S.; Nagasaka, M. Spotlight on Tepotinib and Capmatinib for Non-Small Cell Lung Cancer with MET Exon 14 Skipping Mutation. Lung Cancer Targets Ther. 2022, 13, 33–45. [Google Scholar] [CrossRef]
- Shah, R.; Alex, D.; Xu, Z. MET Exon 14 Skipping Alterations in Non-small Cell Lung Carcinoma—Current Understanding and Therapeutic Advances. Oncol. Hematol. Rev. 2021, 16, 100. [Google Scholar] [CrossRef]
- Majeed, U.; Manochakian, R.; Zhao, Y.; Lou, Y. Targeted therapy in advanced non-small cell lung cancer: Current advances and future trends. J. Hematol. Oncol. 2021, 14, 108. [Google Scholar] [CrossRef]
- Cui, J.J.; McTigue, M.; Nambu, M.; Tran-Dubé, M.; Pairish, M.; Shen, H.; Jia, L.; Cheng, H.; Hoffman, J.; Le, P.; et al. Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (c-MET) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-ylmethyl)-1H-[1,2,3]triazolo[4,5-b]pyrazin-6-yl)-1H-pyrazol-1-y. J. Med. Chem. 2012, 55, 8091–8109. [Google Scholar] [CrossRef]
- Batra, U.; Nathany, S. MET: A narrative review of exon 14 skipping mutation in non-small-cell lung carcinoma. Cancer Res. Stat. Treat. 2022, 5, 284. [Google Scholar] [CrossRef]
- Prins, P.; Al-Hajeili, M.; Kim, K.; Hwang, J.; Hartley, M.; He, A. MET inhibition and merestinib (LY-2801653) for cancer treatment. Drugs Futur. 2016, 41, 0607. [Google Scholar] [CrossRef]
- Puccini, A.; Marín-Ramos, N.I.; Bergamo, F.; Schirripa, M.; Lonardi, S.; Lenz, H.-J.; Loupakis, F.; Battaglin, F. Safety and Tolerability of c-MET Inhibitors in Cancer. Drug Saf. 2019, 42, 211–233. [Google Scholar] [CrossRef]
- Yan, S.B.; Peek, V.L.; Ajamie, R.; Buchanan, S.G.; Graff, J.R.; Heidler, S.A.; Hui, Y.-H.; Huss, K.L.; Konicek, B.W.; Manro, J.R.; et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Investig. New Drugs 2013, 31, 833–844. [Google Scholar] [CrossRef]
- Fujino, T.; Suda, K.; Mitsudomi, T. Emerging MET tyrosine kinase inhibitors for the treatment of non-small cell lung cancer. Expert Opin. Emerg. Drugs 2020, 25, 229–249. [Google Scholar] [CrossRef]
- Fujino, T.; Suda, K.; Koga, T.; Hamada, A.; Ohara, S.; Chiba, M.; Shimoji, M.; Takemoto, T.; Soh, J.; Mitsudomi, T. Foretinib can overcome common on-target resistance mutations after capmatinib/tepotinib treatment in NSCLCs with MET exon 14 skipping mutation. J. Hematol. Oncol. 2022, 15, 79. [Google Scholar] [CrossRef]
- Kosciuczuk, E.M.; Saleiro, D.; Kroczynska, B.; Beauchamp, E.M.; Eckerdt, F.; Blyth, G.T.; Abedin, S.M.; Giles, F.J.; Altman, J.K.; Platanias, L.C. Merestinib blocks Mnk kinase activity in acute myeloid leukemia progenitors and exhibits antileukemic effects in vitro and in vivo. Blood 2016, 128, 410–414. [Google Scholar] [CrossRef]
- Yan, S.-C.B.; Um, S.L.; Peek, V.L.; Thobe, M.N.; Credille, K.M.; Stephens, J.R.; Manro, J.R.; Ballard, D.W.; Baker, J.A.; Cook, J.D.; et al. Abstract 4403: Preclinical evaluation of LY2801653, an orally bioavailable small molecule oncokinase inhibitor, in cholangiocarcinoma models. Cancer Res. 2014, 74, 4403. [Google Scholar] [CrossRef]
- Kawada, I.; Hasina, R.; Arif, Q.; Mueller, J.; Smithberger, E.; Husain, A.N.; Vokes, E.E.; Salgia, R. Dramatic Antitumor Effects of the Dual MET/RON Small-Molecule Inhibitor LY2801653 in Non–Small Cell Lung Cancer. Cancer Res. 2014, 74, 884–895. [Google Scholar] [CrossRef]
- Gan, H.K.; Lickliter, J.; Millward, M.; Gu, Y.; Su, W.; Frigault, M.; Qi, C.; Mu, H. First-in-human phase I study of a selective c-Met inhibitor volitinib (HMP504/AZD6094) in patients with advanced solid tumors. J. Clin. Oncol. 2014, 32, 11111. [Google Scholar] [CrossRef]
- Jones, R.; Cheung, A.; Coleman, T.; Ballard, P.; D’Cruz, C.; Schuller, A.; Frigault, M.; Gu, Y.; Sai, Y.; Weiguo, S.; et al. 392 Using modelling & simulation to integrate mouse PK–PD-efficacy with preliminary human PK data to inform the Phase II doses and schedule for the experimental c-Met inhibitor AZD6094 (Volitinib). Eur. J. Cancer 2014, 50, 125–126. [Google Scholar] [CrossRef]
- Zhou, F.; Ren, Y.; Cui, Y.; Chen, H.; Jiao, L.; Dai, G.; Fan, S.; Sun, J.; Yu, Y.; Sai, Y.; et al. Abstract 971: Synergistic effect of c-Met inhibitor volitinib in combination with EGFR inhibitor Gefitnib on EGFR-TKI resistant NSCLC model HCC827C4R harboring acquired Met gene amplification. Cancer Res. 2013, 73, 971. [Google Scholar] [CrossRef]
- Cui, Y.; Dai, G.; Ren, Y.; Zhou, F.; Fan, S.; Sai, Y.; Gu, Y.; Yan, J.; Li, J.; Qing, W.; et al. Abstract 3612: A novel and selective c-Met inhibitor against subcutaneous xenograft and othotopic brain tumor models. Cancer Res. 2011, 71, 3612. [Google Scholar] [CrossRef]
- Schuller, A.G.; Barry, E.R.; Jones, R.D.O.; Henry, R.E.; Frigault, M.M.; Beran, G.; Linsenmayer, D.; Hattersley, M.; Smith, A.; Wilson, J.; et al. The MET Inhibitor AZD6094 (Savolitinib, HMPL-504) Induces Regression in Papillary Renal Cell Carcinoma Patient–Derived Xenograft Models. Clin. Cancer Res. 2015, 21, 2811–2819. [Google Scholar] [CrossRef]
- Jia, H.; Dai, G.; Weng, J.; Zhang, Z.; Wang, Q.; Zhou, F.; Jiao, L.; Cui, Y.; Ren, Y.; Fan, S.; et al. Discovery of (S)-1-(1-(Imidazo[1,2- a ]pyridin-6-yl)ethyl)-6-(1-methyl-1 H -pyrazol-4-yl)-1 H -[1,2,3]triazolo[4,5- b ]pyrazine (Volitinib) as a Highly Potent and Selective Mesenchymal-Epithelial Transition Factor (c-Met) Inhibitor in Clinical Development. J. Med. Chem. 2014, 57, 7577–7589. [Google Scholar] [CrossRef]
- Tang, Q.; Aronov, A.M.; Deininger, D.D.; Giroux, S.; Lauffer, D.J.; Li, P.; Liang, J.; McGinty, K.; Ronkin, S.; Swett, R.; et al. Discovery of Potent, Selective Triazolothiadiazole-Containing c-Met Inhibitors. ACS Med. Chem. Lett. 2021, 12, 955–960. [Google Scholar] [CrossRef]
- Ayati, A.; Moghimi, S.; Salarinejad, S.; Safavi, M.; Pouramiri, B.; Foroumadi, A. A review on progression of epidermal growth factor receptor (EGFR) inhibitors as an efficient approach in cancer targeted therapy. Bioorg. Chem. 2020, 99, 103811. [Google Scholar] [CrossRef]
- Rosenkranz, A.A.; Slastnikova, T.A. Epidermal Growth Factor Receptor: Key to Selective Intracellular Delivery. Biochemistry 2020, 85, 967–993. [Google Scholar] [CrossRef]
- Abourehab, M.A.S.; Alqahtani, A.M.; Youssif, B.G.M.; Gouda, A.M. Globally Approved EGFR Inhibitors: Insights into Their Syntheses, Target Kinases, Biological Activities, Receptor Interactions, and Metabolism. Molecules 2021, 26, 6677. [Google Scholar] [CrossRef]
- Zhai, X.; Ward, R.A.; Doig, P.; Argyrou, A. Insight into the Therapeutic Selectivity of the Irreversible EGFR Tyrosine Kinase Inhibitor Osimertinib through Enzyme Kinetic Studies. Biochemistry 2020, 59, 1428–1441. [Google Scholar] [CrossRef]
- Lee, J.; Hong, M.H.; Cho, B.C. Lazertinib: On the Way to Its Throne. Yonsei Med. J. 2022, 63, 799–805. [Google Scholar] [CrossRef]
- Yun, J.; Hong, M.H.; Kim, S.-Y.; Park, C.-W.; Yun, M.R.; Na Kang, H.; Pyo, K.-H.; Lee, S.S.; Koh, J.S.; Song, H.-J.; et al. YH25448, an Irreversible EGFR-TKI with Potent Intracranial Activity in EGFR Mutant Non–Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 2575–2587. [Google Scholar] [CrossRef]
- Nagasaka, M.; Zhu, V.W.; Lim, S.M.; Greco, M.; Wu, F.; Ou, S.-H.I. Beyond Osimertinib: The Development of Third-Generation EGFR Tyrosine Kinase Inhibitors For Advanced EGFR+ NSCLC. J. Thorac. Oncol. 2021, 16, 740–763. [Google Scholar] [CrossRef]
- Dhillon, S. Lazertinib: First Approval. Drugs 2021, 81, 1107–1113. [Google Scholar] [CrossRef]
- Colclough, N.; Chen, K.; Johnström, P.; Strittmatter, N.; Yan, Y.; Wrigley, G.L.; Schou, M.; Goodwin, R.; Varnäs, K.; Adua, S.J.; et al. Preclinical Comparison of the Blood-brain barrier Permeability of Osimertinib with Other EGFR TKIs. Clin. Cancer Res. 2021, 27, 189–201. [Google Scholar] [CrossRef]
- Planken, S.; Behenna, D.C.; Nair, S.K.; Johnson, T.O.; Nagata, A.; Almaden, C.; Bailey, S.; Ballard, T.E.; Bernier, L.; Cheng, H.; et al. Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants. J. Med. Chem. 2017, 60, 3002–3019. [Google Scholar] [CrossRef]
- Niggenaber, J.; Heyden, L.; Grabe, T.; Müller, M.P.; Lategahn, J.; Rauh, D. Complex Crystal Structures of EGFR with Third-Generation Kinase Inhibitors and Simultaneously Bound Allosteric Ligands. ACS Med. Chem. Lett. 2020, 11, 2484–2490. [Google Scholar] [CrossRef]
- Pandey, P.; Khan, F.; Upadhyay, T.K.; Seungjoon, M.; Park, M.N.; Kim, B. New insights about the PDGF/PDGFR signaling pathway as a promising target to develop cancer therapeutic strategies. Biomed. Pharmacother. 2023, 161, 114491. [Google Scholar] [CrossRef]
- Winger, B.A.; Cortopassi, W.A.; Ruiz, D.G.; Ding, L.; Jang, K.; Leyte-Vidal, A.; Zhang, N.; Esteve-Puig, R.; Jacobson, M.P.; Shah, N.P. ATP-Competitive Inhibitors Midostaurin and Avapritinib Have Distinct Resistance Profiles in Exon 17–Mutant KIT. Cancer Res. 2019, 79, 4283–4292. [Google Scholar] [CrossRef]
- Evans, E.; Gardino, A.; Hodous, B.; Davis, A.; Zhu, M.J.; Kohl, N.E.; Lengauer, C. Blu-285, a Potent and Selective Inhibitor for Hematologic Malignancies with KIT Exon 17 Mutations. Blood 2015, 126, 568. [Google Scholar] [CrossRef]
- Schneider-Stock, R. BLU-285—The breakthrough in treatment of patients with aggressive systemic mastocytosis and gastrointestinal stromal tumor. Ann. Transl. Med. 2018, 6, 232. [Google Scholar] [CrossRef]
- Dai, S.; Zhou, Z.; Chen, Z.; Xu, G.; Chen, Y. Fibroblast Growth Factor Receptors (FGFRs): Structures and Small Molecule Inhibitors. Cells 2019, 8, 614. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, W.; Li, L.; He, Y.; Wei, Y.; Dang, Y.; Nie, S.; Guo, Z. Signaling Pathway and Small-Molecule Drug Discovery of FGFR: A Comprehensive Review. Front. Chem. 2022, 10, 860985. [Google Scholar] [CrossRef]
- Chae, Y.K.; Hong, F.; Vaklavas, C.; Cheng, H.H.; Hammerman, P.; Mitchell, E.P.; Zwiebel, J.A.; Ivy, S.P.; Gray, R.J.; Li, S.; et al. Phase II Study of AZD4547 in Patients with Tumors Harboring Aberrations in the FGFR Pathway: Results from the NCI-MATCH Trial (EAY131) Subprotocol W. J. Clin. Oncol. 2020, 38, 2407–2417. [Google Scholar] [CrossRef]
- Michael, M.; Bang, Y.-J.; Park, Y.S.; Kang, Y.-K.; Kim, T.M.; Hamid, O.; Thornton, D.; Tate, S.C.; Raddad, E.; Tie, J. A Phase 1 Study of LY2874455, an Oral Selective pan-FGFR Inhibitor, in Patients with Advanced Cancer. Target. Oncol. 2017, 12, 463–474. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Akiyama, N.; Tsukaguchi, T.; Fujii, T.; Sakata, K.; Sase, H.; Isobe, T.; Morikami, K.; Shindoh, H.; Mio, T.; et al. The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor. Mol. Cancer Ther. 2014, 13, 2547–2558. [Google Scholar] [CrossRef]
- Szklener, K.; Chmiel, P.; Michalski, A.; Mańdziuk, S. New Directions and Challenges in Targeted Therapies of Advanced Bladder Cancer: The Role of FGFR Inhibitors. Cancers 2022, 14, 1416. [Google Scholar] [CrossRef]
- Markham, A. Erdafitinib: First Global Approval. Drugs 2019, 79, 1017–1021. [Google Scholar] [CrossRef]
- Roskoski, R. The role of fibroblast growth factor receptor (FGFR) protein-tyrosine kinase inhibitors in the treatment of cancers including those of the urinary bladder. Pharmacol. Res. 2020, 151, 104567. [Google Scholar] [CrossRef]
- Perera, T.P.S.; Jovcheva, E.; Mevellec, L.; Vialard, J.; De Lange, D.; Verhulst, T.; Paulussen, C.; Van De Ven, K.; King, P.; Freyne, E.; et al. Discovery and Pharmacological Characterization of JNJ-42756493 (Erdafitinib), a Functionally Selective Small-Molecule FGFR Family Inhibitor. Mol. Cancer Ther. 2017, 16, 1010–1020. [Google Scholar] [CrossRef]
- Wang, X.; Ye, C.-H.; Li, E.-M.; Xu, L.-Y.; Lin, W.-Q.; Chen, G.-H. Discovery of octahydropyrrolo [3,2-b] pyridin derivative as a highly selective Type I inhibitor of FGFR3 over VEGFR2 by high-throughput virtual screening. J. Cell. Biochem. 2023, 124, 221–238. [Google Scholar] [CrossRef]
- Feng, W.; Zhang, M.; Wu, Z.-X.; Wang, J.-Q.; Dong, X.-D.; Yang, Y.; Teng, Q.-X.; Chen, X.-Y.; Cui, Q.; Yang, D.-H. Erdafitinib Antagonizes ABCB1-Mediated Multidrug Resistance in Cancer Cells. Front. Oncol. 2020, 10, 955. [Google Scholar] [CrossRef]
- Moccia, M.; Frett, B.; Zhang, L.; Lakkaniga, N.R.; Briggs, D.; Chauhan, R.; Brescia, A.; Federico, G.; Yan, W.; Santoro, M.; et al. Bioisosteric Discovery of NPA101.3, a Second-Generation RET/VEGFR2 Inhibitor Optimized for Single-Agent Polypharmacology. J. Med. Chem. 2020, 63, 4506–4516. [Google Scholar] [CrossRef]
- Subbiah, V.; Gainor, J.F.; Rahal, R.; Brubaker, J.D.; Kim, J.L.; Maynard, M.; Hu, W.; Cao, Q.; Sheets, M.P.; Wilson, D.; et al. Precision Targeted Therapy with BLU-667 for RET-Driven Cancers. Cancer Discov. 2018, 8, 836–849. [Google Scholar] [CrossRef]
- Kim, J.; Bradford, D.; Larkins, E.; Pai-Scherf, L.H.; Chatterjee, S.; Mishra-Kalyani, P.S.; Wearne, E.; Helms, W.S.; Ayyoub, A.; Bi, Y.; et al. FDA Approval Summary: Pralsetinib for the Treatment of Lung and Thyroid Cancers With RET Gene Mutations or Fusions. Clin. Cancer Res. 2021, 27, 5452–5456. [Google Scholar] [CrossRef]
- Subbiah, V.; Shen, T.; Terzyan, S.; Liu, X.; Hu, X.; Patel, K.; Hu, M.; Cabanillas, M.; Behrang, A.; Meric-Bernstam, F.; et al. Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations. Ann. Oncol. 2021, 32, 261–268. [Google Scholar] [CrossRef]
- Shen, T.; Hu, X.; Liu, X.; Subbiah, V.; Mooers, B.H.M.; Wu, J. The L730V/I RET roof mutations display different activities toward pralsetinib and selpercatinib. Npj Precis. Oncol. 2021, 5, 48. [Google Scholar] [CrossRef]
- Liu, D.; Mamorska-Dyga, A. Syk inhibitors in clinical development for hematological malignancies. J. Hematol. Oncol. 2017, 10, 145. [Google Scholar] [CrossRef]
- Yu, J.; Huck, J.; Theisen, M.; He, H.; Tirrell, S.; Zhang, M.; Kannan, K. Anti-tumor activity of TAK-659, a dual inhibitor of SYK and FLT-3 kinases, in AML models. J. Clin. Oncol. 2016, 34, e14091. [Google Scholar] [CrossRef]
- Gomez, E.B.; Ebata, K.; Randeria, H.S.; Rosendahl, M.S.; Cedervall, E.P.; Morales, T.H.; Hanson, L.M.; Brown, N.; Gong, X.; Stephens, J.R.; et al. Pirtobrutinib preclinical characterization: A highly selective, non-covalent (reversible) BTK inhibitor. Blood 2023, 142, 62–72. [Google Scholar] [CrossRef]
- Thompson, P.; Tam, C. Pirtobrutinib: A New Hope for patients with BTK-inhibitor refractory lymphoproliferative disorders. Blood 2023, 141, 3137–3142. [Google Scholar] [CrossRef]
- Aslan, B.; Kismali, G.; Iles, L.R.; Manyam, G.C.; Ayres, M.L.; Chen, L.S.; Gagea, M.; Bertilaccio, M.T.S.; Wierda, W.G.; Gandhi, V. Pirtobrutinib inhibits wild-type and mutant Bruton’s tyrosine kinase-mediated signaling in chronic lymphocytic leukemia. Blood Cancer J. 2022, 12, 80. [Google Scholar] [CrossRef]
- Schnute, M.E.; Benoit, S.E.; Buchler, I.P.; Caspers, N.; Grapperhaus, M.L.; Han, S.; Hotchandani, R.; Huang, N.; Hughes, R.O.; Juba, B.M.; et al. Aminopyrazole Carboxamide Bruton’s Tyrosine Kinase Inhibitors. Irreversible to Reversible Covalent Reactive Group Tuning. ACS Med. Chem. Lett. 2019, 10, 80–85. [Google Scholar] [CrossRef]
- Keam, S.J. Pirtobrutinib: First Approval. Drugs 2023, 83, 547–553. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nitulescu, G.M.; Stancov, G.; Seremet, O.C.; Nitulescu, G.; Mihai, D.P.; Duta-Bratu, C.G.; Barbuceanu, S.F.; Olaru, O.T. The Importance of the Pyrazole Scaffold in the Design of Protein Kinases Inhibitors as Targeted Anticancer Therapies. Molecules 2023, 28, 5359. https://doi.org/10.3390/molecules28145359
Nitulescu GM, Stancov G, Seremet OC, Nitulescu G, Mihai DP, Duta-Bratu CG, Barbuceanu SF, Olaru OT. The Importance of the Pyrazole Scaffold in the Design of Protein Kinases Inhibitors as Targeted Anticancer Therapies. Molecules. 2023; 28(14):5359. https://doi.org/10.3390/molecules28145359
Chicago/Turabian StyleNitulescu, George Mihai, Gheorghe Stancov, Oana Cristina Seremet, Georgiana Nitulescu, Dragos Paul Mihai, Cosmina Gabriela Duta-Bratu, Stefania Felicia Barbuceanu, and Octavian Tudorel Olaru. 2023. "The Importance of the Pyrazole Scaffold in the Design of Protein Kinases Inhibitors as Targeted Anticancer Therapies" Molecules 28, no. 14: 5359. https://doi.org/10.3390/molecules28145359
APA StyleNitulescu, G. M., Stancov, G., Seremet, O. C., Nitulescu, G., Mihai, D. P., Duta-Bratu, C. G., Barbuceanu, S. F., & Olaru, O. T. (2023). The Importance of the Pyrazole Scaffold in the Design of Protein Kinases Inhibitors as Targeted Anticancer Therapies. Molecules, 28(14), 5359. https://doi.org/10.3390/molecules28145359






