Application of Band-Selective HSQC NMR in Species Discrimination and Adulteration Identification of Panax Linn
Abstract
1. Introduction
2. Results and Discussion
2.1. The Establishment of Bs-HSQC NMR Coupled with NUS for Ginsenosides
2.2. Performance Evaluation
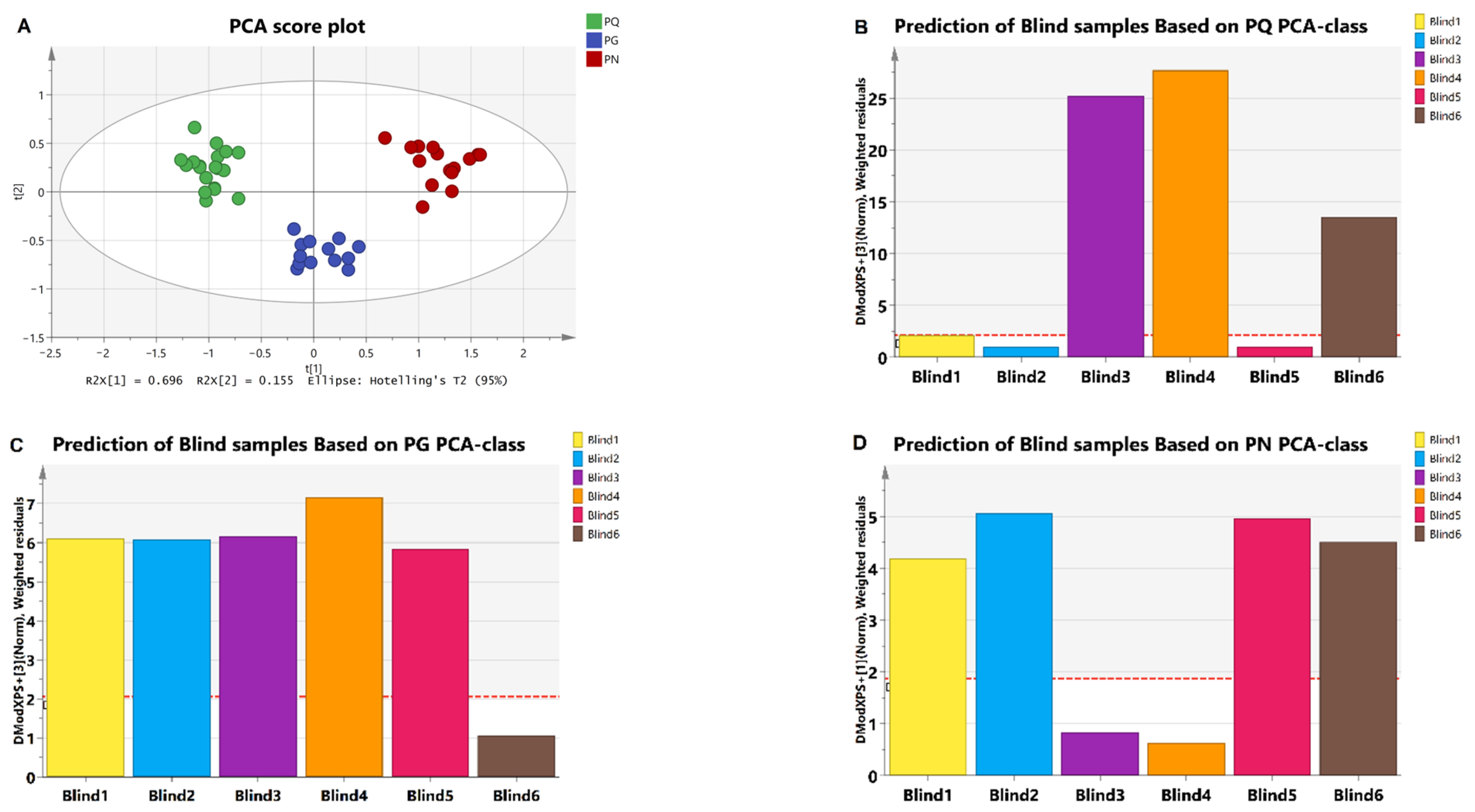
2.3. Species Distinguish
2.4. Identification of Adulteration
2.4.1. Simulative Adulterated Samples at the Signal Mixing Level
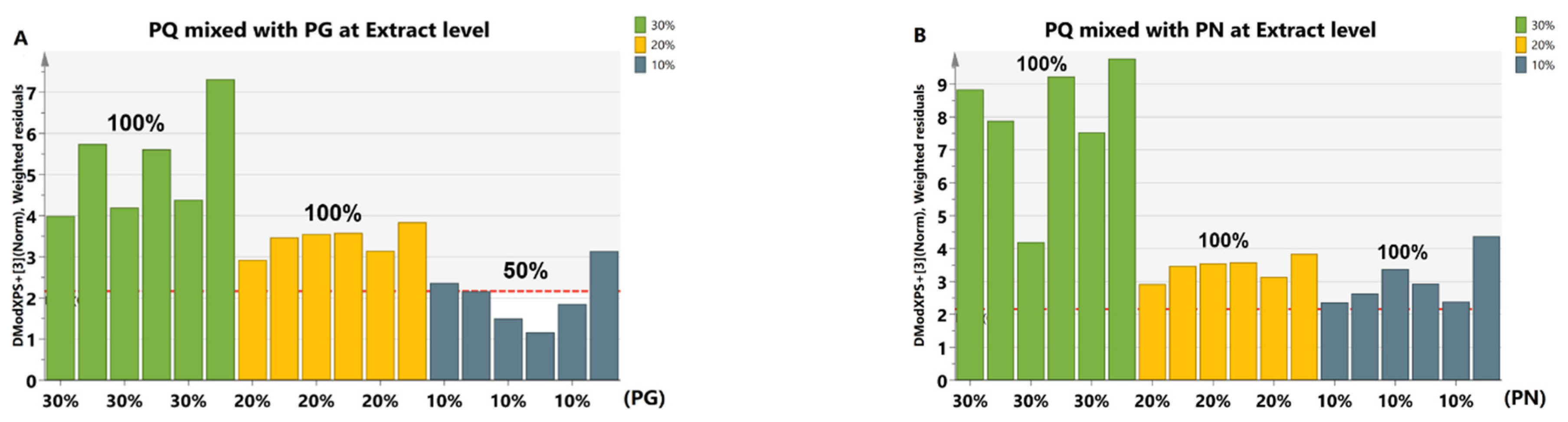
2.4.2. Adulterated Samples at the Saponins Extract Level
2.4.3. Adulterated Samples at the Herb Level
3. Materials and Methods
3.1. Materials
3.1.1. Samples Collection
3.1.2. Extraction of Saponins
3.1.3. Adulterated Samples Preparation
- (1)
- Digitally diluted adulterated data: We took 6 groups of NMR signals from PQ, PG, and PN separately. The integration was multiplied by scale factors according to the desired relative content of saponins extract, and then added the obtained data correspondingly to generate simulative adulterated samples at the signal mixing level. For example, in order to obtain the samples of PQ mixed with 10% PG, the integration from pure PQ and PG was multiplied by 90% and 10%, respectively. Then, the obtained values were added correspondingly to obtain the theoretical integration of 90% PQ + 10% PG. In this way, we obtained PQ adulteration mixed with PG and PN, respectively. The proportions of adulterant were 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, and 90%. There were 36 samples for each proportion (Table S5). PG and PN adulteration was obtained in the same way;
- (2)
- Physically diluted adulterated extract: We took 4 samples of saponins extract of PQ, PG, and PN separately. They were prepared in 10 mg/mL mother liquor with Pyr-d5, respectively. Then, according to the desired relative content, we took the corresponding volume of mother liquor and mixed it to obtain adulterated samples at the saponins extract level. For example, in order to obtain the PQ extracts mixed with that of 10% PG, 720 μL PQ extract solution and 80 μL PG were mixed to obtain 90% PQ + 10% PG. In this way, we obtained the PQ adulteration at extract level mixed with PG in 3 proportions of 10%, 20%, and 30%, respectively. There were 6 samples for each proportion. A total of 18 samples were obtained (Table S6). The PQ adulteration mixed with PN at extract level was obtained in the same way. The proportions of PN were 10%, 20%, and 30%, respectively. A total of 18 samples were obtained;
- (3)
- Physically diluted adulterated herb: Take 1 sample of PQ and 2 of PG. They were crushed and mixed in a glass bottle according to the desired relative mass ratio to obtain adulterated samples at the herb level. For example, to obtain a sample of PQ mixed with 10% PG, 9 g PQ and 1 g PG were mixed. In this way, we obtained the mixtures of PQ mixed with PG in 9 proportions of 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, and 90%, respectively. A total of 18 samples were obtained with 2 in each proportion (Table S7).
3.2. Methods
3.2.1. NMR Sample Preparation
3.2.2. Data Processing
3.2.3. Multivariate Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, C.-F.; Chiou, W.-F.; Zhang, J.-T. Comparison of the pharmacological effects of Panax ginseng and Panax quinquefolium. Acta Pharmacol. Sin. 2008, 29, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.-Y.; Liu, X.-C.; Sun, S.; Shi, C.-C.; Du, X.; Han, K.; Yang, B.-R.; Fu, Y.-Y.; Liu, M.-H.; Seim, I.; et al. The Chromosome Level Genome and Genome-wide Association Study for the Agronomic Traits of Panax Notoginseng. iScience 2020, 23, 101538. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.-H.; Liu, C.-K.; Yang, Z.-Y.; Yang, L.-F.; He, Z.-S.; Wang, H.-C.; Yang, J.-B.; Yi, T.-S. Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae). Mol. Ecol. Resour. 2019, 19, 1333–1345. [Google Scholar] [CrossRef]
- Yang, W.-Z.; Shi, X.-J.; Yao, C.-L.; Huang, Y.; Hou, J.-J.; Han, S.-M.; Feng, Z.-J.; Wei, W.-L.; Wu, W.-Y.; Guo, D.-A. A novel neutral loss/product ion scan-incorporated integral approach for the untargeted characterization and comparison of the carboxyl-free ginsenosides from Panax ginseng, Panax quinquefolius, and Panax notoginseng. J. Pharm. Biomed. Anal. 2020, 177, 112813. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-D.; Zhang, C.-X.; Zuo, T.-T.; Li, W.-W.; Jia, L.; Wang, X.-Y.; Qian, Y.-X.; Guo, D.; Yang, W.-Z. In-depth profiling, characterization, and comparison of the ginsenosides among three different parts (the root, stem leaf, and flower bud) of Panax quinquefolius L. by ultra-high performance liquid chromatography/quadrupole-Orbitrap mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 7817–7829. [Google Scholar] [CrossRef]
- Dai, Y.-L.; Qiao, M.-D.; Yu, P.; Zheng, F.; Yue, H.; Liu, S.-Y. Comparing eight types of ginsenosides in ginseng of different plant ages and regions using RRLC-Q-TOF MS/MS. J. Ginseng Res. 2020, 44, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.-C.; Wu, Q.-S.; Wang, R.; Li, S.-P.; Lin, L.-G.; Chen, P.; Zhang, Q.-W. A Novel Strategy for Quantitative Analysis of Major Ginsenosides in Panacis Japonici Rhizoma with a Standardized Reference Fraction. Molecules 2017, 22, 2067. [Google Scholar] [CrossRef]
- Ouyang, L.-F.; Wang, Z.-L.; Dai, J.-G.; Chen, L.; Zhao, Y.-N. Determination of total ginsenosides in ginseng extracts using charged aerosol detection with post-column compensation of the gradient. Chin. J. Nat. Med. 2014, 12, 857–868. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Sakaridis, I.; Argiriou, A.; Tsaftaris, A. Advances of DNA-based methods for tracing the botanical origin of food products. Food Res. Int. 2014, 60, 163–172. [Google Scholar] [CrossRef]
- Wan, J.-B.; Li, S.P.; Chen, J.-M.; Wang, Y.-T. Chemical characteristics of three medicinal plants of the Panax genus determined by HPLC-ELSD. J. Sep. Sci. 2007, 30, 825–832. [Google Scholar] [CrossRef]
- Convertini, R.; Patz, C.; Kumar, K.; May, B.; Andlauer, W.; Schweiggert, R. 1H NMR spectrometry for methanol quantification in apple wines and ciders as optimised by comparison to SIDA-HS-GC-MS. Food Chem. 2022, 387, 132912. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Hatzakis, E. NMR-Based Analysis of Pomegranate Juice Using Untargeted Metabolomics Coupled with Nested and Quantitative Approaches. Anal. Chem. 2020, 92, 11177–11185. [Google Scholar] [CrossRef] [PubMed]
- Scettri, A.; Schievano, E. Quantification of polyols in sugar-free foodstuffs by qNMR. Food Chem. 2022, 390, 133125. [Google Scholar] [CrossRef] [PubMed]
- Belmonte-Sánchez, E.; Romero-González, R.; Garrido Frenich, A. Applicability of high-resolution NMR in combination with chemometrics for the compositional analysis and quality control of spices and plant-derived condiments. J. Sci. Food Agric. 2021, 101, 3541–3550. [Google Scholar] [CrossRef]
- Babenko, M.; Peron, J.-R.; Kaialy, W.; Calabrese, G.; Alany, R.-G.; ElShaer, A. 1H NMR quantification of spray dried and spray freeze-dried saccharide carriers in dry powder inhaler formulations. Int. J. Pharm. 2019, 564, 318–328. [Google Scholar] [CrossRef]
- Huang, Y.-J.; Li, X.; Peng, X.-R.; Adegoke, A.-T.; Chen, J.-C.; Su, H.-G.; Hu, G.-L.; Wei, G.; Qiu, M.-H. NMR-based Structural Classification, Identification, and Quantification of Triterpenoids from Edible Mushroom Ganoderma resinaceum. J. Agric. Food Chem. 2020, 68, 2816–2825. [Google Scholar] [CrossRef]
- Wang, T.-T.; Liu, Q.-H.; Wang, M.; Zhou, J.; Yang, M.-R.; Chen, G.; Tang, F.-F.; Hatzakis, E.; Zhang, L.-M. Quantitative Measurement of a Chiral Drug in a Complex Matrix: A J-Compensated Quantitative HSQC NMR Method. Anal. Chem. 2020, 92, 3636–3642. [Google Scholar] [CrossRef]
- Peez, N.; Janiska, M.-C.; Imhof, W. The first application of quantitative 1H NMR spectroscopy as a simple and fast method of identification and quantification of microplastic particles (PE, PET, and PS). Anal. Bioanal. Chem. 2019, 411, 823–833. [Google Scholar] [CrossRef]
- Crestini, C.; Melone, F.; Sette, M.; Saladino, R. Milled wood lignin: A linear oligomer. Biomacromolecules 2011, 12, 3928–3935. [Google Scholar] [CrossRef]
- Mauri, L.; Boccardi, G.; Torri, G.; Karfunkle, M.; Macchi, E.; Muzi, L.; Keire, D.; Guerrini, M. Qualification of HSQC methods for quantitative composition of heparin and low molecular weight heparins. J. Pharm. Biomed. Anal. 2017, 136, 92–105. [Google Scholar] [CrossRef]
- Liu, F.-C.; Su, C.-R.; Wu, T.-Y.; Su, S.-G.; Yang, H.-L.; Lin, J.-H.; Wu, T.-S. Efficient H-NMR quantitation and investigation of N-acetyl-d-glucosamine (GlcNAc) and N, N’-diacetylchitobiose (GlcNAc)(2) from chitin. Int. J. Mol. Sci. 2011, 12, 5828–5843. [Google Scholar] [CrossRef] [PubMed]
- Kunc, F.; Nirmalananthan-Budau, N.; Rühle, B.; Sun, Y.; Johnston, L.-J.; Resch-Genger, U. Interlaboratory Comparison on the Quantification of Total and Accessible Amine Groups on Silica Nanoparticles with qNMR and Optical Assays. Anal. Chem. 2021, 93, 15271–15278. [Google Scholar] [CrossRef] [PubMed]
- Çiçek, S.-S.; Pfeifer Barbosa, A.-L.; Girreser, U. Quantification of diterpene acids in Copaiba oleoresin by UHPLC-ELSD and heteronuclear two-dimensional qNMR. J. Pharm. Biomed. Anal. 2018, 160, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Çiçek, S.-S.; Ugolini, T.; Girreser, U. Two-dimensional qNMR of anthraquinones in Frangula alnus (Rhamnus frangula) using surrogate standards and delay time adaption. Anal. Chim. Acta 2019, 1081, 131–137. [Google Scholar] [CrossRef]
- Gaillet, C.; Lequart, C.; Debeire, P.; Nuzillard, J.M. Band-selective HSQC and HMBC experiments using excitation sculpting and PFGSE. J. Magn. Reson. 1999, 139, 454–459. [Google Scholar] [CrossRef]
- Poppe, L.; van Halbeek, H. 13C-selective, 1H-detected 2D {1H, 13C} correlation spectra of oligosaccharides. Magn. Reson. Chem. 1991, 29, 848–851. [Google Scholar] [CrossRef]
- Mobli, M.; Hoch, J.-C. Nonuniform sampling and non-Fourier signal processing methods in multidimensional NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 83, 21–41. [Google Scholar] [CrossRef]
- Gołowicz, D.; Kasprzak, P.; Orekhov, V.; Kazimierczuk, K. Fast time-resolved NMR with non-uniform sampling. Prog. Nucl. Magn. Reson. Spectrosc. 2020, 116, 40–55. [Google Scholar] [CrossRef]
- Mohanan, P.; Subramaniyam, S.; Mathiyalagan, R.; Yang, D.-C. Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. J. Ginseng Res. 2018, 42, 123–132. [Google Scholar] [CrossRef]
- Zhang, B.; Powers, R.; O’Day, E.M. Evaluation of Non-Uniform Sampling 2D 1H-13C HSQC Spectra for Semi-Quantitative Metabolomics. Metabolites 2020, 10, 203. [Google Scholar] [CrossRef]
- Fiamegos, Y.; Dumitrascu, C.; Papoci, S.; de la Calle, M.-B. Authentication of PDO paprika powder (Pimentón de la Vera) by multivariate analysis of the elemental fingerprint determined by ED-XRF. A feasibility study. Food Control 2021, 120, 107496. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-Q.; Deng, A.-J.; Qin, H.-L. Distinctive features of chemical composition of Bupleurum chinense applicable to original authentication. Anal. Methods 2014, 6, 1067–1075. [Google Scholar] [CrossRef]
- Van den Berg, R.-A.; Hoefsloot, H.-C.; Westerhuis, J.-A.; Smilde, A.-K.; van der Werf, M.-J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]

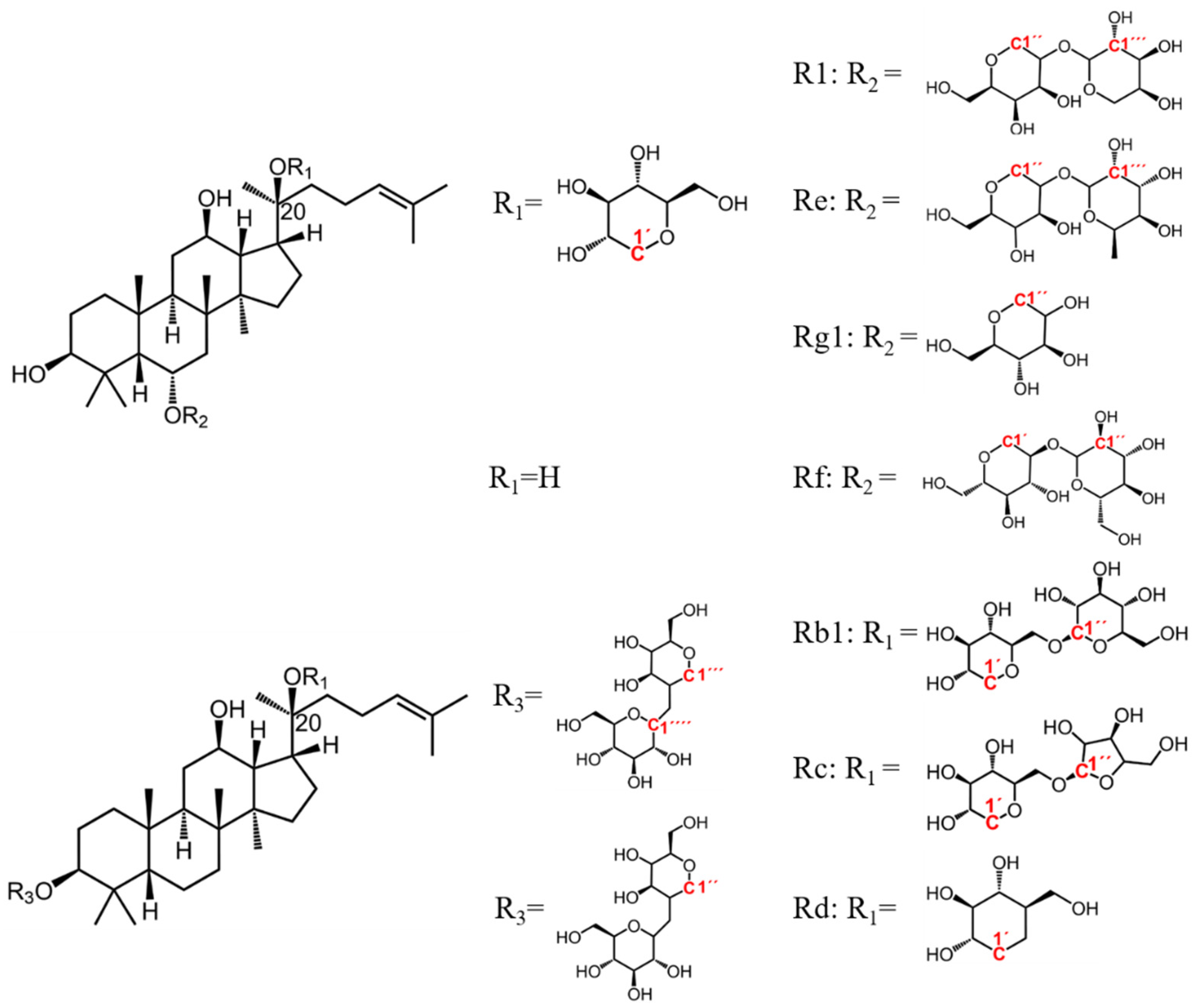




| No. | δC | δH | Attribution | PQ | PG | PN | Statistical Variable |
|---|---|---|---|---|---|---|---|
| 1 | 101.75 | 6.50 | Re-1‴ | 248.78 | 63.29 | 31.15 | Y |
| 2 | 103.55 | 5.93 | Rf-1″ | - | 23.71 | - | Y |
| 3 | 104.55 | 5.78 | R1-1‴ | - | 19.70 | 51.10 | Y |
| 4 | 109.94 | 5.66 | Rc-1″ | 17.37 | 31.04 | - | N |
| 5 | 105.79 | 5.37 | Rc-1′′′′ + Rb1-1′′′′ | 140.38 | 146.42 | 120.68 | Y |
| 6 | 101.64 | 5.25 | Re-1″ | 163.80 | 39.03 | 16.64 | N |
| 7 | 97.99 | 5.14 | 1H/13C-1′ | 265.80 | 260.36 | 332.82 | Y |
| 8 | 105.16 | 5.09 | Rb1-1″ | 118.73 | 75.64 | 91.10 | Y |
| 9 | 105.72 | 5.01 | Rg1-1″ | 15.98 | 116.42 | 248.25 | Y |
| 10 | 104.52 | 4.99 | Unknown1 | - | 16.45 | - | N |
| 11 | 106.78 | 4.93 | Unknown2 | 21.38 | - | - | N |
| 12 | 103.33 | 4.91 | R1-1″ | - | - | 29.71 | N |
| 13 | 103.66 | 4.91 | Rf-1′ | - | 13.43 | - | N |
| 14 | 104.89 | 4.90 | Rd-1″ +Rc-1‴ +Rb1-1‴ | 128.07 | 118.61 | 106.73 | Y |
| Ginsenosides | Precision (%) | Repeatability (%) | Stability (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PQ | PG | PN | PQ | PG | PN | PQ | PG | PN | |
| Re-1‴ | 3.84 | 12.99 | 23.77 | 1.63 | 6.36 | 8.21 | 0.85 | 7.28 | 22.27 |
| Rf-1″ | - | 11.92 | - | - | 17.89 | - | - | 14.57 | - |
| R1-1‴ | - | - | 7.77 | - | - | 7.40 | - | 0.00 | 0.00 |
| Rc-1″ | 22.27 | 8.45 | - | 33.47 | 12.65 | - | 10.65 | 0.00 | - |
| Rc-1′′′′ + Rb1-1′′′′ | 5.73 | 1.45 | 6.51 | 1.86 | 4.13 | 3.74 | 2.26 | 1.70 | 1.68 |
| 1H/13C-1′ | 3.00 | 3.26 | 5.03 | 1.59 | 2.39 | 2.84 | 1.09 | 6.64 | 0.83 |
| Rb1-1″ | 1.86 | 1.50 | 9.25 | 1.60 | 7.97 | 5.32 | 2.67 | 11.89 | 1.93 |
| Rg1-1″ | - | 3.19 | 9.32 | 22.27 | 2.53 | 3.32 | 14.41 | 2.35 | 0.85 |
| Unknown1 | - | 0.00 | - | - | 31.05 | - | - | 22.13 | - |
| Unknown2 | 16.85 | - | - | 9.96 | - | - | 41.06 | - | 24.49 |
| Rd-1″ +Rc-1‴ +Rb1-1‴ | 1.90 | 1.64 | 8.66 | 0.71 | 4.47 | 5.12 | 1.03 | 2.07 | 3.96 |
| Average | 6.45 | 5.57 | 4.78 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, C.; Dong, J.; Deng, L.; Cheng, K.; Xu, Y.; Zhu, H.; Deng, A.; Zhou, X.; Qin, H.; Wang, Y. Application of Band-Selective HSQC NMR in Species Discrimination and Adulteration Identification of Panax Linn. Molecules 2023, 28, 4332. https://doi.org/10.3390/molecules28114332
Guo C, Dong J, Deng L, Cheng K, Xu Y, Zhu H, Deng A, Zhou X, Qin H, Wang Y. Application of Band-Selective HSQC NMR in Species Discrimination and Adulteration Identification of Panax Linn. Molecules. 2023; 28(11):4332. https://doi.org/10.3390/molecules28114332
Chicago/Turabian StyleGuo, Congcong, Jiyang Dong, Lingli Deng, Kiankai Cheng, Yue Xu, Haowen Zhu, Anjun Deng, Xia Zhou, Hailin Qin, and Yinghong Wang. 2023. "Application of Band-Selective HSQC NMR in Species Discrimination and Adulteration Identification of Panax Linn" Molecules 28, no. 11: 4332. https://doi.org/10.3390/molecules28114332
APA StyleGuo, C., Dong, J., Deng, L., Cheng, K., Xu, Y., Zhu, H., Deng, A., Zhou, X., Qin, H., & Wang, Y. (2023). Application of Band-Selective HSQC NMR in Species Discrimination and Adulteration Identification of Panax Linn. Molecules, 28(11), 4332. https://doi.org/10.3390/molecules28114332









