Bisphenol A Analogues Inhibit Human and Rat 11β-Hydroxysteroid Dehydrogenase 1 Depending on Its Lipophilicity
Abstract
1. Introduction
2. Results
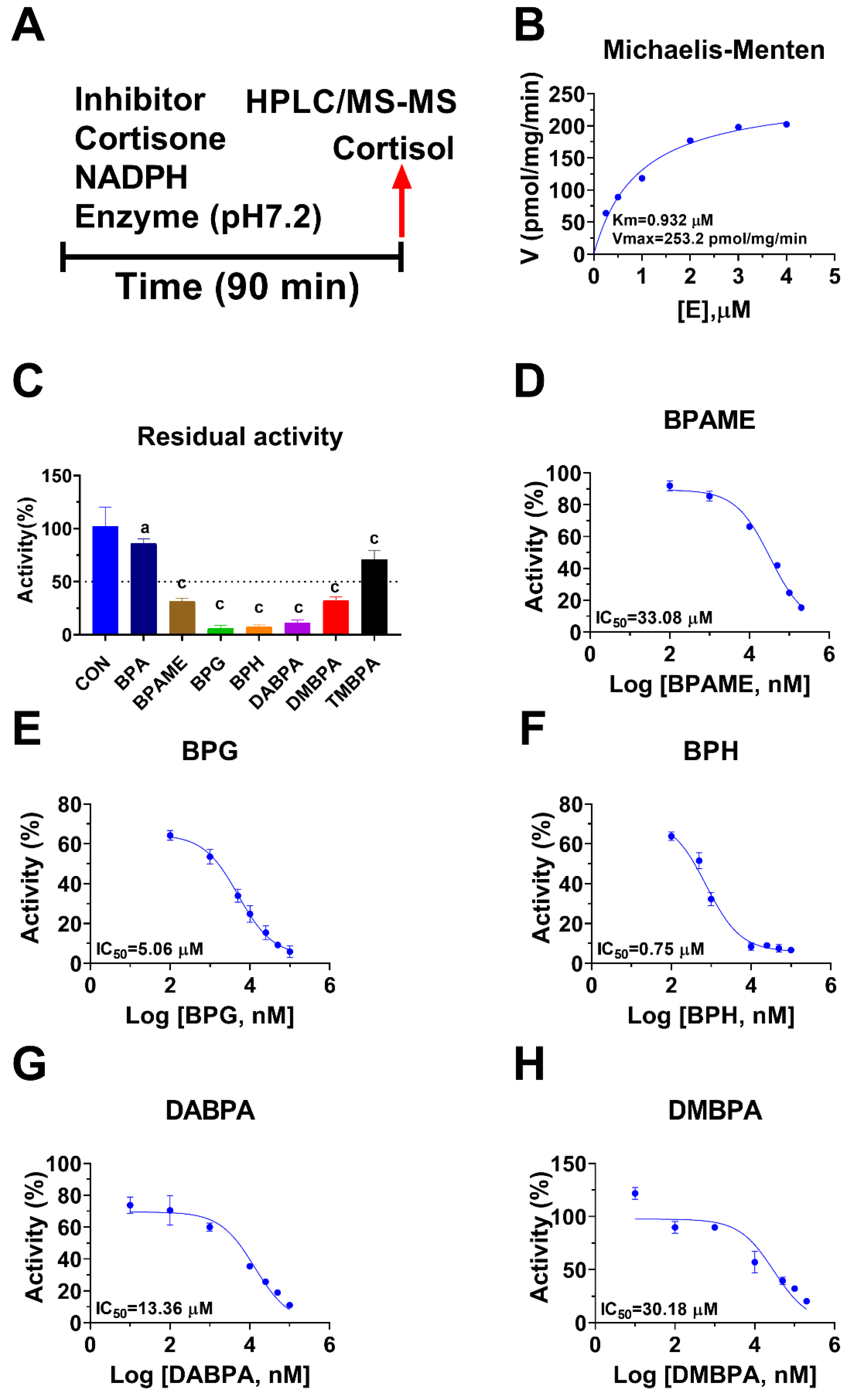
2.1. BPA Analogues Inhibit Human 11β-HSD1A Activity
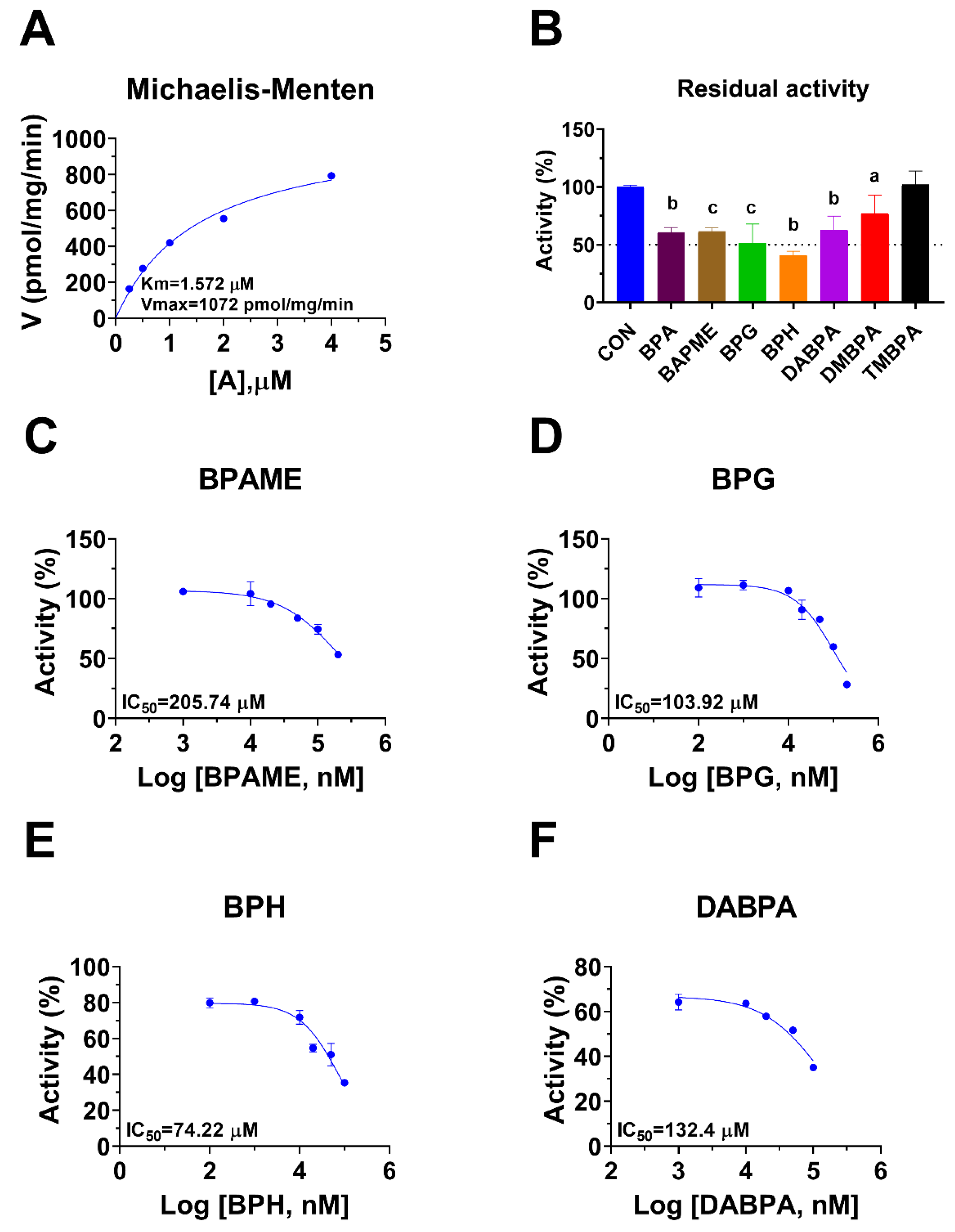
2.2. BPA Analogues Inhibit Rat 11β-HSD1 Activity
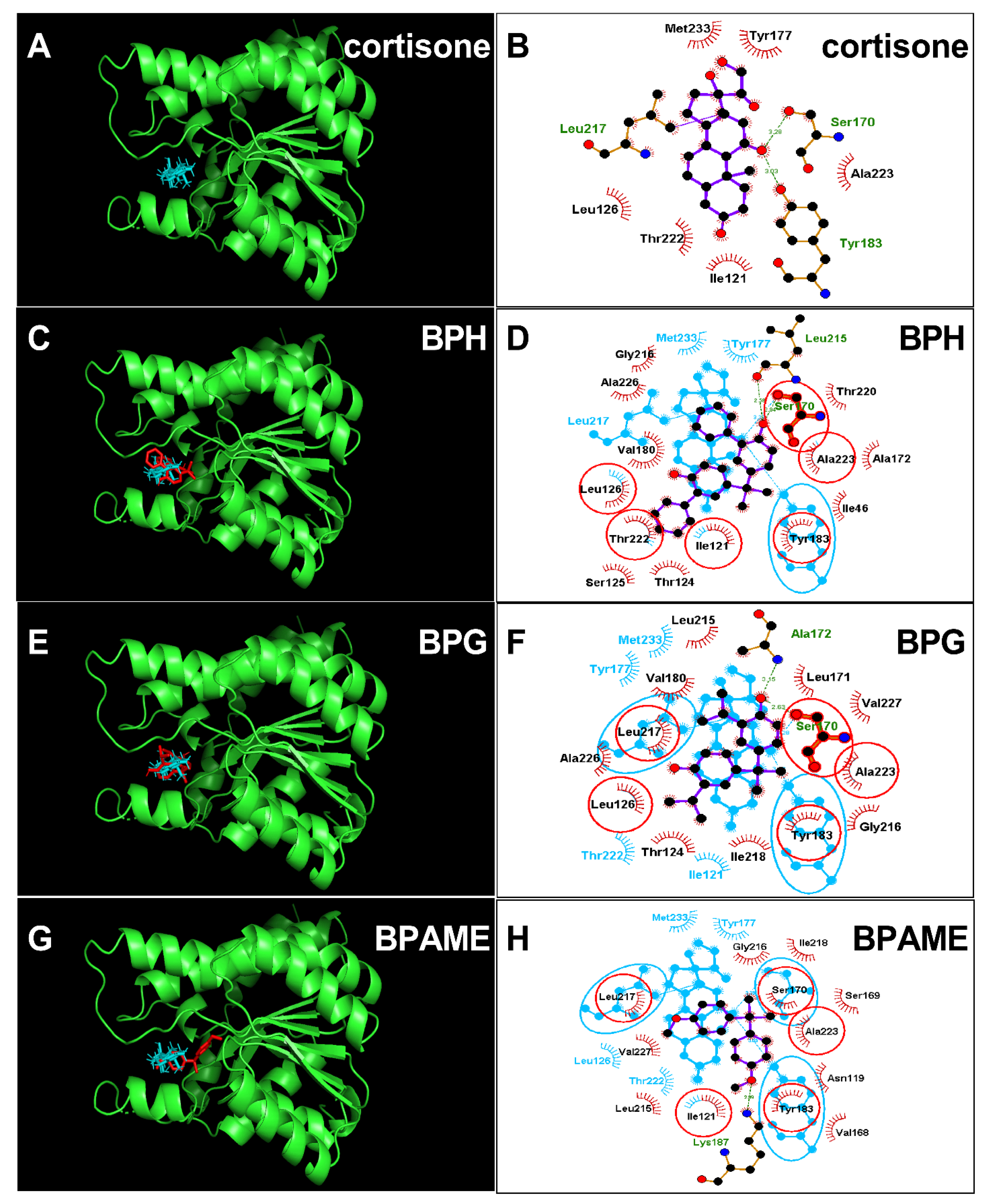
2.3. Molecular Docking Analysis of BPA Analogues with Human 11β-HSD1
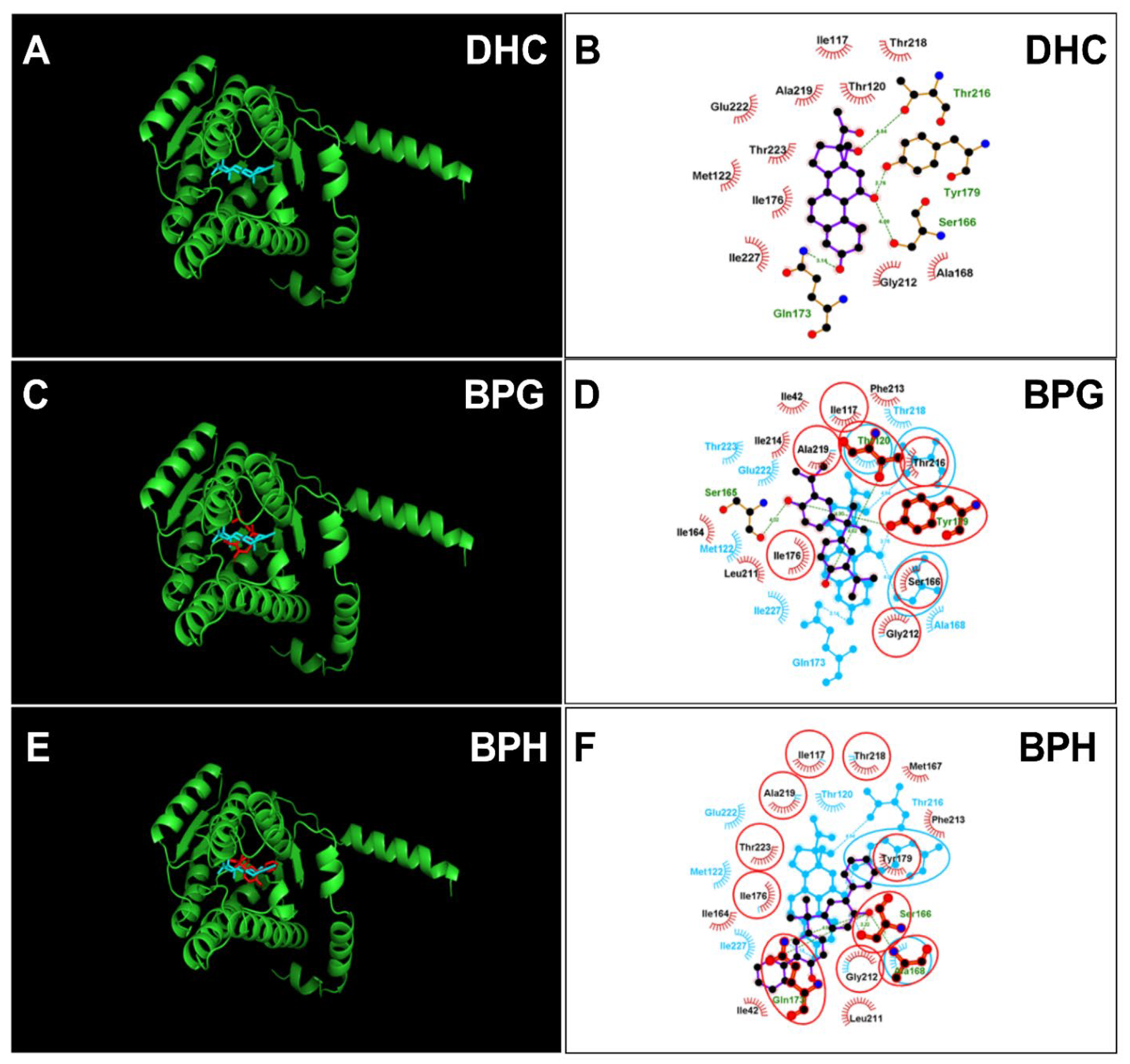
2.4. Molecular Docking Analysis of BPA Analogues with Rat 11β-HSD1
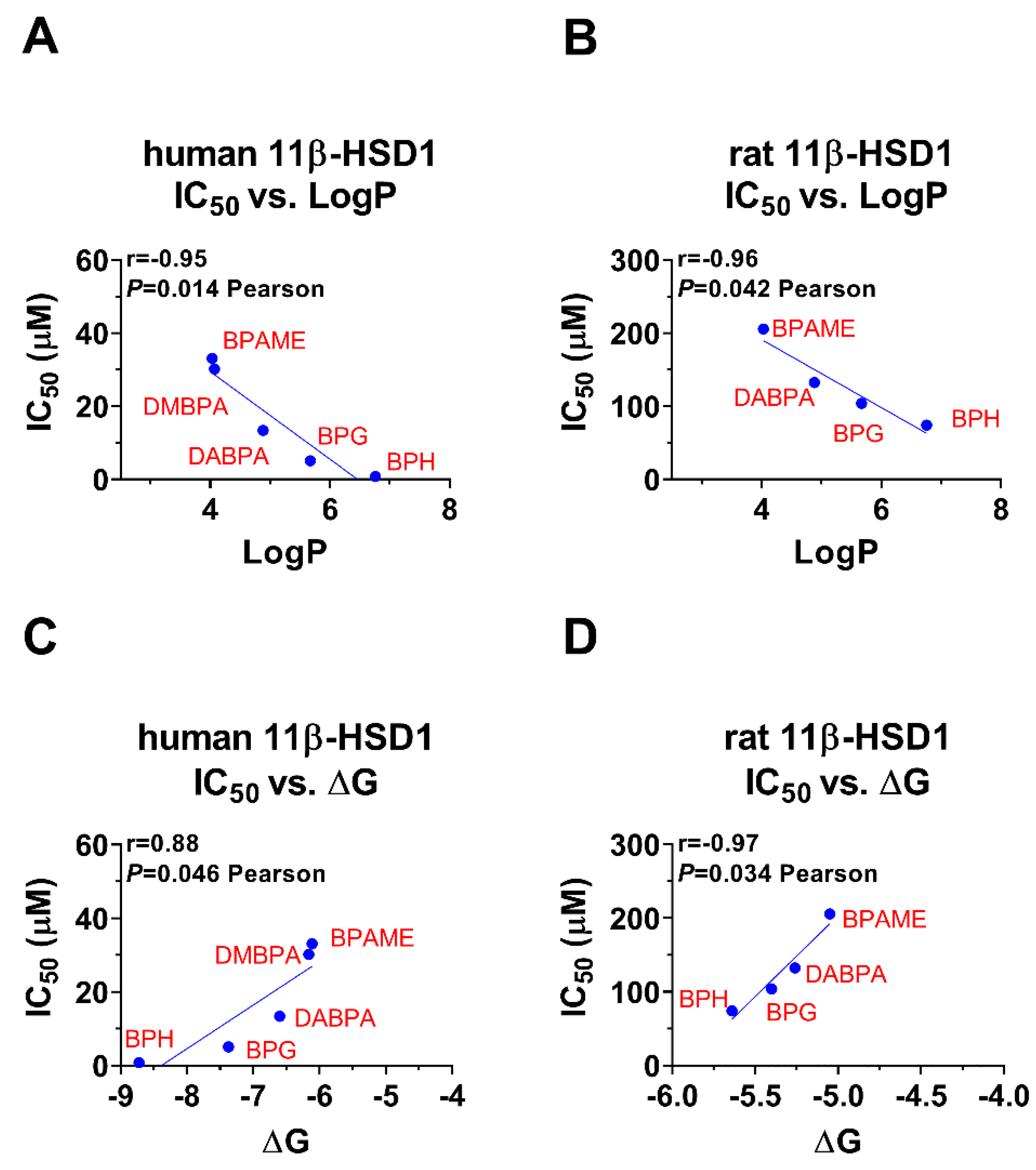
2.5. Bivariate Correlation Analysis for Inhibitory Strength BPA Analogues with Structural Features
2.6. ADMET Prediction of BPA Analogues
3. Discussion
4. Materials and Methods
4.1. Materials and Chemicals
4.2. Preparation of Rat Liver Microsomes
4.3. 11β-HSD1 Assay
4.4. Determination of Cortisol and CORT by HPLC-MS/MS Method
4.5. Analysis of 11β-HSD1 Kinetics and Inhibitory Parameters of BPA Analogues
4.6. Molecular Docking Analysis of Human and Rat 11β-HSD1
4.7. Bivariate Correlation Analysis for Structural Features of BPA Analogues with IC50 Values
4.8. Drug Metabolism and Pharmacokinetics Prediction
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Li, X.; Wen, Z.; Wang, Y.; Mo, J.; Zhong, Y.; Ge, R.S. Bisphenols and Leydig Cell Development and Function. Front. Endocrinol. 2020, 11, 447. [Google Scholar] [CrossRef] [PubMed]
- Patisaul, H.B. Achieving CLARITY on bisphenol A, brain and behaviour. J. Neuroendocrinol. 2020, 32, e12730. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Park, Y.J. Bisphenols and Thyroid Hormone. Endocrinol. Metab. 2019, 34, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Kumar, S.; Kumar, V.; Lee, Y.M.; Kim, Y.S.; Kumar, V. Bisphenols as a Legacy Pollutant, and Their Effects on Organ Vulnerability. Int. J. Environ. Res. Public Health 2019, 17, 112. [Google Scholar] [CrossRef]
- Santoro, A.; Chianese, R.; Troisi, J.; Richards, S.; Nori, S.L.; Fasano, S.; Guida, M.; Plunk, E.; Viggiano, A.; Pierantoni, R.; et al. Neuro-toxic and Reproductive Effects of BPA. Curr. Neuropharmacol. 2019, 17, 1109–1132. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, H.; Wu, J.; Yuan, L.; Wang, Y.; Du, X.; Wang, R.; Marwa, P.W.; Petlulu, P.; Chen, X.; et al. The adverse health effects of bisphenol A and related toxicity mechanisms. Environ. Res. 2019, 176, 108575. [Google Scholar] [CrossRef]
- Dallio, M.; Diano, N.; Masarone, M.; Gravina, A.G.; Patane, V.; Romeo, M.; Di Sarno, R.; Errico, S.; Nicolucci, C.; Abenavoli, L.; et al. Chemical Effect of Bisphenol A on Non-Alcoholic Fatty Liver Disease. Int. J. Environ. Res. Public Health 2019, 16, 3134. [Google Scholar] [CrossRef]
- Cesen, M.; Lenarcic, K.; Mislej, V.; Levstek, M.; Kovacic, A.; Cimrmancic, B.; Uranjek, N.; Kosjek, T.; Heath, D.; Dolenc, M.S.; et al. The occurrence and source identification of bisphenol compounds in wastewaters. Sci. Total Environ. 2018, 616–617, 744–752. [Google Scholar] [CrossRef]
- Shi, L.; Li, J.; Tian, F.; Tang, Y.; Wang, S.; Li, Q.; Zhu, Y.; Zhu, Q.; Ge, R.S. Dimethylbisphenol A inhibits the differentiation of stem Leydig cells in adult male rats by androgen receptor (NR3C4) antagonism. Toxicol. Lett. 2022, 366, 58–71. [Google Scholar] [CrossRef]
- Xue, J.; Kannan, K. Mass flows and removal of eight bisphenol analogs, bisphenol A diglycidyl ether and its derivatives in two wastewater treatment plants in New York State, USA. Sci. Total Environ. 2019, 648, 442–449. [Google Scholar] [CrossRef]
- Grimaldi, M.; Boulahtouf, A.; Toporova, L.; Balaguer, P. Functional profiling of bisphenols for nuclear receptors. Toxicology 2019, 420, 39–45. [Google Scholar] [CrossRef]
- Zhang, H.C.; Hu, X.L.; Yin, D.Q.; Lin, Z.F. Development of molecular docking-based binding energy to predict the joint effect of BPA and its analogs. Hum. Exp. Toxicol. 2011, 30, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Champagne, F.A. Early environments, glucocorticoid receptors, and behavioral epigenetics. Behav. Neurosci. 2013, 127, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, M.; Ohno, S.; Nakajin, S. Inhibitory effects of some possible endocrine-disrupting chemicals on the isozymes of human 11beta-hydroxysteroid dehydrogenase and expression of their mRNA in gonads and adrenal glands. Environ. Sci. 2005, 12, 219–230. [Google Scholar] [PubMed]
- Zhou, H.Y.; Chen, X.X.; Lin, H.; Fei, A.L.; Ge, R.S. 11beta-hydroxysteroid dehydrogenase types 1 and 2 in postnatal development of rat testis: Gene expression, localization and regulation by luteinizing hormone and androgens. Asian J. Androl. 2014, 16, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.; Holmes, M.; Seckl, J. 11beta-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 2013, 93, 1139–1206. [Google Scholar] [CrossRef]
- Guo, J.; Yuan, X.; Qiu, L.; Zhu, W.; Wang, C.; Hu, G.; Chu, Y.; Ye, L.; Xu, Y.; Ge, R.S. Inhibition of human and rat 11beta-hydroxysteroid dehydrogenases activities by bisphenol A. Toxicol. Lett. 2012, 215, 126–130. [Google Scholar] [CrossRef]
- Hewitt, K.N.; Walker, E.A.; Stewart, P.M. Minireview: Hexose-6-phosphate dehydrogenase and redox control of 11{beta}-hydroxysteroid dehydrogenase type 1 activity. Endocrinology 2005, 146, 2539–2543. [Google Scholar] [CrossRef]
- Tannin, G.M.; Agarwal, A.K.; Monder, C.; New, M.I.; White, P.C. The human gene for 11 beta-hydroxysteroid dehydrogenase. Structure, tissue distribution, and chromosomal localization. J. Biol. Chem. 1991, 266, 16653–16658. [Google Scholar] [CrossRef]
- Ge, R.S.; Gao, H.B.; Nacharaju, V.L.; Gunsalus, G.L.; Hardy, M.P. Identification of a kinetically distinct activity of 11beta-hydroxysteroid dehydrogenase in rat Leydig cells. Endocrinology 1997, 138, 2435–2442. [Google Scholar] [CrossRef]
- Arampatzis, S.; Kadereit, B.; Schuster, D.; Balazs, Z.; Schweizer, R.A.; Frey, F.J.; Langer, T.; Odermatt, A. Comparative enzymology of 11beta-hydroxysteroid dehydrogenase type 1 from six species. J. Mol. Endocrinol. 2005, 35, 89–101. [Google Scholar] [CrossRef]
- Obeid, J.; White, P.C. Tyr-179 and Lys-183 are essential for enzymatic activity of 11 beta-hydroxysteroid dehydrogenase. Biochem. Biophys. Res. Commun. 1992, 188, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, B.; Ying, Y.; Tang, Y.; Wang, S.; Zhu, Y.; Li, H.; Ge, R.S.; Liu, Y. Halogen atoms determine the inhibitory potency of halogenated bisphenol A derivatives on human and rat placental 11beta-hydroxysteroid dehydrogenase 2. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2023, 175, 113739. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, S.; Tang, Y.; Hu, Z.; Shi, L.; Lu, J.; Li, H.; Wang, Y.; Zhu, Y.; Lin, H.; et al. Direct inhibition of bisphenols on human and rat 11beta-hydroxysteroid dehydrogenase 2: Structure-activity relationship and docking analysis. Ecotoxicol. Environ. Saf. 2023, 254, 114715. [Google Scholar] [CrossRef]
- Tang, L.; Shi, L.; Tang, Y.; Ying, Y.; Dong, Y.; Li, H.; Ge, R.S. Chemicals of environmental concern as inhibitors of human placental 3beta-hydroxysteroid dehydrogenase 1 and aromatase: Screening and docking analysis. Chem. Biol. Interact. 2022, 368, 110243. [Google Scholar] [CrossRef]
- Ye, L.; Zhao, B.; Hu, G.; Chu, Y.; Ge, R.S. Inhibition of human and rat testicular steroidogenic enzyme activities by bisphenol A. Toxicol. Lett. 2011, 207, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, B.; Zhai, Y.; Tang, Y.; Lou, Y.; Zhu, Y.; Wang, Y.; Ge, R.S.; Li, H. Structure-activity relationship analysis of perfluoroalkyl carbonic acids on human and rat placental 3beta-hydroxysteroid dehydrogenase activity. Toxicology 2022, 480, 153334. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ji, M.; Wen, X.; Chen, D.; Huang, F.; Guan, X.; Tian, J.; Xie, J.; Shao, J.; Wang, J.; et al. Effects of Midazolam on the Development of Adult Leydig Cells from Stem Cells In Vitro. Front. Endocrinol. 2021, 12, 765251. [Google Scholar] [CrossRef]
- Goldberg, F.W.; Dossetter, A.G.; Scott, J.S.; Robb, G.R.; Boyd, S.; Groombridge, S.D.; Kemmitt, P.D.; Sjogren, T.; Gutierrez, P.M.; de Schoolmeester, J.; et al. Optimization of brain penetrant 11beta-hydroxysteroid dehydrogenase type I inhibitors and in vivo testing in diet-induced obese mice. J. Med. Chem. 2014, 57, 970–986. [Google Scholar] [CrossRef]
- Zhang, J.; Osslund, T.D.; Plant, M.H.; Clogston, C.L.; Nybo, R.E.; Xiong, F.; Delaney, J.M.; Jordan, S.R. Crystal structure of murine 11 beta-hydroxysteroid dehydrogenase 1: An important therapeutic target for diabetes. Biochemistry 2005, 44, 6948–6957. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Ongtanasup, T.; Wanmasae, S.; Srisang, S.; Manaspon, C.; Net-Anong, S.; Eawsakul, K. In silico investigation of ACE2 and the main protease of SARS-CoV-2 with phytochemicals from Myristica fragrans (Houtt.) for the discovery of a novel COVID-19 drug. Saudi J. Biol. Sci. 2022, 29, 103389. [Google Scholar] [CrossRef] [PubMed]
- Ongtanasup, T.; Mazumder, A.; Dwivedi, A.; Eawsakul, K. Homology Modeling, Molecular Docking, Molecular Dynamic Simulation, and Drug-Likeness of the Modified Alpha-Mangostin against the beta-Tubulin Protein of Acanthamoeba Keratitis. Molecules 2022, 27, 6338. [Google Scholar] [CrossRef] [PubMed]
- Schoning-Stierand, K.; Diedrich, K.; Ehrt, C.; Flachsenberg, F.; Graef, J.; Sieg, J.; Penner, P.; Poppinga, M.; Ungethum, A.; Rarey, M. ProteinsPlus: A comprehensive collection of web-based molecular modeling tools. Nucleic Acids Res. 2022, 50, W611–W615. [Google Scholar] [CrossRef]
- Thomas, J.L.; Mason, J.I.; Brandt, S.; Spencer, B.R., Jr.; Norris, W. Structure/function relationships responsible for the kinetic differences between human type 1 and type 2 3beta-hydroxysteroid dehydrogenase and for the catalysis of the type 1 activity. J. Biol. Chem. 2002, 277, 42795–42801. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, F.W.; Leach, A.G.; Scott, J.S.; Snelson, W.L.; Groombridge, S.D.; Donald, C.S.; Bennett, S.N.; Bodin, C.; Gutierrez, P.M.; Gyte, A.C. Free-Wilson and structural approaches to co-optimizing human and rodent isoform potency for 11beta-hydroxysteroid dehydrogenase type 1 (11beta-HSD1) inhibitors. J. Med. Chem. 2012, 55, 10652–10661. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]



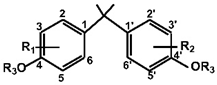
| Compound | R1(C3) | R2(C3′) | R3(C4,4′ -O-) | |
|---|---|---|---|---|
| BPA | H | H | H | 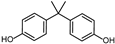 |
| BPG | 3-CH2(CH3)2 | 3′-CH2(CH3)2 | H | 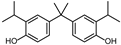 |
| BPH | 3-C6H5 | 3′-C6H5 | H | 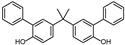 |
| BPAME | H | H | CH3 | 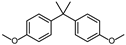 |
| DABPA | 3-CH2CH2CH2 | 3′-CH2CH2CH2 | H | 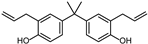 |
| DMBPA | 3-CH3 | 3′-CH3 | H | 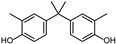 |
| TMBPA | 3-CH3,5-CH3 | 3′-CH3,5′-CH3 | H | 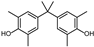 |
| Compound | IC50 μM | Ki μM | Cal. Ki μM | LBE (kcal/mol) | Mode Action | Binding Site |
|---|---|---|---|---|---|---|
| Human 11β-HSD1 (PDB id: 4C7K) | ||||||
| BPA | 297.4 ± 2.99 | ND | ND | ND | ND | ND |
| BPAME | 33.08 ± 0.57 | 33.81 | 33.34 | −6.11 | Competitive | Steroid |
| BPG | 5.06 ± 0.07 | 5.33 | 3.96 | −7.37 | Competitive | Steroid |
| BPH | 0.75 ± 0.02 | 0.44 | 0.40 | −8.72 | Competitive | Steroid |
| DABPA | 13.36 ± 0.19 | 14.04 | 14.52 | −6.60 | Competitive | Steroid |
| DMBPA | 30.18 ± 0.71 | 30.97 | 30.81 | −6.16 | Competitive | Steroid |
| TMBPA | 319.16 ± 2.78 | ND | ND | ND | ND | ND |
| Rat 11β-HSD1 (AlphaFold id: AF-P16232-F1-model_v4) | ||||||
| BPA | 36.76 ± 1.09 | ND | ||||
| BPAME | 205.74 ± 2.16 | ND | 204.2 | −5.05 | ND | Steroid |
| BPG | 103.92 ± 1.52 | 88.65 | 106.97 | −5.40 | Mixed | Steroid |
| BPH | 74.22 ± 0.98 | 74.56 | 73.73 | −5.64 | Competitive | Steroid |
| DABPA | 132.4 ± 1.66 | ND | 133.51 | −5.26 | ND | Steroid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Sang, J.; Ji, Z.; Yu, Y.; Wang, S.; Zhu, Y.; Li, H.; Wang, Y.; Zhu, Q.; Ge, R. Bisphenol A Analogues Inhibit Human and Rat 11β-Hydroxysteroid Dehydrogenase 1 Depending on Its Lipophilicity. Molecules 2023, 28, 4894. https://doi.org/10.3390/molecules28134894
Wang H, Sang J, Ji Z, Yu Y, Wang S, Zhu Y, Li H, Wang Y, Zhu Q, Ge R. Bisphenol A Analogues Inhibit Human and Rat 11β-Hydroxysteroid Dehydrogenase 1 Depending on Its Lipophilicity. Molecules. 2023; 28(13):4894. https://doi.org/10.3390/molecules28134894
Chicago/Turabian StyleWang, Hong, Jianmin Sang, Zhongyao Ji, Yang Yu, Shaowei Wang, Yang Zhu, Huitao Li, Yiyan Wang, Qiqi Zhu, and Renshan Ge. 2023. "Bisphenol A Analogues Inhibit Human and Rat 11β-Hydroxysteroid Dehydrogenase 1 Depending on Its Lipophilicity" Molecules 28, no. 13: 4894. https://doi.org/10.3390/molecules28134894
APA StyleWang, H., Sang, J., Ji, Z., Yu, Y., Wang, S., Zhu, Y., Li, H., Wang, Y., Zhu, Q., & Ge, R. (2023). Bisphenol A Analogues Inhibit Human and Rat 11β-Hydroxysteroid Dehydrogenase 1 Depending on Its Lipophilicity. Molecules, 28(13), 4894. https://doi.org/10.3390/molecules28134894







