The Versatile Biocatalyst of Cytochrome P450 CYP102A1: Structure, Function, and Engineering
Abstract
1. Introduction
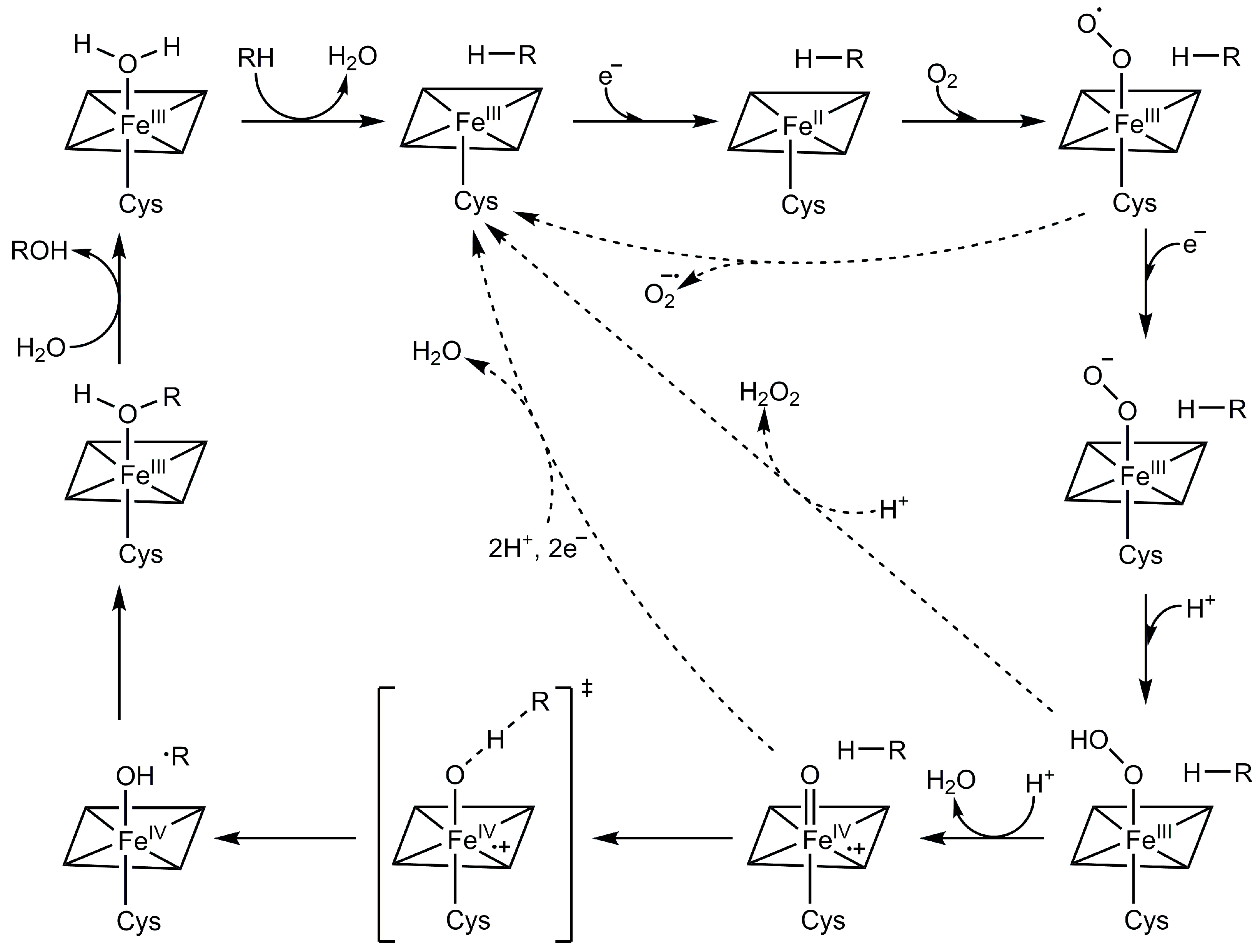
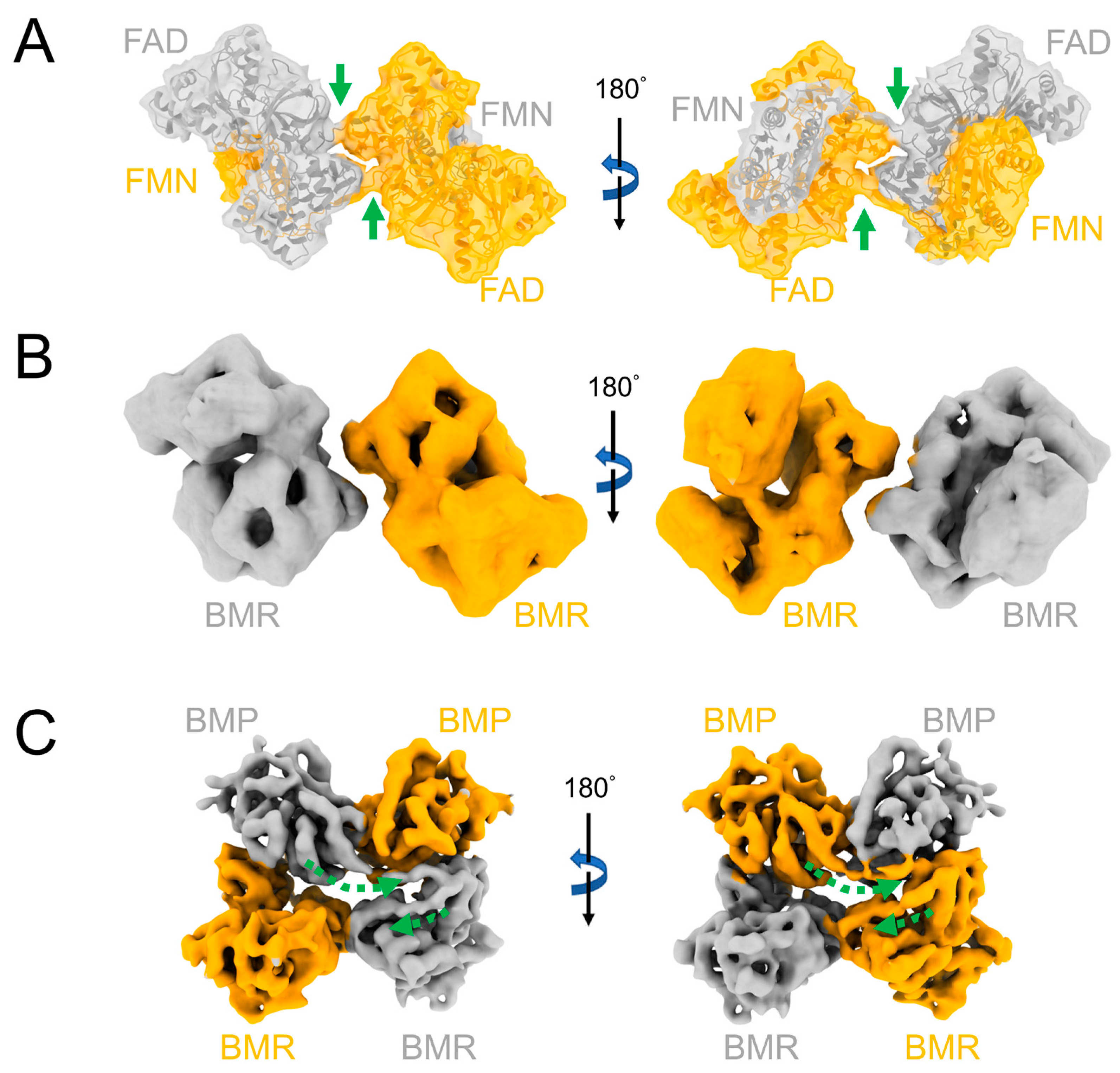
2. Structure and Function of CYP102A1
3. Engineering and Design of CYP102A1
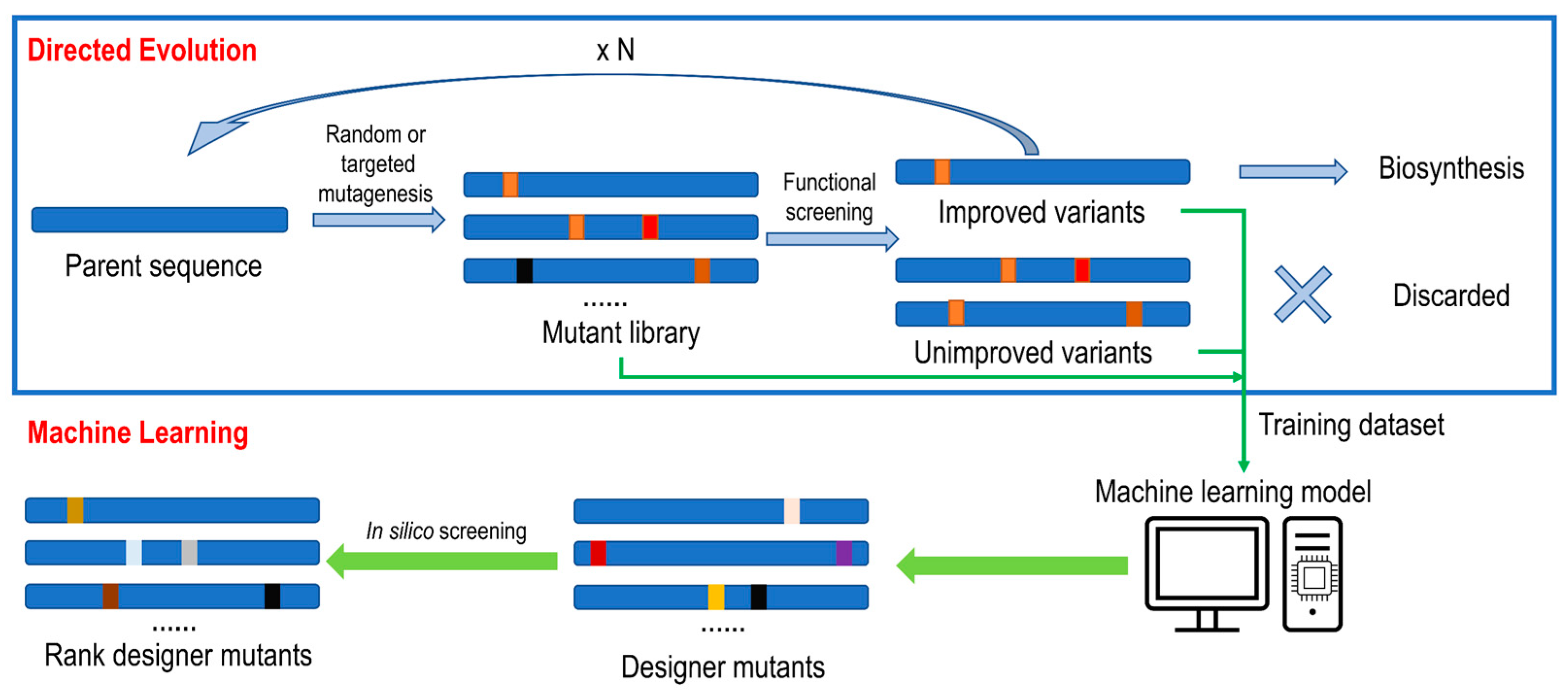
3.1. Laboratory Directed Evolution (LDE)
3.2. Rational Design
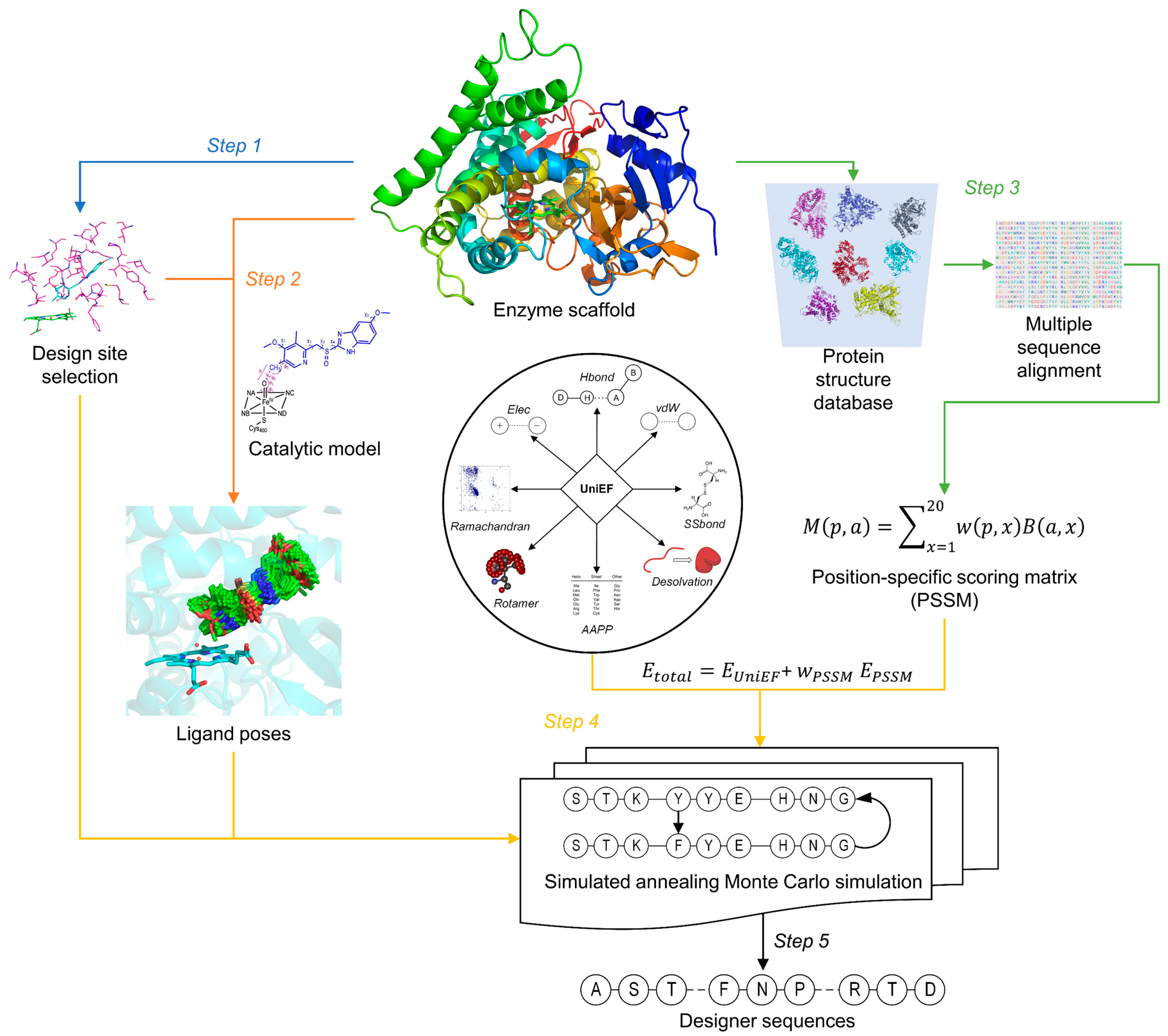
3.3. Computational Protein Design (CPD)
3.4. Machine Learning (ML)
3.5. Chemical Engineering
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Brodie, B.B.; Axelrod, J.; Cooper, J.R.; Gaudette, L.; La Du, B.N.; Mitoma, C.; Udenfriend, S. Detoxication of drugs and other foreign compounds by liver microsomes. Science 1955, 121, 603–604. [Google Scholar] [CrossRef]
- Klingenberg, M. Pigments of rat liver microsomes. Arch. Biochem. Biophys. 1958, 75, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Omura, T.; Sato, R. Fractional solubilization of haemoproteins and partial purification of carbon monoxide-binding cytochrome from liver microsomes. Biochim. Biophys. Acta 1963, 71, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Human Cytochrome P450 Enzyme. In Cytochrome P450: Structure, Mechanism, and Biochemistry; de Montellano, P.R., Ed.; Plenum: New York, NY, USA, 1995; pp. 473–535. [Google Scholar]
- Dunham, N.P.; Arnold, F.H. Nature’s Machinery, Repurposed: Expanding the Repertoire of Iron-Dependent Oxygenases. ACS Catal. 2020, 10, 12239–12255. [Google Scholar] [CrossRef]
- White, R.E.; Coon, M.J. Oxygen activation by cytochrome P-450. Annu. Rev. Biochem. 1980, 49, 315–356. [Google Scholar] [CrossRef] [PubMed]
- Sligar, S.G. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry 1976, 15, 5399–5406. [Google Scholar] [CrossRef]
- Sligar, S.G.; Cinti, D.L.; Gibson, G.G.; Schenkman, J.B. Spin state control of the hepatic cytochrome P450 redox potential. Biochem. Biophys. Res. Commun. 1979, 90, 925–932. [Google Scholar] [CrossRef]
- Zhang, H.; Gruenke, L.; Arscott, D.; Shen, A.; Kasper, C.; Harris, D.L.; Glavanovich, M.; Johnson, R.; Waskell, L. Determination of the rate of reduction of oxyferrous cytochrome P450 2B4 by 5-deazariboflavin adenine dinucleotide T491V cytochrome P450 reductase. Biochemistry 2003, 42, 11594–11603. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Ballou, D.P.; Coon, M.J. Purified liver microsomal cytochrome P-450. Electron-accepting properties and oxidation-reduction potential. J. Biol. Chem. 1975, 250, 7405–7414. [Google Scholar] [CrossRef]
- Rittle, J.; Green, M.T. Cytochrome P450 compound I: Capture, characterization, and C-H bond activation kinetics. Science 2010, 330, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Groves, J.T.; McClusky, G.A. Aliphatic hydroxylation via oxygen rebound. Oxygen transfer catalyzed by iron. J. Am. Chem. Soc. 1976, 98, 859–861. [Google Scholar] [CrossRef]
- Koppenol, W.H. Oxygen activation by cytochrome p450: A thermodynamic analysis. J. Am. Chem. Soc. 2007, 129, 9686–9690. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Meyer, M.M.; Meinhold, P.; Otey, C.R.; MacMillan, D.; Arnold, F.H. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 2005, 15, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Caswell, J.M.; O’Neill, M.; Taylor, S.J.; Moody, T.S. Engineering and application of P450 monooxygenases in pharmaceutical and metabolite synthesis. Curr. Opin. Chem. Biol. 2013, 17, 271–275. [Google Scholar] [CrossRef]
- Narhi, L.O.; Fulco, A.J. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 1986, 261, 7160–7169. [Google Scholar] [CrossRef]
- Whitehouse, C.J.; Bell, S.G.; Wong, L.L. P450(BM3) (CYP102A1): Connecting the dots. Chem. Soc. Rev. 2012, 41, 1218–1260. [Google Scholar] [CrossRef]
- Thistlethwaite, S.; Jeffreys, L.N.; Girvan, H.M.; McLean, K.J.; Munro, A.W. A Promiscuous Bacterial P450: The Unparalleled Diversity of BM3 in Pharmaceutical Metabolism. Int. J. Mol. Sci. 2021, 22, 1380. [Google Scholar] [CrossRef]
- Munro, A.W.; Girvan, H.M.; Mason, A.E.; Dunford, A.J.; McLean, K.J. What makes a P450 tick? Trends Biochem. Sci. 2013, 38, 140–150. [Google Scholar] [CrossRef]
- Black, S.D.; Martin, S.T. Evidence for conformational dynamics and molecular aggregation in cytochrome P450 102 (BM-3). Biochemistry 1994, 33, 12056–12062. [Google Scholar] [CrossRef]
- Kitazume, T.; Haines, D.C.; Estabrook, R.W.; Chen, B.; Peterson, J.A. Obligatory intermolecular electron-transfer from FAD to FMN in dimeric P450BM-3. Biochemistry 2007, 46, 11892–11901. [Google Scholar] [CrossRef] [PubMed]
- Neeli, R.; Girvan, H.M.; Lawrence, A.; Warren, M.J.; Leys, D.; Scrutton, N.S.; Munro, A.W. The dimeric form of flavocytochrome P450 BM3 is catalytically functional as a fatty acid hydroxylase. FEBS Lett. 2005, 579, 5582–5588. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Chakraborty, S.; Osawa, Y.; Zhang, H. Cryo-EM reveals the architecture of the dimeric cytochrome P450 CYP102A1 enzyme and conformational changes required for redox partner recognition. J. Biol. Chem. 2020, 295, 1637–1645. [Google Scholar] [CrossRef]
- Sevrioukova, I.F.; Li, H.; Zhang, H.; Peterson, J.A.; Poulos, T.L. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. USA 1999, 96, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Urban, P.; Pompon, D. Confrontation of AlphaFold models with experimental structures enlightens conformational dynamics supporting CYP102A1 functions. Sci. Rep. 2022, 12, 15982. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Kim, S.Y.; Kim, D.; Kim, D.H.; Shin, S.M.; Park, S.H.; Kim, K.H.; Jung, H.C.; Pan, J.G.; Joung, Y.H.; et al. Characterization of diverse natural variants of CYP102A1 found within a species of Bacillus megaterium. AMB Express 2011, 1, 1. [Google Scholar] [CrossRef]
- Fasan, R.; Chen, M.M.; Crook, N.C.; Arnold, F.H. Engineered alkane-hydroxylating cytochrome P450(BM3) exhibiting nativelike catalytic properties. Angew. Chem. Int. Ed. Engl. 2007, 46, 8414–8418. [Google Scholar] [CrossRef]
- Farinas, E.T.; Schwaneberg, U.; Glieder, A.; Arnold, F.H. Directed evolution of a cytochrome P450 monooxygenase for alkane oxidation. Adv. Synth. Catal. 2001, 343, 601–606. [Google Scholar] [CrossRef]
- Glieder, A.; Farinas, E.T.; Arnold, F.H. Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat. Biotechnol. 2002, 20, 1135–1139. [Google Scholar] [CrossRef]
- Farinas, E.T.; Alcalde, M.; Arnold, F. Alkene epoxidation catalyzed by cytochrome P450BM-3 139-3. Tetrahedron 2004, 60, 525–528. [Google Scholar] [CrossRef]
- Sawayama, A.M.; Chen, M.M.; Kulanthaivel, P.; Kuo, M.S.; Hemmerle, H.; Arnold, F.H. A panel of cytochrome P450 BM3 variants to produce drug metabolites and diversify lead compounds. Chemistry 2009, 15, 11723–11729. [Google Scholar] [CrossRef] [PubMed]
- van Vugt-Lussenburg, B.M.; Stjernschantz, E.; Lastdrager, J.; Oostenbrink, C.; Vermeulen, N.P.; Commandeur, J.N. Identification of critical residues in novel drug metabolizing mutants of cytochrome P450 BM3 using random mutagenesis. J. Med. Chem. 2007, 50, 455–461. [Google Scholar] [CrossRef]
- Kim, K.H.; Kang, J.Y.; Kim, D.H.; Park, S.H.; Park, S.H.; Kim, D.; Park, K.D.; Lee, Y.J.; Jung, H.C.; Pan, J.G.; et al. Generation of human chiral metabolites of simvastatin and lovastatin by bacterial CYP102A1 mutants. Drug Metab. Dispos. 2011, 39, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.H.; Park, B.Y.; Kim, S.Y.; Park, S.H.; Jung, H.J.; Park, M.; Park, K.D.; Ahn, T.; Kang, H.S.; Yun, C.H. Regioselective hydroxylation of omeprazole enantiomers by bacterial CYP102A1 mutants. Drug Metab. Dispos. 2014, 42, 1493–1497. [Google Scholar] [CrossRef]
- Butler, C.F.; Peet, C.; Mason, A.E.; Voice, M.W.; Leys, D.; Munro, A.W. Key mutations alter the cytochrome P450 BM3 conformational landscape and remove inherent substrate bias. J. Biol. Chem. 2013, 288, 25387–25399. [Google Scholar] [CrossRef]
- Noble, M.A.; Miles, C.S.; Chapman, S.K.; Lysek, D.A.; MacKay, A.C.; Reid, G.A.; Hanzlik, R.P.; Munro, A.W. Roles of key active-site residues in flavocytochrome P450 BM3. Biochem. J. 1999, 339 Pt 2, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Do, T.A.; Schmid, R.D.; Pleiss, J.; Urlacher, V.B. Altering the regioselectivity of the subterminal fatty acid hydroxylase P450 BM-3 towards gamma- and delta-positions. J. Biotechnol. 2009, 139, 115–117. [Google Scholar] [CrossRef]
- Ost, T.W.; Miles, C.S.; Murdoch, J.; Cheung, Y.; Reid, G.A.; Chapman, S.K.; Munro, A.W. Rational re-design of the substrate binding site of flavocytochrome P450 BM3. FEBS Lett. 2000, 486, 173–177. [Google Scholar] [CrossRef]
- Yamazaki, H.; Inoue, K.; Shaw, P.M.; Checovich, W.J.; Guengerich, F.P.; Shimada, T. Different contributions of cytochrome P450 2C19 and 3A4 in the oxidation of omeprazole by human liver microsomes: Effects of contents of these two forms in individual human samples. J. Pharmacol. Exp. Ther. 1997, 283, 434–442. [Google Scholar]
- Li, X.Q.; Weidolf, L.; Simonsson, R.; Andersson, T.B. Enantiomer/enantiomer interactions between the S- and R- isomers of omeprazole in human cytochrome P450 enzymes: Major role of CYP2C19 and CYP3A4. J. Pharmacol. Exp. Ther. 2005, 315, 777–787. [Google Scholar] [CrossRef]
- Butler, C.F.; Peet, C.; McLean, K.J.; Baynham, M.T.; Blankley, R.T.; Fisher, K.; Rigby, S.E.; Leys, D.; Voice, M.W.; Munro, A.W. Human P450-like oxidation of diverse proton pump inhibitor drugs by ‘gatekeeper’ mutants of flavocytochrome P450 BM3. Biochem. J. 2014, 460, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, J.; Yang, D.; Zhang, J.; Xia, X.; Chen, Y.E.; Xu, J. Decoding CRISPR–Cas PAM recognition with UniDesign. Brief. Bioinform. 2023, 24, bbad133. [Google Scholar] [CrossRef]
- Jang, H.H.; Ryu, S.H.; Le, T.K.; Doan, T.T.; Nguyen, T.H.; Park, K.D.; Yim, D.E.; Kim, D.H.; Kang, C.K.; Ahn, T.; et al. Regioselective C-H hydroxylation of omeprazole sulfide by Bacillus megaterium CYP102A1 to produce a human metabolite. Biotechnol. Lett. 2017, 39, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, M.A.; Bombino, E.; de Jong, R.M.; Wijma, H.J.; Janssen, D.B.; McLean, K.J.; Munro, A.W. Computation-Aided Engineering of Cytochrome P450 for the Production of Pravastatin. ACS Catal. 2022, 12, 15028–15044. [Google Scholar] [CrossRef]
- McLean, K.J.; Hans, M.; Meijrink, B.; van Scheppingen, W.B.; Vollebregt, A.; Tee, K.L.; van der Laan, J.M.; Leys, D.; Munro, A.W.; van den Berg, M.A. Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum. Proc. Natl. Acad. Sci. USA 2015, 112, 2847–2852. [Google Scholar] [CrossRef]
- Li, D.; Ma, Y.; Zhou, Y.; Gou, J.; Zhong, Y.; Zhao, L.; Han, L.; Ovchinnikov, S.; Ma, L.; Huang, S.; et al. A structural and data-driven approach to engineering a plant cytochrome P450 enzyme. Sci. China Life Sci. 2019, 62, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Zidek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Athavale, S.V.; Gao, S.; Das, A.; Mallojjala, S.C.; Alfonzo, E.; Long, Y.; Hirschi, J.S.; Arnold, F.H. Enzymatic Nitrogen Insertion into Unactivated C-H Bonds. J. Am. Chem. Soc. 2022, 144, 19097–19105. [Google Scholar] [CrossRef]
- Wu, Z.; Kan, S.B.J.; Lewis, R.D.; Wittmann, B.J.; Arnold, F.H. Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl. Acad. Sci. USA 2019, 116, 8852–8858. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef]
- Mazurenko, S.; Prokop, Z.; damborsky, J. Machine learning in enxyme engineering. ACS Catal. 2020, 10, 1210–1223. [Google Scholar] [CrossRef]
- Yang, Y.; Niroula, A.; Shen, B.; Vihinen, M. PON-Sol: Prediction of effects of amino acid substitutions on protein solubility. Bioinformatics 2016, 32, 2032–2034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, J.; Hu, H.; Gong, H.; Chen, L.; Cheng, C.; Zeng, J. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic Acids Res. 2016, 44, e32. [Google Scholar] [CrossRef] [PubMed]
- Defresne, M.; Barbe, S.; Schiex, T. Protein Design with Deep Learning. Int. J. Mol. Sci. 2021, 22, 1741. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.A.; Krause, A.; Arnold, F.H. Navigating the protein fitness landscape with Gaussian processes. Proc. Natl. Acad. Sci. USA 2013, 110, E193–E201. [Google Scholar] [CrossRef]
- Salazar, O.; Cirino, P.C.; Arnold, F.H. Thermostabilization of a cytochrome p450 peroxygenase. ChemBioChem 2003, 4, 891–893. [Google Scholar] [CrossRef]
- Xu, Y.T.; Verma, D.; Sheridan, R.P.; Liaw, A.; Ma, J.S.; Marshall, N.M.; McIntosh, J.; Sherer, E.C.; Svetnik, V.; Johnston, J.M. Deep Dive into Machine Learning Models for Protein Engineering. J. Chem. Inf. Model. 2020, 60, 2773–2790. [Google Scholar] [CrossRef]
- Fox, R.J.; Davis, S.C.; Mundorff, E.C.; Newman, L.M.; Gavrilovic, V.; Ma, S.K.; Chung, L.M.; Ching, C.; Tam, S.; Muley, S.; et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat. Biotechnol. 2007, 25, 338–344. [Google Scholar] [CrossRef]
- Liao, J.; Warmuth, M.K.; Govindarajan, S.; Ness, J.E.; Wang, R.P.; Gustafsson, C.; Minshull, J. Engineering proteinase K using machine learning and synthetic genes. BMC Biotechnol. 2007, 7, 16. [Google Scholar] [CrossRef]
- Du, S.; Wang, X.; Wang, R.; Lu, L.; Luo, Y.; You, G.; Wu, S. Machine-learning-assisted molecular design of phenylnaphthylamine-type antioxidants. Phys. Chem. Chem. Phys. 2022, 24, 13399–13410. [Google Scholar] [CrossRef]
- Han, X.; Ning, W.; Ma, X.; Wang, X.; Zhou, K. Improving protein solubility and activity by introducing small peptide tags designed with machine learning models. Metab. Eng. Commun. 2020, 11, e00138. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, J.K.; Shoji, O. The Power of Deception: Using Decoy Molecules to Manipulate P450BM3 Biotransformations. Chem. Lett. 2021, 50, 2025–2031. [Google Scholar] [CrossRef]
- Zilly, F.E.; Acevedo, J.P.; Augustyniak, W.; Deege, A.; Hausig, U.W.; Reetz, M.T. Tuning a P450 Enzyme for Methane Oxidation. Angew. Chem. Int. Edit 2011, 50, 2720–2724. [Google Scholar] [CrossRef] [PubMed]
- Shoji, O.; Kunimatsu, T.; Kawakami, N.; Watanabe, Y. Highly Selective Hydroxylation of Benzene to Phenol by Wild-type Cytochrome P450BM3 Assisted by Decoy Molecules. Angew. Chem. Int. Edit 2013, 52, 6606–6610. [Google Scholar] [CrossRef]
- Ariyasu, S.; Yonemura, K.; Kasai, C.; Aiba, Y.; Onoda, H.; Shisaka, Y.; Sugimoto, H.; Tosha, T.; Kubo, M.; Kamachi, T.; et al. Catalytic oxidation of methane by wild-type cytochrome P450BM3 with chemicallly evolved decoy molecules. ACS Catal. 2023, 13, 8613–8623. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Huang, X.; Osawa, Y.; Chen, Y.E.; Zhang, H. The Versatile Biocatalyst of Cytochrome P450 CYP102A1: Structure, Function, and Engineering. Molecules 2023, 28, 5353. https://doi.org/10.3390/molecules28145353
Sun Y, Huang X, Osawa Y, Chen YE, Zhang H. The Versatile Biocatalyst of Cytochrome P450 CYP102A1: Structure, Function, and Engineering. Molecules. 2023; 28(14):5353. https://doi.org/10.3390/molecules28145353
Chicago/Turabian StyleSun, Yudong, Xiaoqiang Huang, Yoichi Osawa, Yuqing Eugene Chen, and Haoming Zhang. 2023. "The Versatile Biocatalyst of Cytochrome P450 CYP102A1: Structure, Function, and Engineering" Molecules 28, no. 14: 5353. https://doi.org/10.3390/molecules28145353
APA StyleSun, Y., Huang, X., Osawa, Y., Chen, Y. E., & Zhang, H. (2023). The Versatile Biocatalyst of Cytochrome P450 CYP102A1: Structure, Function, and Engineering. Molecules, 28(14), 5353. https://doi.org/10.3390/molecules28145353






