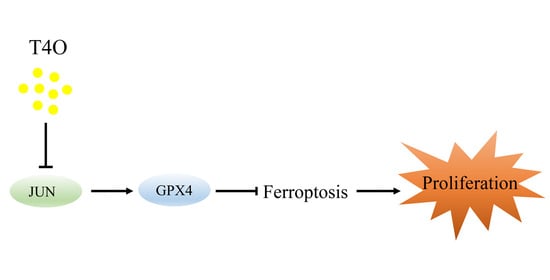
Terpinen-4-ol Induces Ferroptosis of Glioma Cells via Downregulating JUN Proto-Oncogene
Abstract
1. Introduction
2. Results
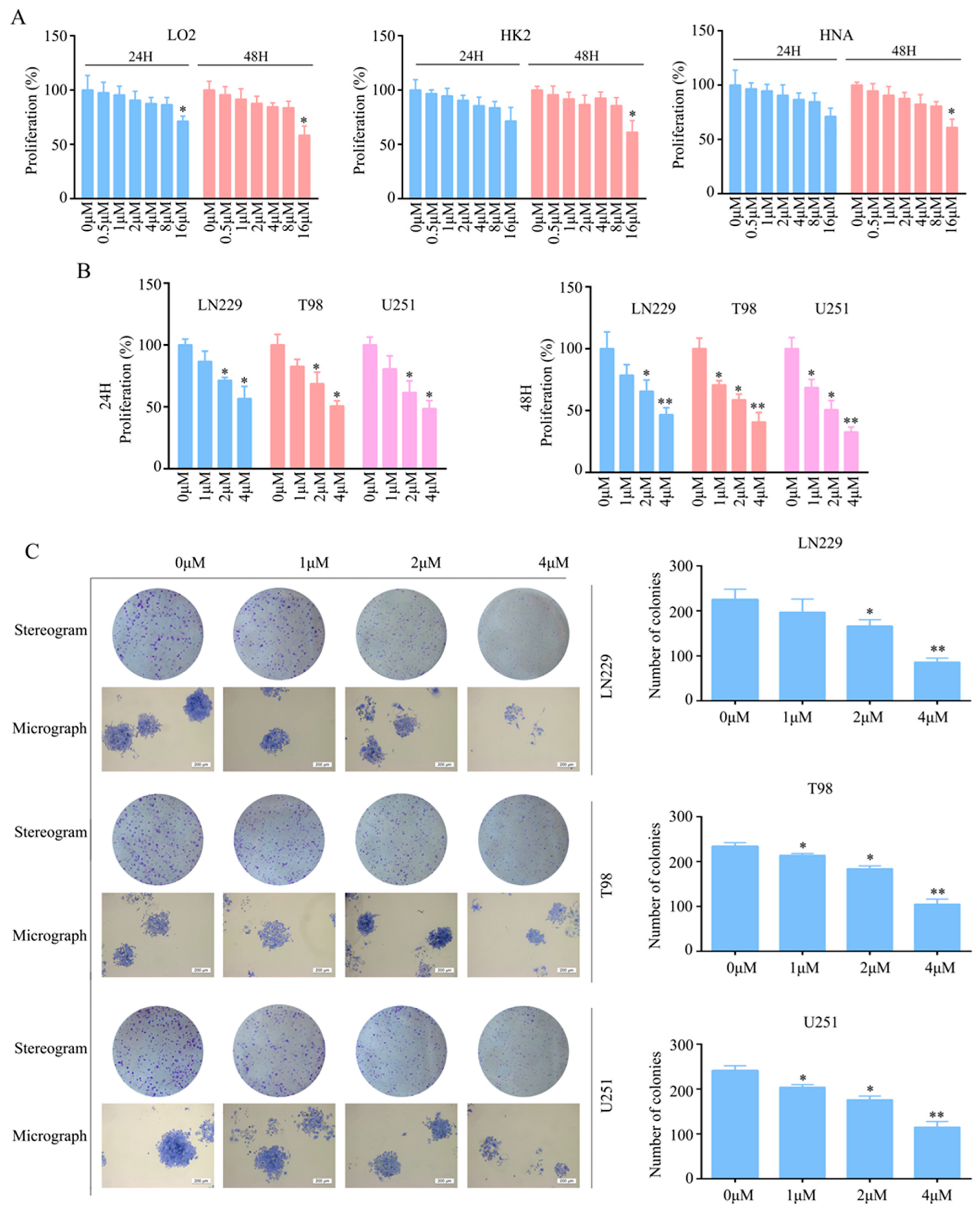
2.1. T4O Inhibited Glioma Cell Proliferation and Colony Formation
2.2. T4O Suppressed the Proliferation of Glioma Cells In Vivo
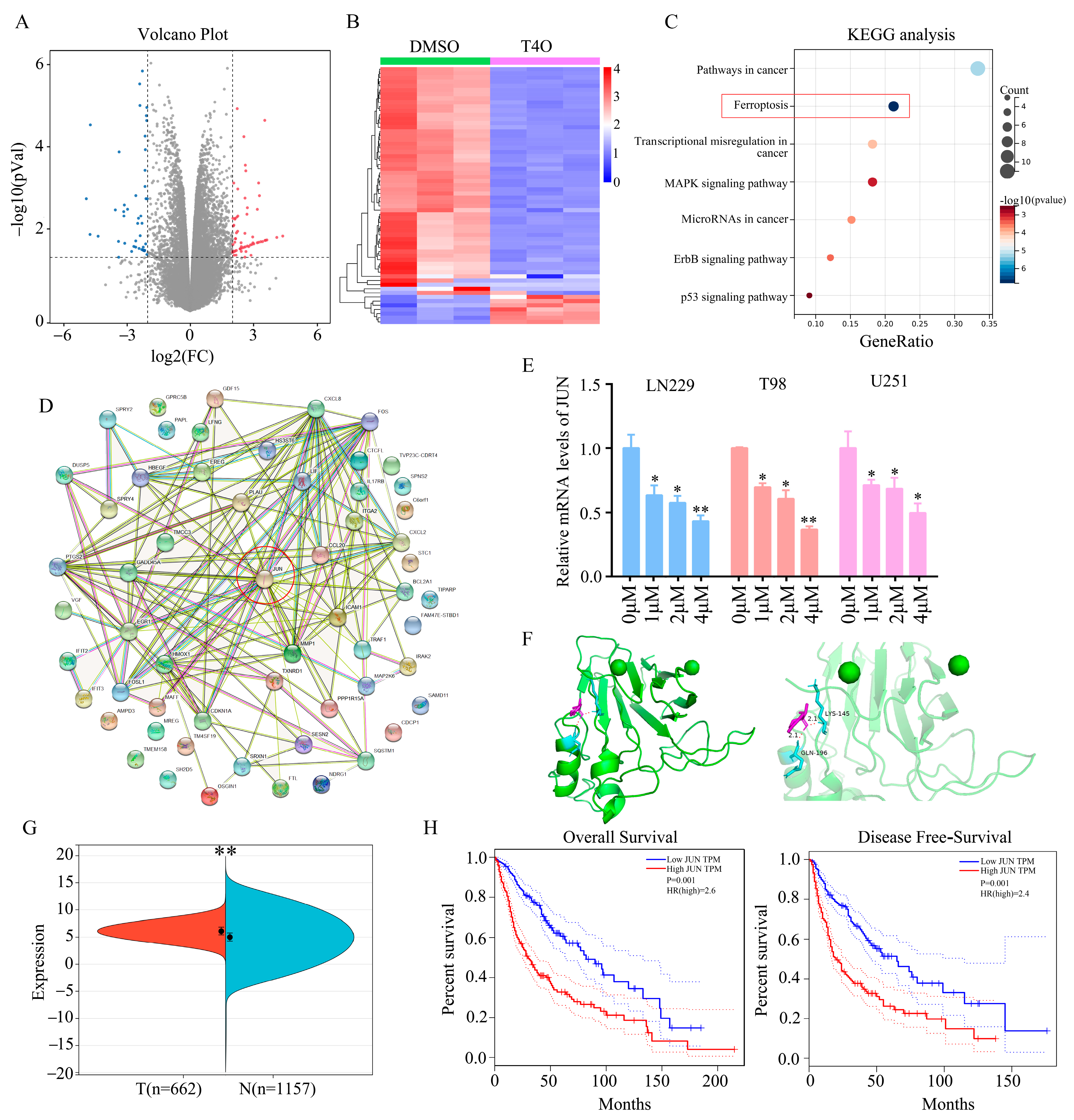
2.3. JUN Was Identified as a Key Target of T4O
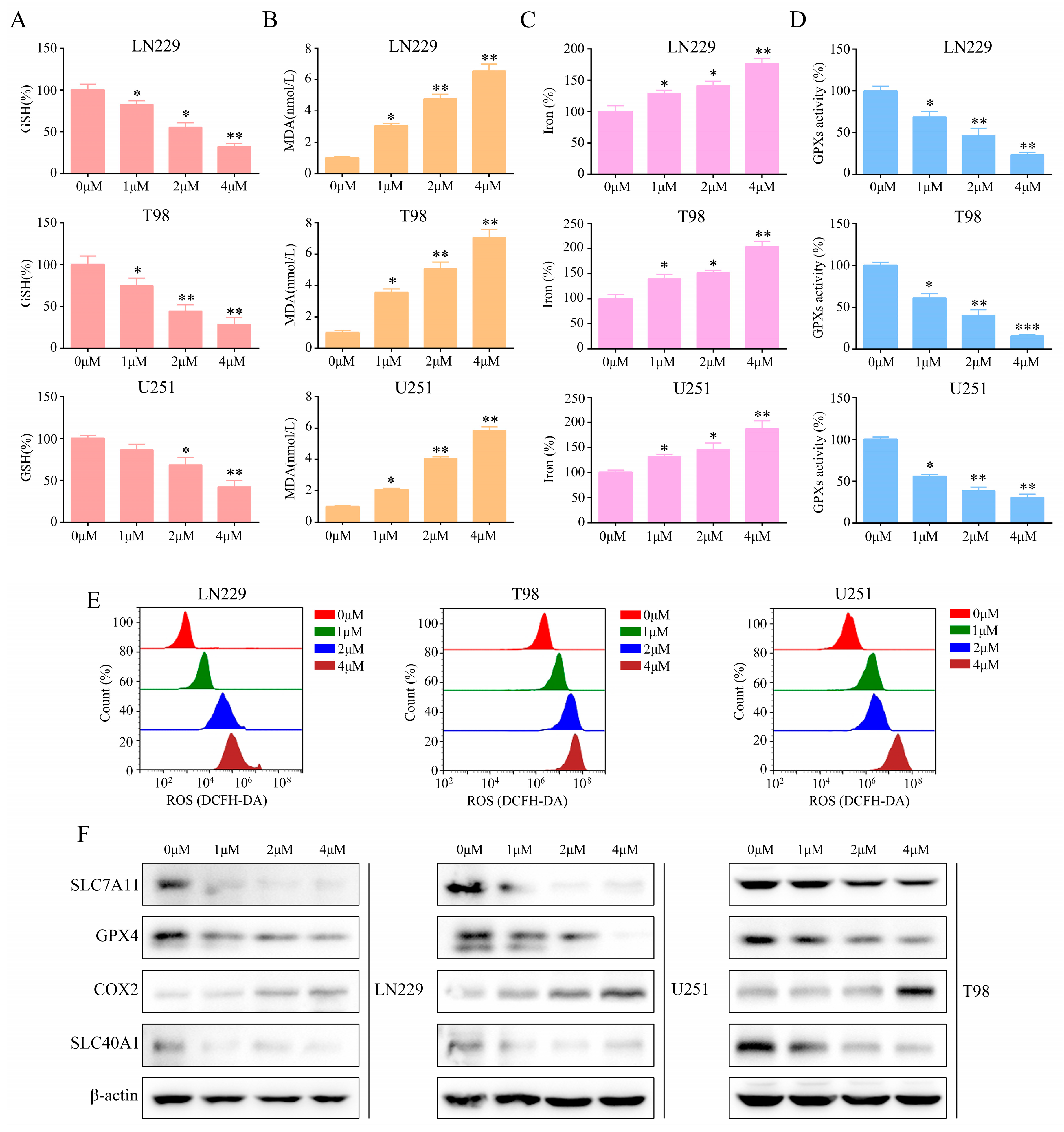
2.4. T4O Induced Glioma Cell Ferroptosis
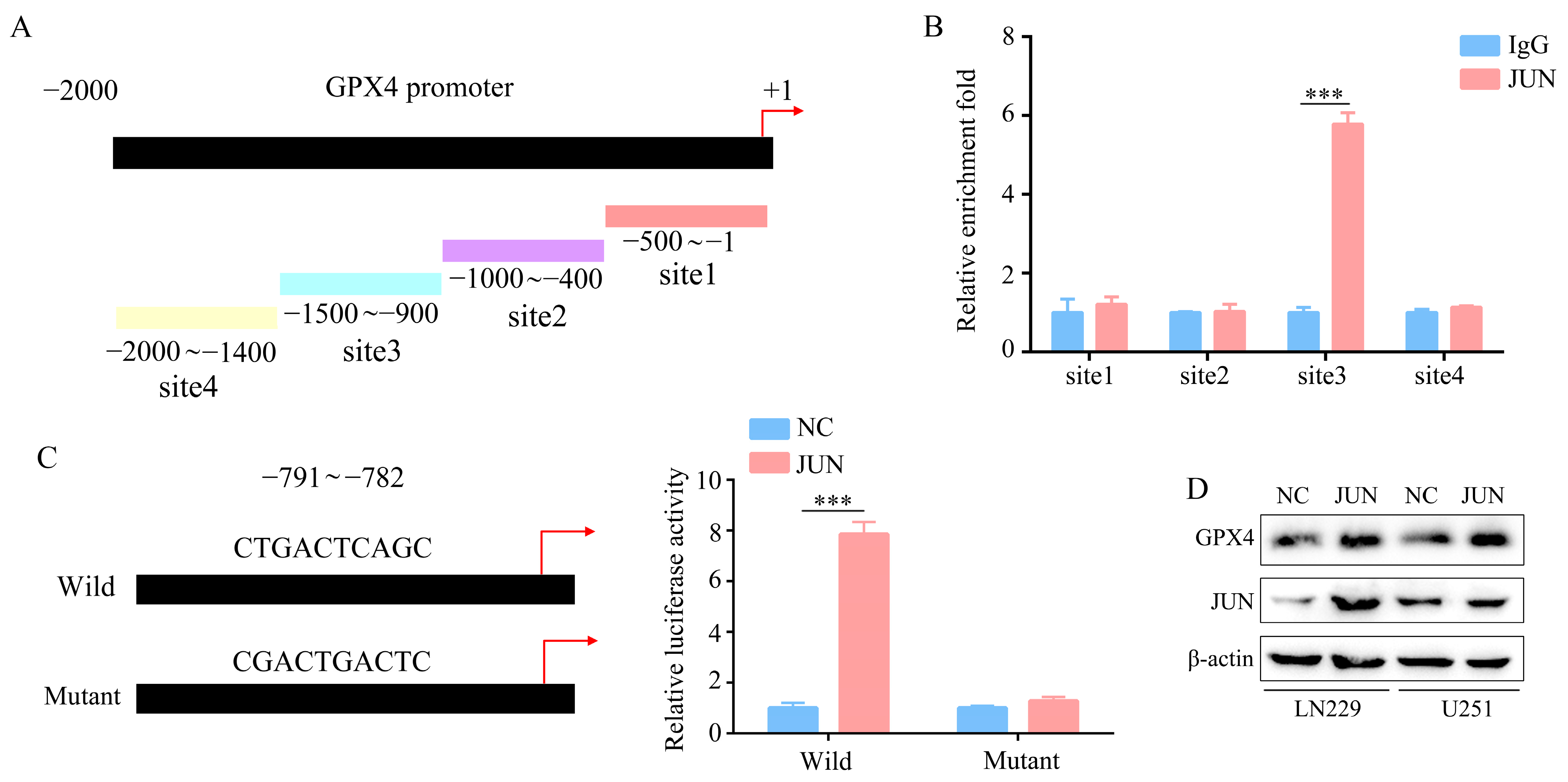
2.5. GPX4 Is a Downstream Gene of JUN
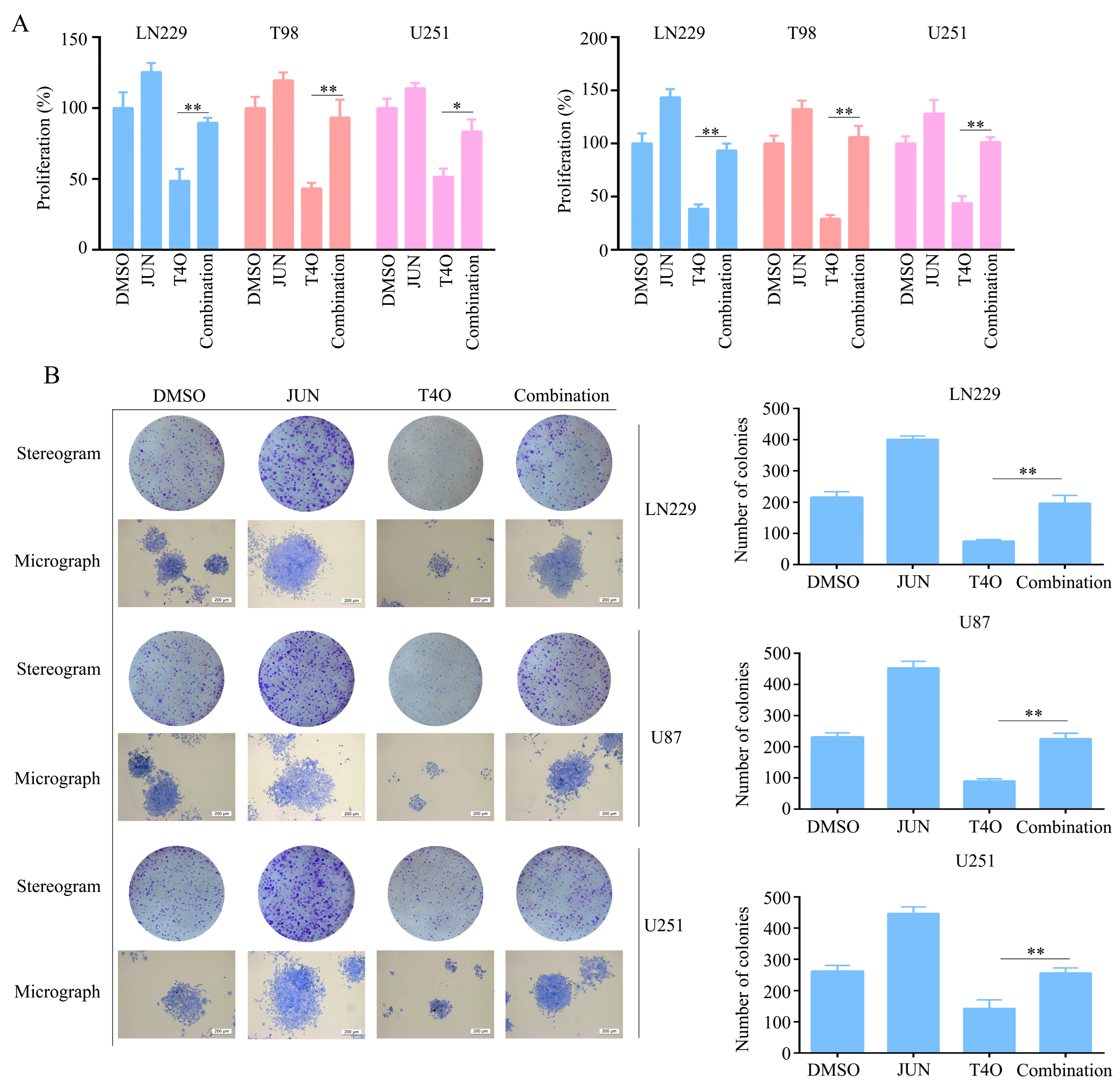
2.6. Overexpression of JUN Attenuated the Inhibitory Effect of T4O on Glioma Cell Proliferation
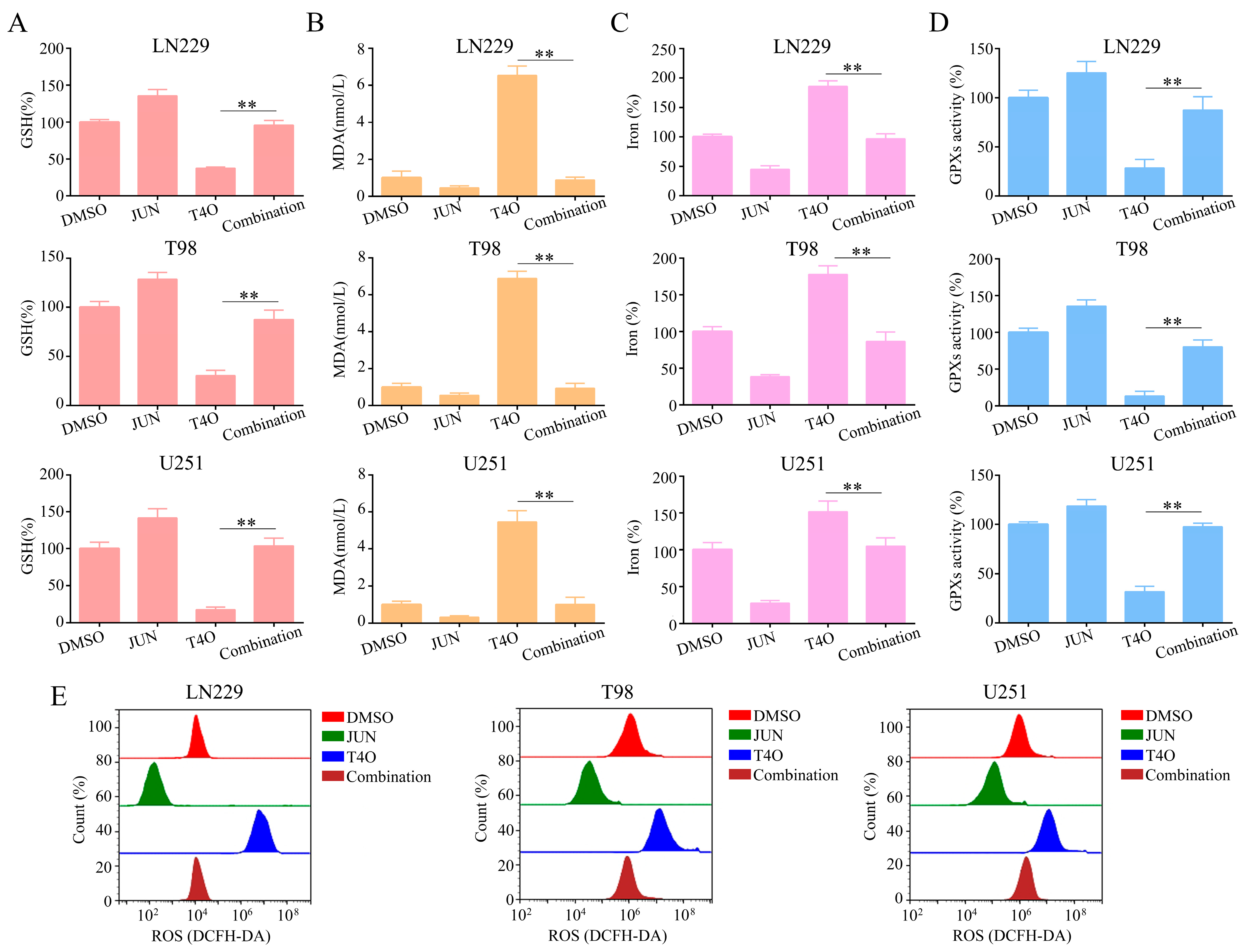
2.7. Overexpression of JUN Attenuated the Promotion of Ferroptosis by T4O in Glioma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. CCK-8 Assay
4.3. Colony Formation Assay
4.4. Western Blot
4.5. Subcutaneous Tumorigenesis Experiments
4.6. Measurement of Cellular Ferroptosis Levels
4.7. RNA Sequencing
4.8. GEPIA Data Analysis
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| T4O | Terpinen-4-ol |
| JUN | JUN proto-oncogene |
| ROS | Reactive oxygen species |
| MDA | Malondialdehyde |
| GSH | Glutathione |
| GPX4 | Glutathione peroxidase 4 |
| RT-qPCR | Real-time quantitative polymerase chain reaction |
References
- Liu, X.-P.; Jin, X.; Ahmadian, S.S.; Yang, X.; Tian, S.-F.; Cai, Y.-X.; Chawla, K.; Snijders, A.M.; Xia, Y.; van Diest, P.J.; et al. Clinical significance and molecular annotation of cellular morphometric subtypes in lower-grade gliomas discovered by machine learning. Neuro Oncol. 2023, 25, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Karimi, E.; Yu, M.W.; Maritan, S.M.; Perus, L.J.M.; Rezanejad, M.; Sorin, M.; Dankner, M.; Fallah, P.; Doré, S.; Zuo, D.; et al. Single-cell spatial immune landscapes of primary and metastatic brain tumours. Nature 2023, 614, 555–563. [Google Scholar] [CrossRef]
- Schaff, L.R.; Mellinghoff, I.K. Glioblastoma and Other Primary Brain Malignancies in Adults: A Review. JAMA 2023, 329, 574–587. [Google Scholar] [CrossRef]
- Tong, S.; Hong, Y.; Xu, Y.; Sun, Q.; Ye, L.; Cai, J.; Ye, Z.; Chen, Q.; Tian, D. TFR2 regulates ferroptosis and enhances temozolomide chemo-sensitization in gliomas. Exp. Cell Res. 2023, 424, 113474. [Google Scholar] [CrossRef]
- Tornero-Martínez, A.; Silva-Lucero, M.D.C.; Sampedro, E.C.; Ramón-Gallegos, E.; Pérez-Cruz, C.; Pérez-Grijalva, B.; Mora-Escobedo, R. Aloe vera and Fermented Extracts Exhibit an Anti-Inflammatory Effect on Human Glioblastoma/Astrocytoma U373 MG Cells. Plant Foods Hum. Nutr. 2022, 77, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Tian, R.; Pan, R.; Sun, B.; Xiao, C.; Chen, Y.; Zeng, Z.; Lei, S. Terpinen-4-ol inhibits the proliferation and mobility of pancreatic cancer cells by downregulating Rho-associated coiled-coil containing protein kinase 2. Bioengineered 2022, 13, 8643–8656. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Murata, S.; Ito, H.; Iwasaki, K.; Villareal, M.O.; Zheng, Y.W.; Matsui, H.; Isoda, H.; Ohkohchi, N. Terpinen-4-ol inhibits colorectal cancer growth via reactive oxygen species. Oncol. Lett. 2017, 14, 2015–2024. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, X.; Hu, C.; Li, H.; Xu, Z.; Lei, P.; Luo, X.; Hao, Y. JUN and PDGFRA as Crucial Candidate Genes for Childhood Autism Spectrum Disorder. Front. Neuroinform. 2022, 16, 800079. [Google Scholar] [CrossRef]
- Chen, H.; Padia, R.; Li, T.; Li, Y.; Li, B.; Jin, L.; Huang, S. Signaling of MK2 sustains robust AP1 activity for triple negative breast cancer tumorigenesis through direct phosphorylation of JAB1. NPJ Breast Cancer 2021, 7, 91. [Google Scholar] [CrossRef]
- Njouendou, A.J.; Szarvas, T.; Tiofack, A.A.Z.; Kenfack, R.N.; Tonouo, P.D.; Ananga, S.N.; Bell, E.; Simo, G.; Hoheisel, J.D.; Siveke, J.T.; et al. SOX2 dosage sustains tumor-promoting inflammation to drive disease aggressiveness by modulating the FOSL2/IL6 axis. Mol. Cancer 2023, 22, 52. [Google Scholar] [CrossRef]
- Shi, H.; Duan, L.; Lan, Y.; He, Q.; Pu, P.; Tang, H. RING Finger Protein 10 Regulates AP-1/Meox2 to Mediate Pirarubicin-Induced Cardiomyocyte Apoptosis. Oxid. Med. Cell. Longev. 2023, 2023, 7872193. [Google Scholar] [CrossRef] [PubMed]
- Sarangthem, V.; Yi, A.; Kim, Y.; Rehemtulla, A.; Lee, B.-H.; Jeon, Y.H.; Singh, T.D.; Park, R.-W. Therapeutic Effect of IL-4 Receptor-Targeting Pro-Apoptotic Peptide (AP1-ELP-KLAK) in Glioblastoma Tumor Model. Int. J. Nanomed. 2021, 16, 5039–5052. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Suzuki, A.; Leland, P.; Joshi, B.H.; Puri, R.K. Identification of a novel role of IL-13Ralpha2 in human Glioblastoma multiforme: Interleukin-13 mediates signal transduction through AP-1 pathway. J. Transl. Med. 2018, 16, 369. [Google Scholar] [CrossRef] [PubMed]
- Dang, S.; Zhang, R.; Tian, S.; Hou, P.; Li, G.; Ji, M. MicroRNA-218 inhibits the malignant phenotypes of glioma by modulating the TNC/AKT/AP-1/TGFbeta1 feedback signaling loop. Int. J. Mol. Med. 2021, 48, 205. [Google Scholar] [CrossRef] [PubMed]
- Kamide, D.; Yamashita, T.; Araki, K.; Tomifuji, M.; Tanaka, Y.; Tanaka, S.; Shiozawa, S.; Shiotani, A. Selective activator protein-1 inhibitor T-5224 prevents lymph node metastasis in an oral cancer model. Cancer Sci. 2016, 107, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Manguinhas, R.; Fernandes, A.S.; Costa, J.G.; Saraiva, N.; Camões, S.P.; Gil, N.; Rosell, R.; Castro, M.; Miranda, J.P.; Oliveira, N.G. Impact of the APE1 Redox Function Inhibitor E3330 in Non-Small Cell Lung Cancer Cells Exposed to Cisplatin: Increased Cytotoxicity and Impairment of Cell Migration and Invasion. Antioxidants 2020, 9, 550. [Google Scholar] [CrossRef]
- Mou, Y.; Wang, J.; Wu, J.; He, D.; Zhang, C.; Duan, C.; Li, B. Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J. Hematol. Oncol. 2019, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, T.; Muanprasat, C. Deoxyelephantopin and Its Isomer Isodeoxyelephantopin: Anti-Cancer Natural Products with Multiple Modes of Action. Molecules 2022, 27, 2086. [Google Scholar] [CrossRef]
- Tewari, D.; Patni, P.; Bishayee, A.; Sah, A.N.; Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 2022, 80, 1–17. [Google Scholar] [CrossRef]
- Wang, X.; Hamann, M.T. Marine natural products in the discovery and development of potential pancreatic cancer therapeutics. Adv. Cancer Res. 2019, 144, 299–314. [Google Scholar]
- Chen, C.J.; Shang, H.S.; Huang, Y.L.; Tien, N.; Chen, Y.L.; Hsu, S.Y.; Wu, R.S.; Tang, C.L.; Lien, J.C.; Lee, M.H.; et al. Bisdemethoxycurcumin suppresses human brain glioblastoma multiforme GBM 8401 cell migration and invasion via affecting NF-kappaB and MMP-2 and MMP-9 signaling pathway in vitro. Environ. Toxicol. 2022, 37, 2388–2397. [Google Scholar] [CrossRef] [PubMed]
- Kocaadam, B.; Sanlier, N. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit. Rev. Food Sci. Nutr. 2017, 57, 2889–2895. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Baidoo, J.N.E.; Sampat, S.; Mancuso, A.; David, L.; Cohen, L.S.; Zhou, S.; Banerjee, P. Liposomal TriCurin, A Synergistic Combination of Curcumin, Epicatechin Gallate and Resveratrol, Repolarizes Tumor-Associated Microglia/Macrophages, and Eliminates Glioblastoma (GBM) and GBM Stem Cells. Molecules 2018, 23, 201. [Google Scholar] [CrossRef] [PubMed]
- Su, R.Y.; Hsueh, S.C.; Chen, C.Y.; Hsu, M.J.; Lu, H.F.; Peng, S.F.; Chen, P.Y.; Lien, J.C.; Chen, Y.L.; Chueh, F.S.; et al. Demethoxycurcumin Suppresses Proliferation, Migration, and Invasion of Human Brain Glioblastoma Multiforme GBM 8401 Cells via PI3K/Akt Pathway. Anticancer Res. 2021, 41, 1859–1870. [Google Scholar] [CrossRef]
- Liu, S.; Zhou, L.; Zhao, Y.; Yuan, Y. β-elemene enhances both radiosensitivity and chemosensitivity of glioblastoma cells through the inhibition of the ATM signaling pathway. Oncol. Rep. 2015, 34, 943–951. [Google Scholar] [CrossRef]
- Majchrzak-Celinska, A.; Kleszcz, R.; Stasilowicz-Krzemien, A.; Cielecka-Piontek, J. Sodium Butyrate Enhances Curcuminoids Permeability through the Blood–Brain Barrier, Restores Wnt/beta-Catenin Pathway Antagonists Gene Expression and Reduces the Viability of Glioblastoma Cells. Int. J. Mol. Sci. 2021, 22, 11285. [Google Scholar] [CrossRef]
- Tu, M.; Klein, L.; Espinet, E.; Georgomanolis, T.; Wegwitz, F.; Li, X.; Urbach, L.; Danieli-Mackay, A.; Küffer, S.; Bojarczuk, K.; et al. TNF-α-producing macrophages determine subtype identity and prognosis via AP1 enhancer reprogramming in pancreatic cancer. Nat. Cancer 2021, 2, 1185–1203. [Google Scholar] [CrossRef]
- Jia, Y.; Yan, Q.; Zheng, Y.; Li, L.; Zhang, B.; Chang, Z.; Wang, Z.; Tang, H.; Qin, Y.; Guan, X.-Y. Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. J. Exp. Clin. Cancer Res. 2022, 41, 287. [Google Scholar] [CrossRef]
- Paerhati, P.; Liu, J.; Jin, Z.; Jakoš, T.; Zhu, S.; Qian, L.; Zhu, J.; Yuan, Y. Advancements in Activating Transcription Factor 5 Function in Regulating Cell Stress and Survival. Int. J. Mol. Sci. 2022, 23, 7129. [Google Scholar] [CrossRef]
- Shi, D.; Mu, S.; Hu, B.; Zhang, S.; Liu, J.; Zhang, Z.; Shao, Z. Prognostic role of c-Jun activation domain-binding protein-1 in cancer: A systematic review and meta-analysis. J. Cell. Mol. Med. 2021, 25, 2750–2763. [Google Scholar] [CrossRef]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef]
- Kar, F.; Hacioğlu, C.; Kaçar, S. The dual role of boron in vitro neurotoxication of glioblastoma cells via SEMA3F/NRP2 and ferroptosis signaling pathways. Environ. Toxicol. 2023, 38, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.; Lu, L.; Pan, S.S.; Wei, X.Q.; Miao, R.R.; Liu, X.H.; Xue, M.; Lin, X.K.; Xu, H.L. Targeting NQO1/GPX4-mediated ferroptosis by plumbagin suppresses in vitro and in vivo glioma growth. Br. J. Cancer 2022, 127, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Ye, Z.; Hu, Y.; Ye, L.; Gao, L.; Wang, Y.; Sun, Q.; Tong, S.; Zhang, S.; Wu, L.; et al. Fatostatin induces ferroptosis through inhibition of the AKT/mTORC1/GPX4 signaling pathway in glioblastoma. Cell Death Dis. 2023, 14, 211. [Google Scholar] [CrossRef]
- Upadhyayula, P.S.; Higgins, D.M.; Mela, A.; Banu, M.; Dovas, A.; Zandkarimi, F.; Patel, P.; Mahajan, A.; Humala, N.; Nguyen, T.T.T.; et al. Dietary restriction of cysteine and methionine sensitizes gliomas to ferroptosis and induces alterations in energetic metabolism. Nat. Commun. 2023, 14, 1187. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, W.; Li, Y.; Zeng, Z.; Lei, S. Terpinen-4-ol Induces Ferroptosis of Glioma Cells via Downregulating JUN Proto-Oncogene. Molecules 2023, 28, 4643. https://doi.org/10.3390/molecules28124643
Cao W, Li Y, Zeng Z, Lei S. Terpinen-4-ol Induces Ferroptosis of Glioma Cells via Downregulating JUN Proto-Oncogene. Molecules. 2023; 28(12):4643. https://doi.org/10.3390/molecules28124643
Chicago/Turabian StyleCao, Wenpeng, Yumei Li, Zhirui Zeng, and Shan Lei. 2023. "Terpinen-4-ol Induces Ferroptosis of Glioma Cells via Downregulating JUN Proto-Oncogene" Molecules 28, no. 12: 4643. https://doi.org/10.3390/molecules28124643
APA StyleCao, W., Li, Y., Zeng, Z., & Lei, S. (2023). Terpinen-4-ol Induces Ferroptosis of Glioma Cells via Downregulating JUN Proto-Oncogene. Molecules, 28(12), 4643. https://doi.org/10.3390/molecules28124643