Potent Antifungal Functions of a Living Modified Organism Protein, CP4-EPSPS, against Pathogenic Fungal Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Expression and Purification of Agrobacterium EPSPS Protein
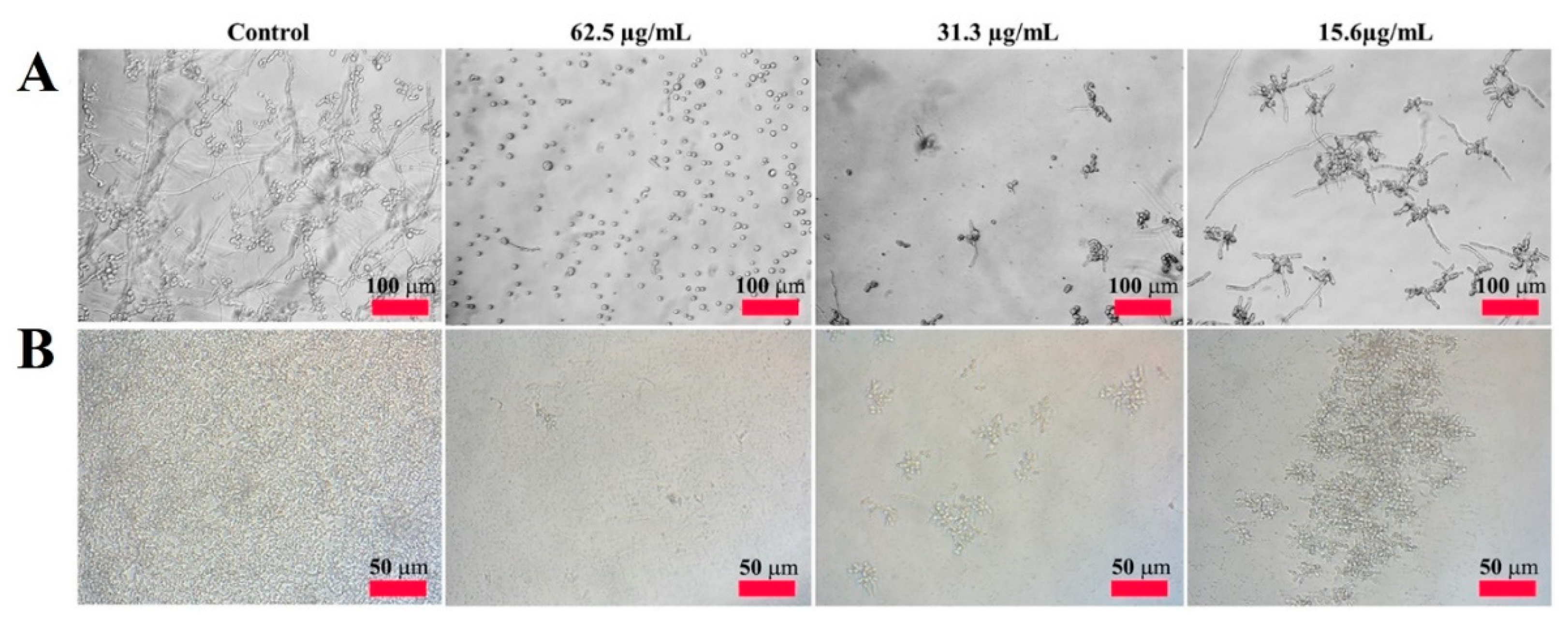
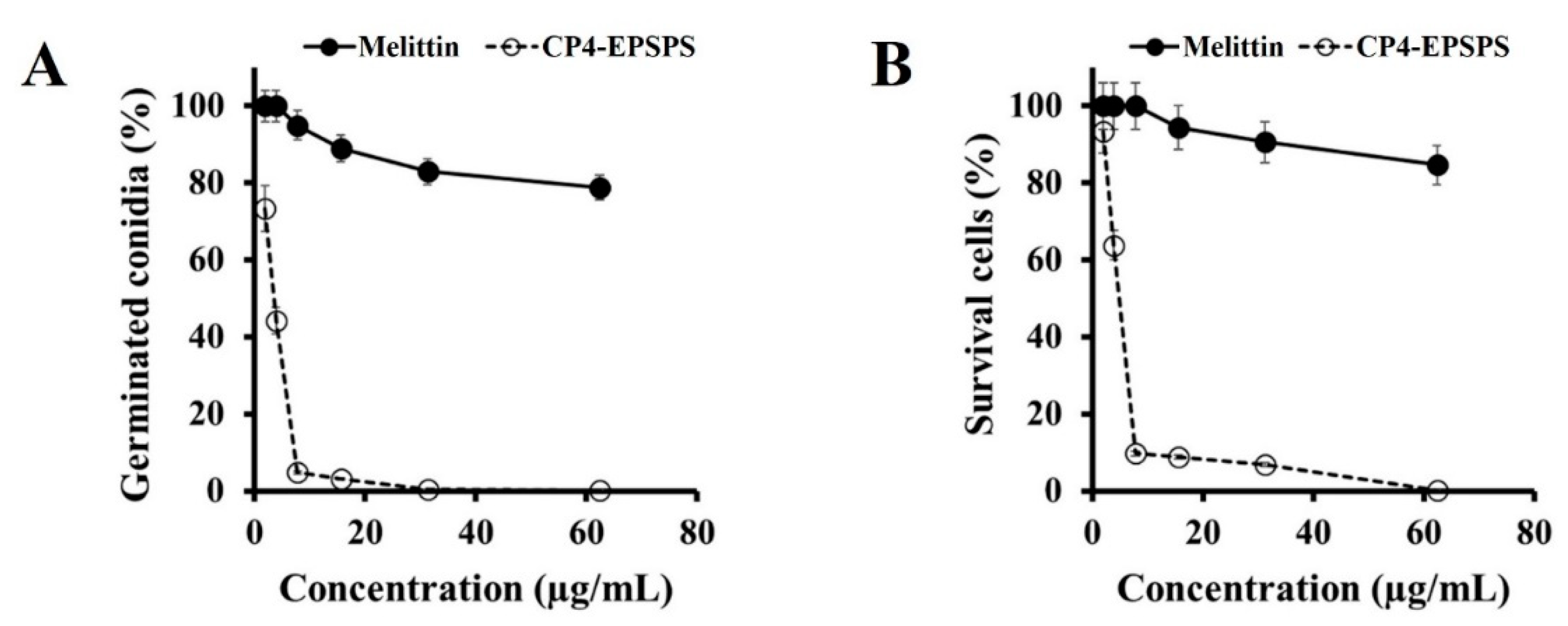
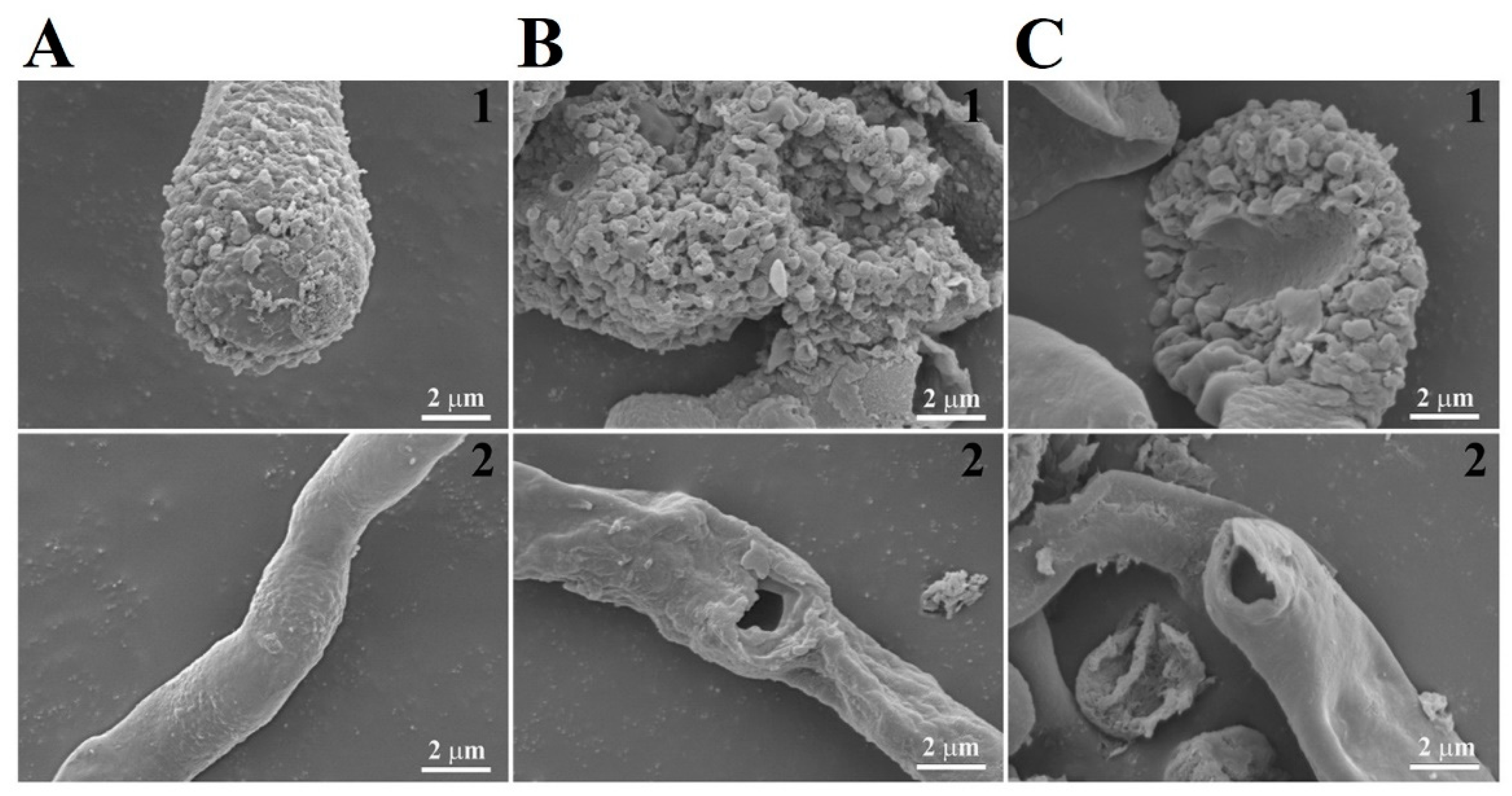
2.2. Antifungal Activity of CP4-EPSPS
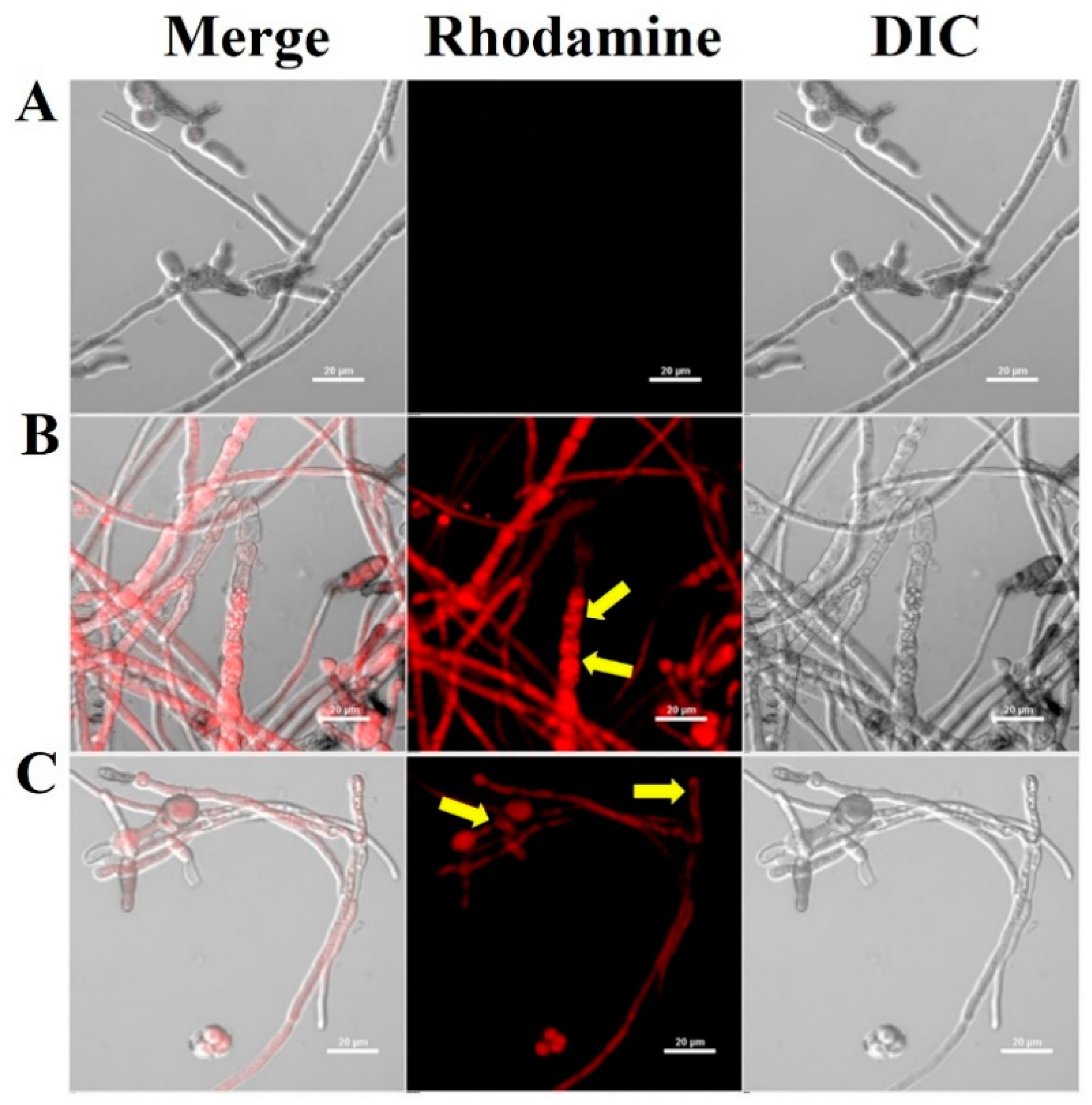
2.3. Subcellular Localization of CP4-EPSPS in Fungal Cells
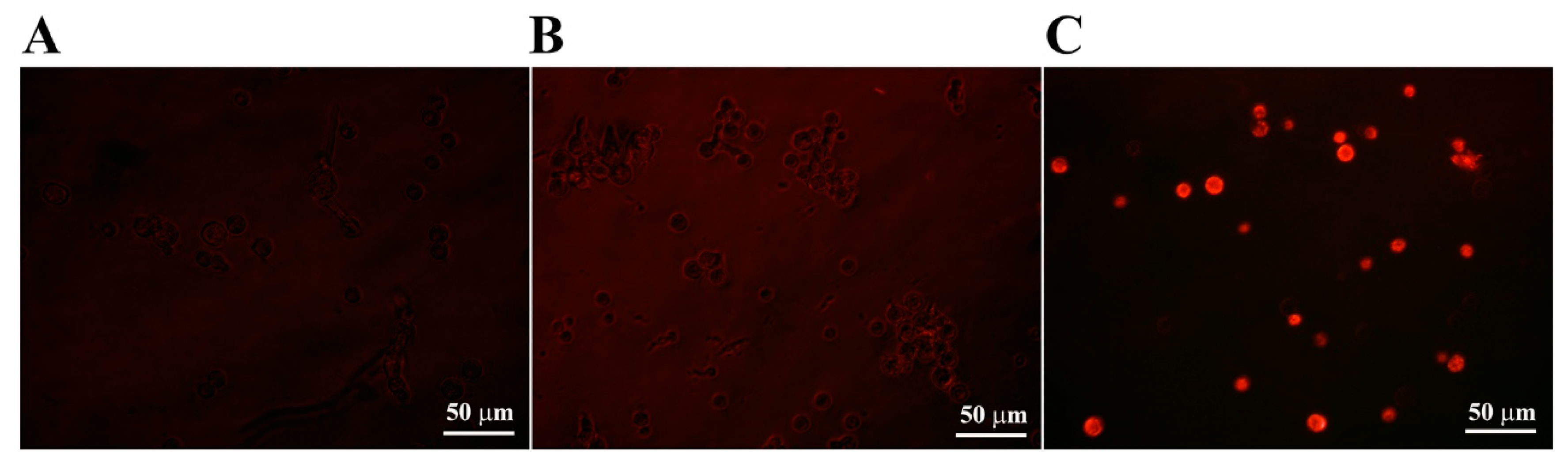
2.4. Cellular Uptake of CP4-EPSPS Protein via Membrane Integrity Change
3. Materials and Methods
3.1. Materials
3.2. Fungal Strains
3.3. Cloning of CP4-EPSPS Gene and Expression of the Protein in Escherichia Coli
3.4. Peptide Synthesis
3.5. SEC and PAGE
3.6. Antifungal Assay
3.7. Analysis Using Confocal Laser Scanning Microscopy (CLSM)
3.8. PI uptake Assay
3.9. SYTOX Green Uptake Assay
3.10. Mitochondrial Superoxide Assay
3.11. Scanning Electron Microscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Gatehouse, A.M.R.; Ferry, N.; Edwards, M.G.; Bell, H.A. Insect-Resistant Biotech Crops and Their Impacts on Beneficial Arthropods. Philos. Trans. R Soc. Lond. B Biol. Sci. 2011, 366, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.S.T. Genetically Engineered (Modified) Crops (Bacillus thuringiensis Crops) and the World Controversy on Their Safety. Egypt. J. Biol. Pest Co. 2018, 28, 52. [Google Scholar] [CrossRef]
- Bonny, S. Genetically Modified Herbicide-Tolerant Crops, Weeds, and Herbicides: Overview and Impact. Environ. Manag. 2016, 57, 31–48. [Google Scholar] [CrossRef]
- Yang, X.; Li, L.; Jiang, X.; Wang, W.; Cai, X.; Su, J.; Wang, F.; Lu, B.R. Genetically Engineered Rice Endogenous 5-Enolpyruvoylshikimate-3-Phosphate Synthase (Epsps) Transgene Alters Phenology and Fitness of Crop-Wild Hybrid Offspring. Sci. Rep. 2017, 7, 6834. [Google Scholar] [CrossRef]
- Carrière, Y.; Fabrick, J.A.; Tabashnik, B.E. Can Pyramids and Seed Mixtures Delay Resistance to Bt Crops? Trends Biotechnol. 2016, 34, 291–302. [Google Scholar] [CrossRef]
- Steinhäuser, K.G. Environmental risks of chemicals and genetically modified organisms: A comparison. Part II: Sustainability and precaution in risk assessment and risk management. Environ. Sci. Pollut. Res. Int. 2001, 8, 222–226. [Google Scholar] [CrossRef]
- Arias, D.M.; Rieseberg, L.H. Gene Flow between Cultivated and Wild Sunflowers. Theor. Appl. Genet. 1994, 89, 655–660. [Google Scholar] [CrossRef]
- Manasse, R.S. Ecological Risks of Transgenic Plants: Effects of Spatial Dispersion on Gene Flow. Ecol. Appl. 1992, 2, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Tsatsakis, A.M.; Nawaz, M.A.; Kouretas, D.; Balias, G.; Savolainen, K.; Tutelyan, V.A.; Golokhvast, K.S.; Lee, J.D.; Yang, S.H.; Chung, G. Environmental Impacts of Genetically Modified Plants: A Review. Environ. Res. 2017, 156, 818–833. [Google Scholar] [CrossRef]
- Singh, A.; Billingsley, K.; Ward, O. Composting: A potentially safe process for disposal of genetically modified organisms. Crit. Rev. Biotechnol. 2006, 26, 1–16. [Google Scholar] [CrossRef]
- Liu, Y.; Stewart, C.N. An Exposure Pathway-based Risk Assessment System for GM Plants. Plant Biotechnol. J. 2019, 17, 1859–1861. [Google Scholar] [CrossRef] [PubMed]
- Funke, T.; Han, H.; Healy-Fried, M.L.; Fischer, M.; Schönbrunn, E. Molecular Basis for the Herbicide Resistance of Roundup Ready Crops. Proc. Natl. Acad. Sci. USA 2006, 103, 13010–13015. [Google Scholar] [CrossRef] [PubMed]
- Center for Environmental Risk Assessment; ILSI Research Foundation. A Review of the Environmental Safety of the CP4 EPSPS Protein. Environ. Biosaf. Res. 2011, 10, 5–25. [Google Scholar]
- Kang, K.; Kim, M. Single Amino Acid Changes in the 5-Enolpyruvylshikimate-3-Phosphate Synthase from Agrobacterium sp. Strain CP4 and Synechocystis sp. PCC6803 Alter Enzyme Activity and Glyphosate Sensitivity. J. Appl. Biol. Chem. 2006, 49, 65–68. [Google Scholar]
- Wen, Z.L.; Yang, M.K.; Du, M.H.; Zhong, Z.Z.; Lu, Y.T.; Wang, G.H.; Hua, X.M.; Fazal, A.; Mu, C.H.; Yan, S.F.; et al. Enrichments of Root-Associated Bacteria Related to Plant Growth and Nutrition Caused by the Growth of an EPSPS-Transgenic Maize Line in the Field. Front. Microbiol. 2019, 10, 1335. [Google Scholar] [CrossRef]
- Dunfield, K.E.; Germida, J.J. Impact of Genetically Modified Crops on Soil- and Plant-Associated Microbial Communities. J. Environ. Qual. 2004, 33, 806–815. [Google Scholar] [CrossRef]
- Giovannetti, M.; Sbrana, C.; Turrini, A. The impact of genetically modified crops on soil microbial communities. Riv. Biol. 2005, 98, 393–417. [Google Scholar]
- Kremer, R.; Means, N.; Kim, S. Glyphosate Affects Soybean Root Exudation and Rhizosphere Microorganisms. Int. J. Environ. Anal. Chem. 2005, 85, 1165–1174. [Google Scholar] [CrossRef]
- Ilyas, H.; Datta, A.; Bhunia, A. An Approach Towards Structure Based Antimicrobial Peptide Design for Use in Development of Transgenic Plants: A Strategy for Plant Disease Management. Curr. Med. Chem. 2017, 24, 1350–1364. [Google Scholar] [CrossRef]
- Lee, I.H.; Jung, Y.J.; Cho, Y.G.; Nou, I.S.; Huq, M.A.; Nogoy, F.M.; Kang, K.K. SP-LL-37, human antimicrobial peptide, enhances disease resistance in transgenic rice. PLoS ONE 2017, 12, e0172936. [Google Scholar] [CrossRef]
- Shanmugaraj, B.; Bulaon, C.J.I.; Malla, A.; Phoolcharoen, W. Biotechnological Insights on the Expression and Production of Antimicrobial Peptides in Plants. Molecules 2021, 26, 4032. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.F.; Qutb, A.M.; Dong, W. Prediction and Activity of a Cationic α-Helix Antimicrobial Peptide ZM-804 from Maize. Int. J. Mol. Sci. 2021, 22, 2643. [Google Scholar] [CrossRef]
- Park, S.C.; Park, Y.; Hahm, K.S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, M. Antimicrobial Peptides: From Design to Clinical Application. Antibiotics 2022, 11, 349. [Google Scholar] [CrossRef] [PubMed]
- Drayton, M.; Deisinger, J.P.; Ludwig, K.C.; Raheem, N.; Müller, A.; Schneider, T.; Straus, S.K. Host Defense Peptides: Dual Antimicrobial and Immunomodulatory Action. Int. J. Mol. Sci. 2021, 22, 11172. [Google Scholar] [CrossRef]
- Das, K.; Datta, K.; Karmakar, S.; Datta, S. Antimicrobial Peptides—Small but Mighty Weapons for Plants in the War against Phytopathogens. Protein Pep. Lett. 2019, 26, 720–742. [Google Scholar] [CrossRef] [PubMed]
- Selitrennikoff, C.P. Antifungal Proteins. Appl. Environ. Microbiol. 2001, 67, 2883–2894. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-Related Proteins and Peptides as Promising Tools for Engineering Plants with Multiple Stress Tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef]
- Parisi, K.; Shafee, T.M.A.; Quimbar, P.; van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. The Evolution, Function and Mechanisms of Action for Plant Defensins. Semin. Cell Dev. Biol. 2019, 88, 107–118. [Google Scholar] [CrossRef]
- Maximiano, M.R.; Franco, O.L. Biotechnological Applications of Versatile Plant Lipid Transfer Proteins (LTPs). Peptides 2021, 140, 170531. [Google Scholar] [CrossRef]
- Jach, G.; Görnhardt, B.; Mundy, J.; Logemann, J.; Pinsdorf, E.; Leah, R.; Schell, J.; Maas, C. Enhanced Quantitative Resistance against Fungal Disease by Combinatorial Expression of Different Barley Antifungal Proteins in Transgenic Tobacco. Plant J. 1995, 8, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.K.; Selitrennikoff, C.P. Zeamatin, an Antifungal Protein from Maize with Membrane-Permeabilizing Activity. Microbiology 1990, 136, 1771–1778. [Google Scholar] [CrossRef]
- Thevissen, K.; Osborn, R.W.; Acland, D.P.; Broekaert, W.F. Specific Binding Sites for an Antifungal Plant Defensin from Dahlia (Dahlia Merckii) on Fungal Cells Are Required for Antifungal Activity. Mol. Plant Microbe Interact. 2000, 13, 54–61. [Google Scholar] [CrossRef]
- Dunfield, K.E.; Germida, J.J. Diversity of Bacterial Communities in the Rhizosphere and Root Interior of Field-Grown Genetically Modified Brassica napus. FEMS Microbiol. Ecol. 2001, 38, 1–9. [Google Scholar] [CrossRef]
- Icoz, I.; Stotzky, G. Fate and Effects of Insect-Resistant Bt Crops in Soil Ecosystems. Soil Biol. Biochem. 2008, 40, 559–586. [Google Scholar] [CrossRef]
- Cho, J.; Lee, D.G. The Antimicrobial Peptide Arenicin-1 Promotes Generation of Reactive Oxygen Species and Induction of Apoptosis. Biochim. Biophys. Acta 2011, 1810, 1246–1251. [Google Scholar] [CrossRef]
- Yun, J.; Hwang, J.-S.; Lee, D.G. The Antifungal Activity of the Peptide, Periplanetasin-2, Derived from American Cockroach Periplaneta americana. Biochem. J. 2017, 474, 3027–3043. [Google Scholar] [CrossRef]
- Rautenbach, M.; Troskie, A.M.; Vosloo, J.A.; Dathe, M.E. Antifungal Membranolytic Activity of the Tyrocidines against Filamentous Plant Fungi. Biochimie 2016, 130, 122–131. [Google Scholar] [CrossRef]
- Mochon, A.B.; Liu, H. The Antimicrobial Peptide Histatin-5 Causes a Spatially Restricted Disruption on the Candida Albicans Surface, Allowing Rapid Entry of the Peptide into the Cytoplasm. PLOS Pathogens 2008, 4, e1000190. [Google Scholar] [CrossRef]
- Park, S.C.; Kim, I.R.; Kim, J.Y.; Lee, Y.; Kim, E.J.; Jung, J.H.; Jung, Y.J.; Jang, M.K.; Lee, J.R. Molecular Mechanism of Arabidopsis Thaliana Profilins as Antifungal Proteins. Biochim. Biophys. Acta 2018, 1862, 2545–2554. [Google Scholar] [CrossRef]
- Park, S.C.; Kim, J.Y.; Lee, J.K.; Lim, H.S.; Son, H.; Yoo, S.H.; Mun, S.E.; Jang, M.K.; Lee, J.R. Antifungal Mechanism of Vip3Aa, a Vegetative Insecticidal Protein, against Pathogenic Fungal Strains. Antibiotics 2021, 10, 1558. [Google Scholar] [CrossRef]
- Son, H.; Jung, Y.J.; Park, S.C.; Kim, I.R.; Park, J.H.; Jang, M.K.; Lee, J.R. Functional Characterization of an Arabidopsis Profilin Protein as a Molecular Chaperone under Heat Shock Stress. Molecules 2022, 27, 5771. [Google Scholar] [CrossRef] [PubMed]
- McLellan, T. Electrophoresis Buffers for Polyacrylamide Gels at Various PH. Anal. Biochem. 1982, 126, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Kim, I.R.; Kim, J.Y.; Lee, Y.; Yoo, S.H.; Jung, J.H.; Cheong, G.W.; Lee, S.Y.; Jang, M.K.; Lee, J.R. Functional Characterization of a Rice Thioredoxin Protein OsTrxm and Its Cysteine Mutant Variant with Antifungal Activity. Antioxidants 2019, 8, 598. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Kim, I.R.; Hwang, J.E.; Kim, J.Y.; Jung, Y.J.; Choi, W.; Lee, Y.; Jang, M.K.; Lee, J.R. Functional Mechanisms Underlying the Antimicrobial Activity of the Oryza Sativa Trx-like Protein. Int. J. Mol. Sci. 2019, 20, 1413. [Google Scholar] [CrossRef] [PubMed]
- Semighini, C.P.; Harris, S.D. Methods to Detect Apoptotic-Like Cell Death in Filamentous Fungi. Methods Mol. Biol. 2010, 638, 269–279. [Google Scholar]
- Park, S.-C.; Son, H.; Kim, Y.-M.; Lee, J.-K.; Park, S.; Lim, H.S.; Lee, J.R.; Jang, M.-K. Design of Antimicrobial Peptides with Cell-Selective Activity and Membrane-Acting Mechanism against Drug-Resistant Bacteria. Antibiotics 2022, 11, 1619. [Google Scholar] [CrossRef]


| Fungal Strains | MIC (μg/mL) a | |
|---|---|---|
| CP4-EPSPS b | Melittin | |
| Yeast | ||
| C. albicans | 125 | 62.5 |
| C. tropicalis | 250 | 125 |
| C. krusei | 62.5 | 62.5 |
| Mold | ||
| C. gloeosporioides | 62.5 | 500 |
| F. solani | 62.5 | 250 |
| F. graminearum | 62.5 | 500 |
| T. virens | 62.5 | 125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-C.; Lim, H.S.; Mun, S.-E.; Jung, Y.J.; Yoon, A.-M.; Son, H.; Kim, C.M.; Choo, Y.-K.; Lee, J.R. Potent Antifungal Functions of a Living Modified Organism Protein, CP4-EPSPS, against Pathogenic Fungal Cells. Molecules 2023, 28, 4289. https://doi.org/10.3390/molecules28114289
Park S-C, Lim HS, Mun S-E, Jung YJ, Yoon A-M, Son H, Kim CM, Choo Y-K, Lee JR. Potent Antifungal Functions of a Living Modified Organism Protein, CP4-EPSPS, against Pathogenic Fungal Cells. Molecules. 2023; 28(11):4289. https://doi.org/10.3390/molecules28114289
Chicago/Turabian StylePark, Seong-Cheol, Hye Song Lim, Seong-Eun Mun, Young Jun Jung, A-Mi Yoon, Hyosuk Son, Chul Min Kim, Young-Kug Choo, and Jung Ro Lee. 2023. "Potent Antifungal Functions of a Living Modified Organism Protein, CP4-EPSPS, against Pathogenic Fungal Cells" Molecules 28, no. 11: 4289. https://doi.org/10.3390/molecules28114289
APA StylePark, S.-C., Lim, H. S., Mun, S.-E., Jung, Y. J., Yoon, A.-M., Son, H., Kim, C. M., Choo, Y.-K., & Lee, J. R. (2023). Potent Antifungal Functions of a Living Modified Organism Protein, CP4-EPSPS, against Pathogenic Fungal Cells. Molecules, 28(11), 4289. https://doi.org/10.3390/molecules28114289





