Author Contributions
Conceptualization, É.A.E.; investigation, T.V.P., F.K., N.I., N.V.M., B.H., L.B., A.R. and G.S.; data curation, T.V.P., N.I., N.V.M., B.H. and L.B.; funding acquisition, É.A.E.; writing—original draft preparation, T.V.P. and É.A.E.; writing—review and editing, T.V.P., É.F., M.K., B.G. and É.A.E. All authors have read and agreed to the published version of the manuscript.
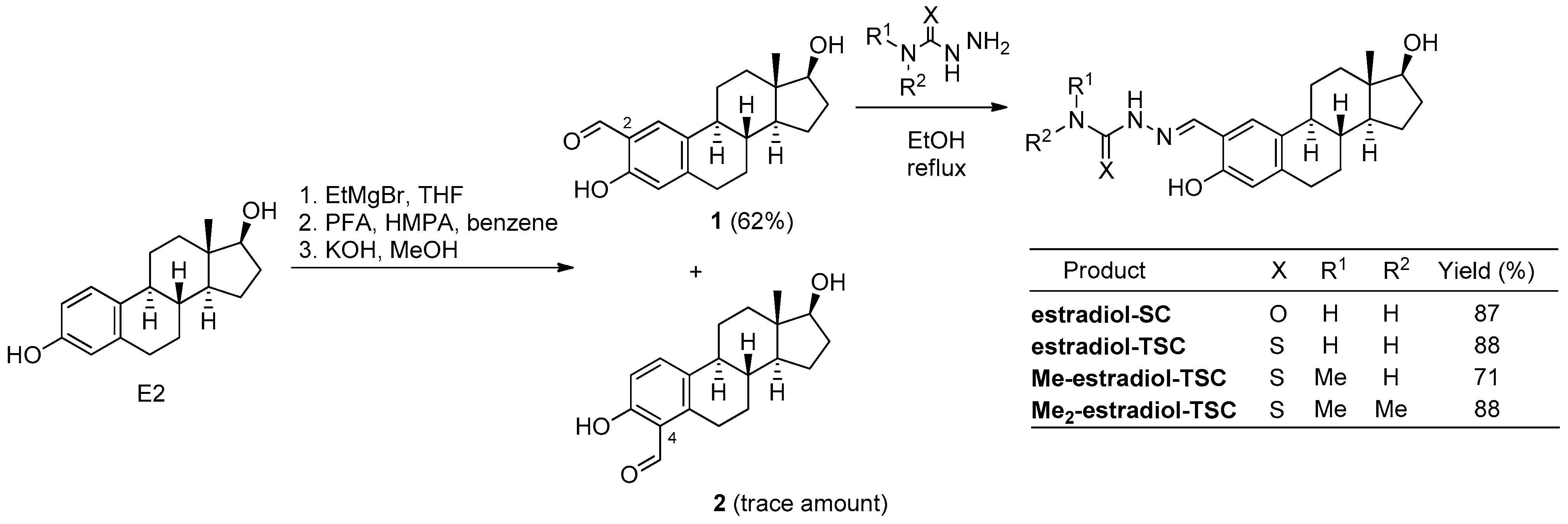
Scheme 1.
Synthesis of 2-formyl-estradiol and its condensation reactions with (thio)semicarbazides (PFA = paraformaldehyde, THF = tetrahydrofuran, HMPA = hexamethylphosphoramide, EtMgBr = ethylmagnesium bromide, EtOH = ethanol).
Scheme 1.
Synthesis of 2-formyl-estradiol and its condensation reactions with (thio)semicarbazides (PFA = paraformaldehyde, THF = tetrahydrofuran, HMPA = hexamethylphosphoramide, EtMgBr = ethylmagnesium bromide, EtOH = ethanol).
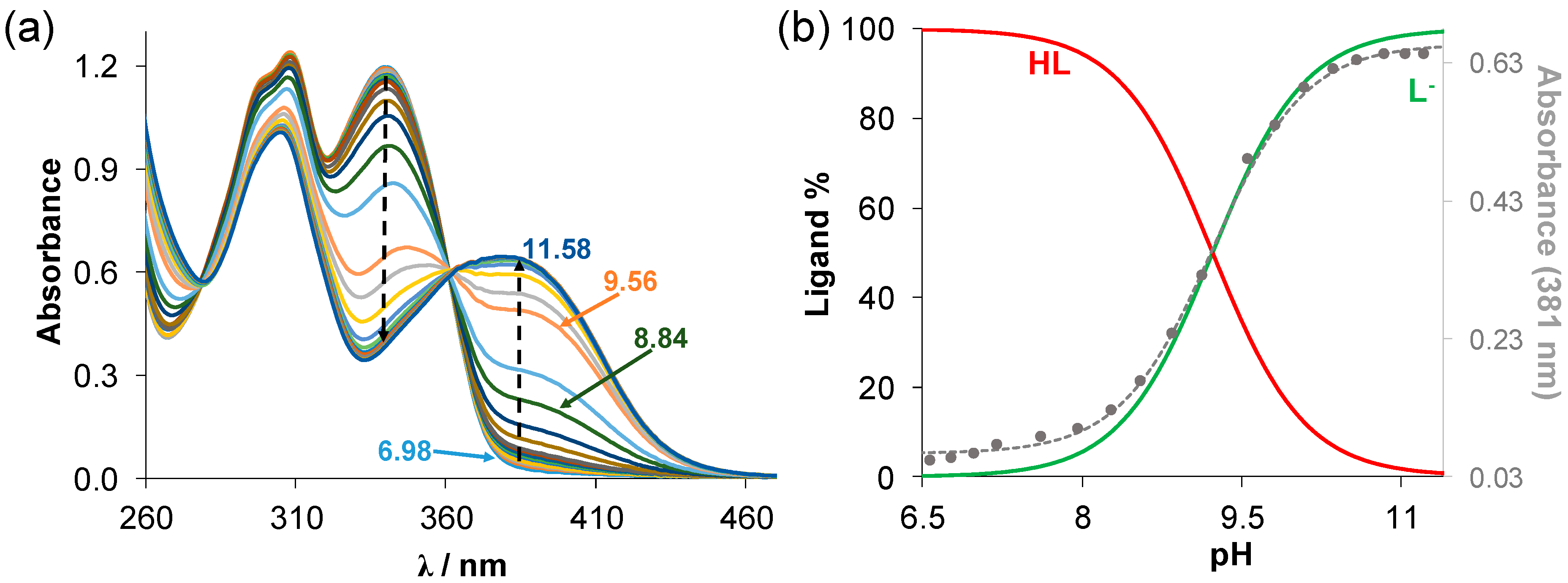
Figure 1.
(a) UV–vis absorption spectra of estradiol-TSC recorded at various pH values in 30% (v/v) DMSO/H2O solvent mixture (cligand = 20 μM; T = 25.0 °C; I = 0.1 M (KCl); ℓ = 2 cm); (b) concentration distribution curves for this ligand with absorbance values measured at 381 nm (●) with the fitted curve. HL denotes the neutral form of the compound.
Figure 1.
(a) UV–vis absorption spectra of estradiol-TSC recorded at various pH values in 30% (v/v) DMSO/H2O solvent mixture (cligand = 20 μM; T = 25.0 °C; I = 0.1 M (KCl); ℓ = 2 cm); (b) concentration distribution curves for this ligand with absorbance values measured at 381 nm (●) with the fitted curve. HL denotes the neutral form of the compound.
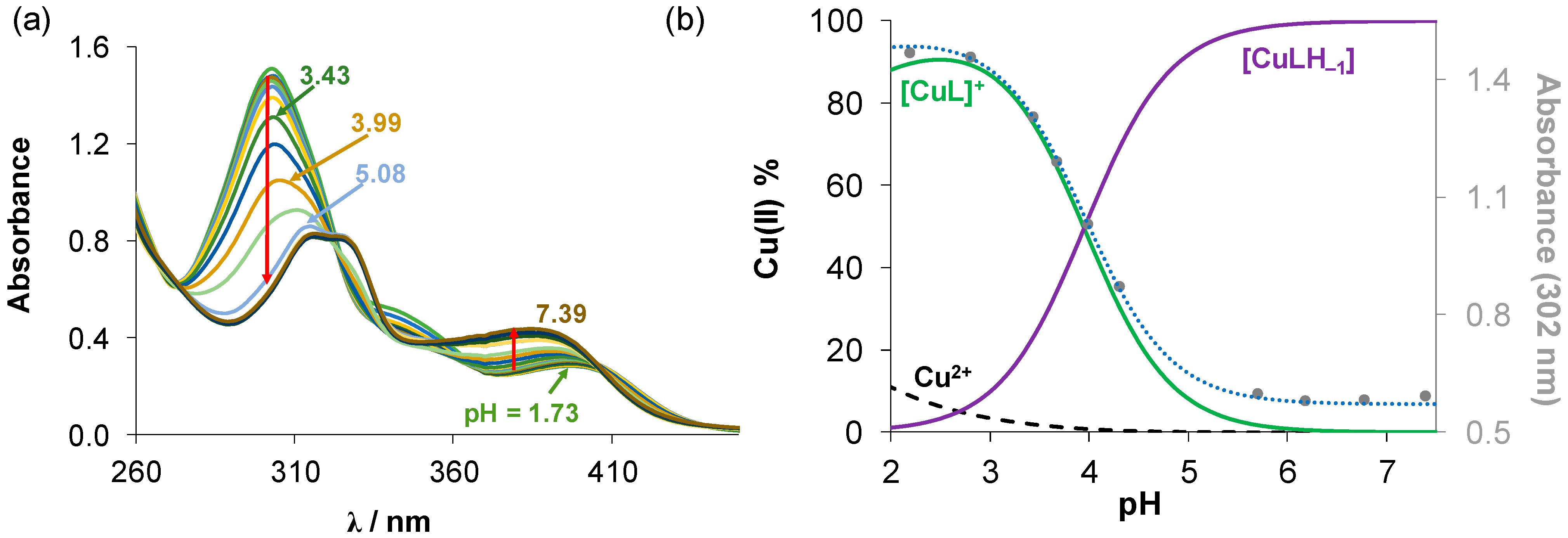
Figure 2.
(a) UV–vis absorption spectra of the Cu(II)-estradiol-TSC (1:1) system in the pH range 1.73–7.39 in 30% (v/v) DMSO/H2O solvent mixture. (b) Concentration distribution curves calculated for the same system plotted together with the absorbance changes at 302 nm (●) with the fitted curve (dotted line). (cligand = 20 µM; cCu(II) = 20 µM; T = 25.0 °C; I = 0.1 M (KCl)).
Figure 2.
(a) UV–vis absorption spectra of the Cu(II)-estradiol-TSC (1:1) system in the pH range 1.73–7.39 in 30% (v/v) DMSO/H2O solvent mixture. (b) Concentration distribution curves calculated for the same system plotted together with the absorbance changes at 302 nm (●) with the fitted curve (dotted line). (cligand = 20 µM; cCu(II) = 20 µM; T = 25.0 °C; I = 0.1 M (KCl)).
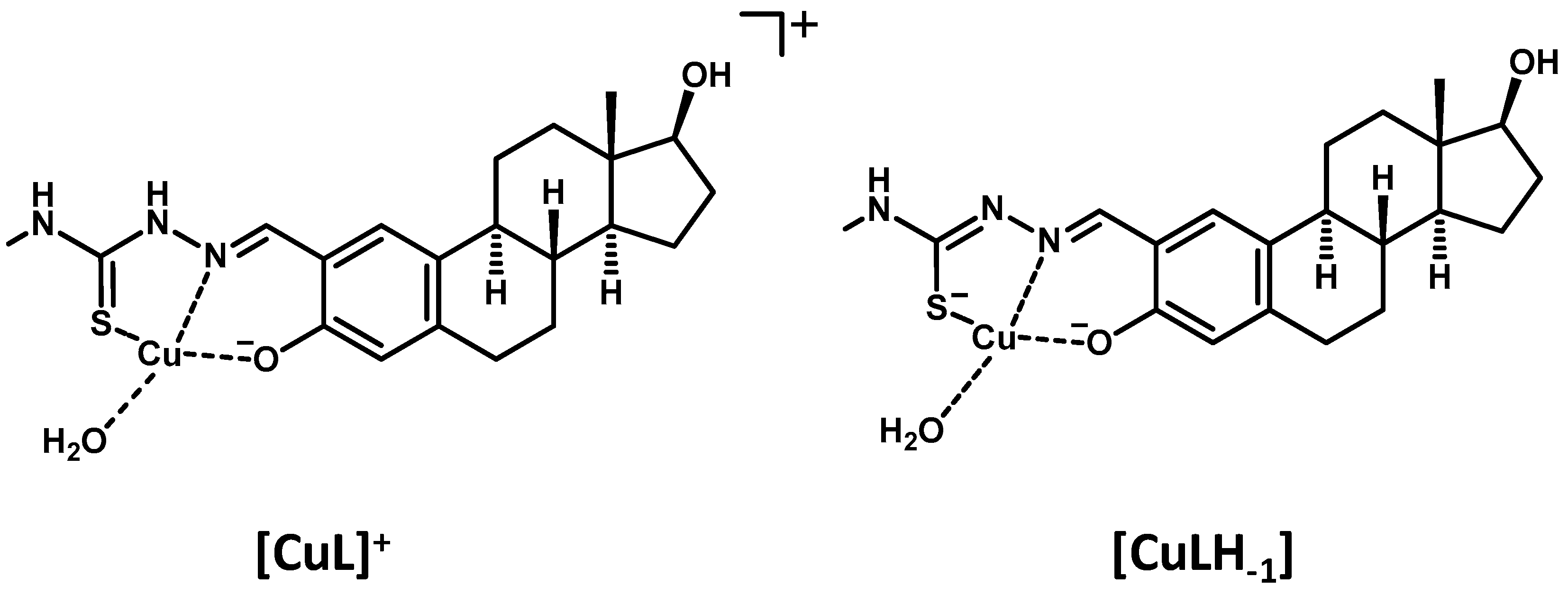
Scheme 2.
Suggested structures for the Cu(II) complexes formed with Me-estadiol-TSC.
Scheme 2.
Suggested structures for the Cu(II) complexes formed with Me-estadiol-TSC.
Figure 3.
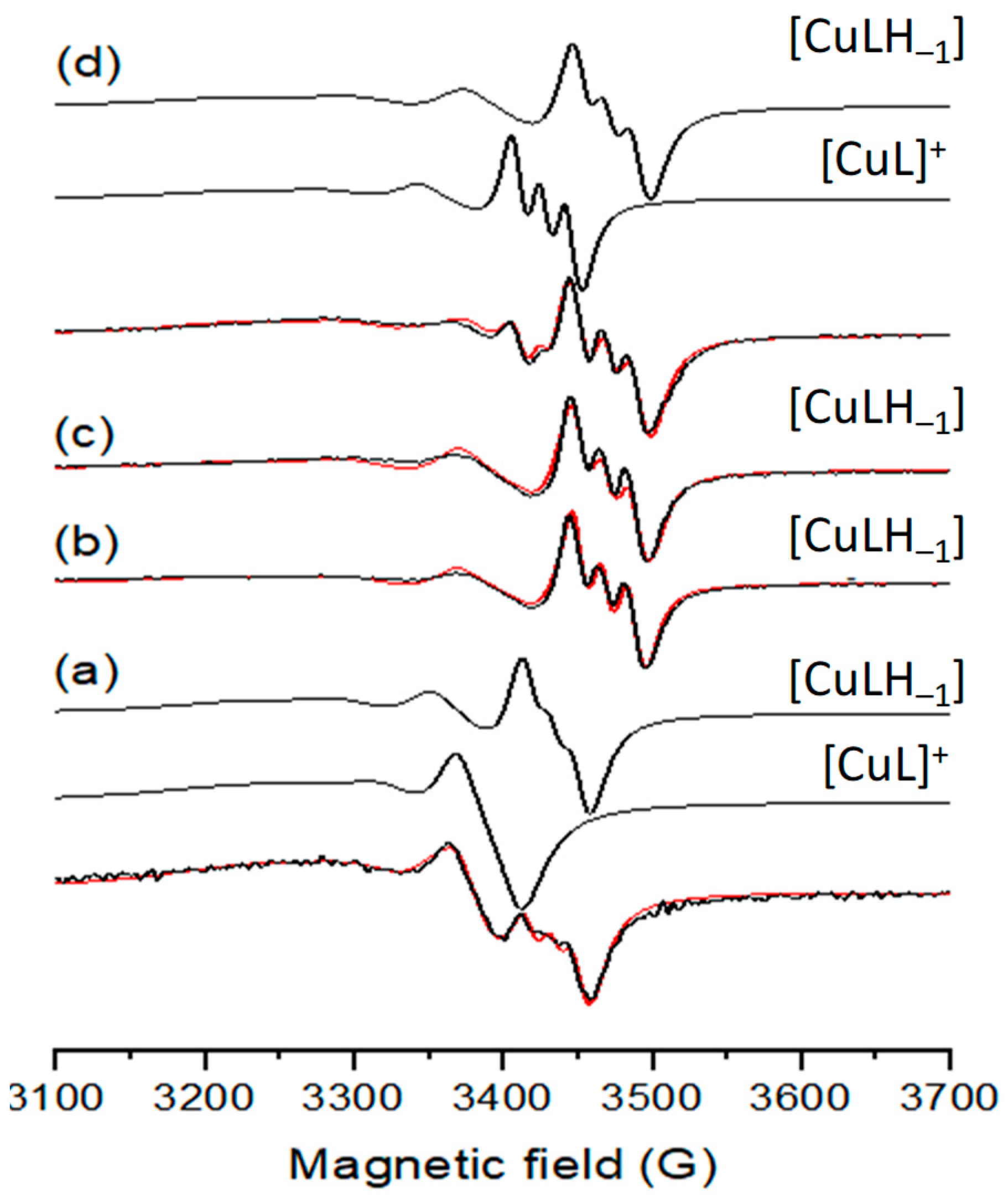
Room temperature solution EPR spectra (black) recorded for Cu(II) complexes dissolved in DMSO of (a) estradiol-SC, (b) estradiol-TSC, (c) Me-estradiol-TSC, (d) Me2-estradiol-TSC and simulated (red) spectra together with the obtained component spectra. Spectrum (a) was simulated with 50% [CuL]+ and 50% [CuLH−1], (b,c) with 100% [CuLH−1] and (d) with 18% [CuL]+ 82% [CuLH−1].
Figure 3.
Room temperature solution EPR spectra (black) recorded for Cu(II) complexes dissolved in DMSO of (a) estradiol-SC, (b) estradiol-TSC, (c) Me-estradiol-TSC, (d) Me2-estradiol-TSC and simulated (red) spectra together with the obtained component spectra. Spectrum (a) was simulated with 50% [CuL]+ and 50% [CuLH−1], (b,c) with 100% [CuLH−1] and (d) with 18% [CuL]+ 82% [CuLH−1].
Figure 4.
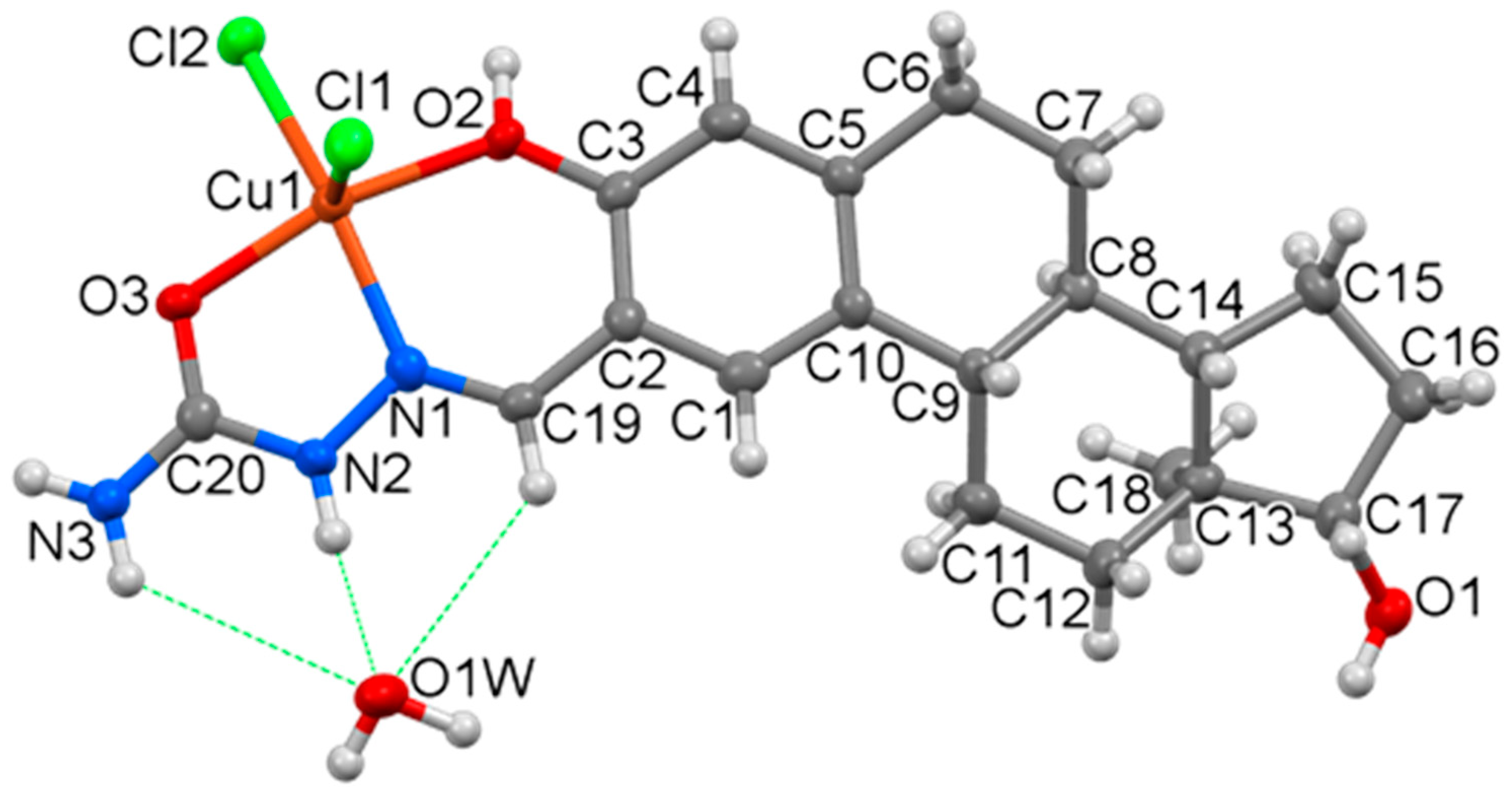
Molecular structure and atom labeling of the Cu(II) complex of estradiol-SC as the [Cu(HL)Cl2] × H2O × 2CH3OH complex molecule. Displacement parameters are drawn at the 50% probability level, and the methanol solvate molecules are omitted for clarity.
Figure 4.
Molecular structure and atom labeling of the Cu(II) complex of estradiol-SC as the [Cu(HL)Cl2] × H2O × 2CH3OH complex molecule. Displacement parameters are drawn at the 50% probability level, and the methanol solvate molecules are omitted for clarity.
Figure 5.
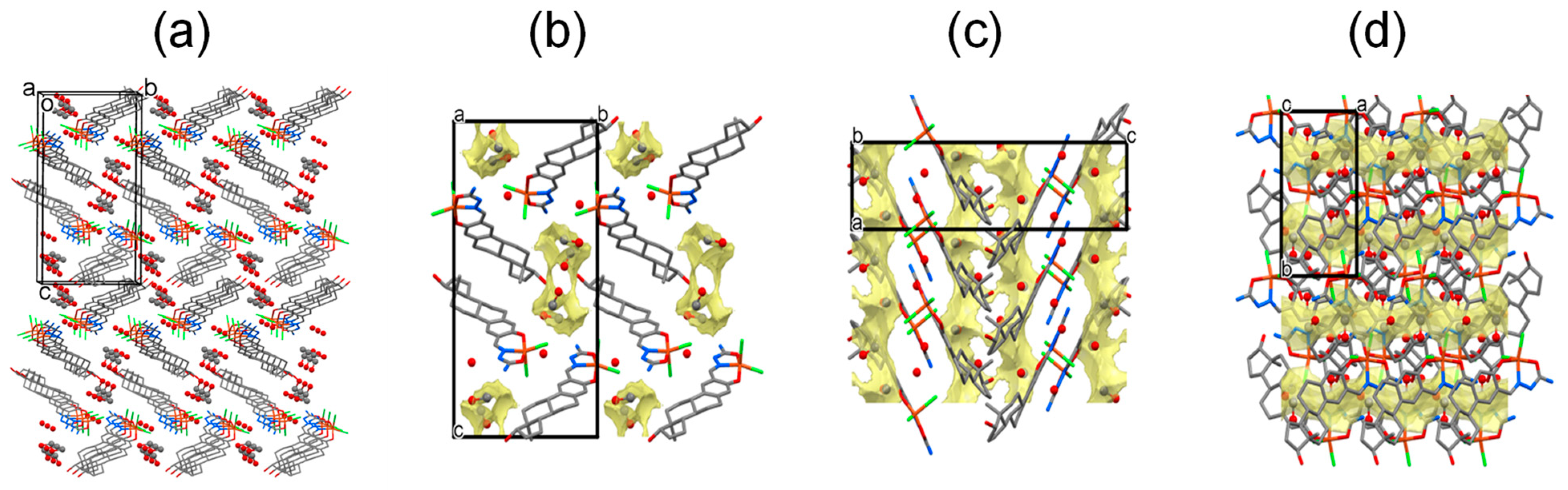
(a) Crystal packing of [Cu(HL)Cl2] × H2O × 2CH3OH complex of estradiol-SC drawn by stick representation and solvate molecules by ball and stick representation (hydrogen atoms are omitted for clarity). (b) View from the a crystallographic axis direction, methanol channels are marked with pale yellow, (c) view from the b crystallographic axis direction, (d) view from the c crystallographic axis direction.
Figure 5.
(a) Crystal packing of [Cu(HL)Cl2] × H2O × 2CH3OH complex of estradiol-SC drawn by stick representation and solvate molecules by ball and stick representation (hydrogen atoms are omitted for clarity). (b) View from the a crystallographic axis direction, methanol channels are marked with pale yellow, (c) view from the b crystallographic axis direction, (d) view from the c crystallographic axis direction.
Figure 6.
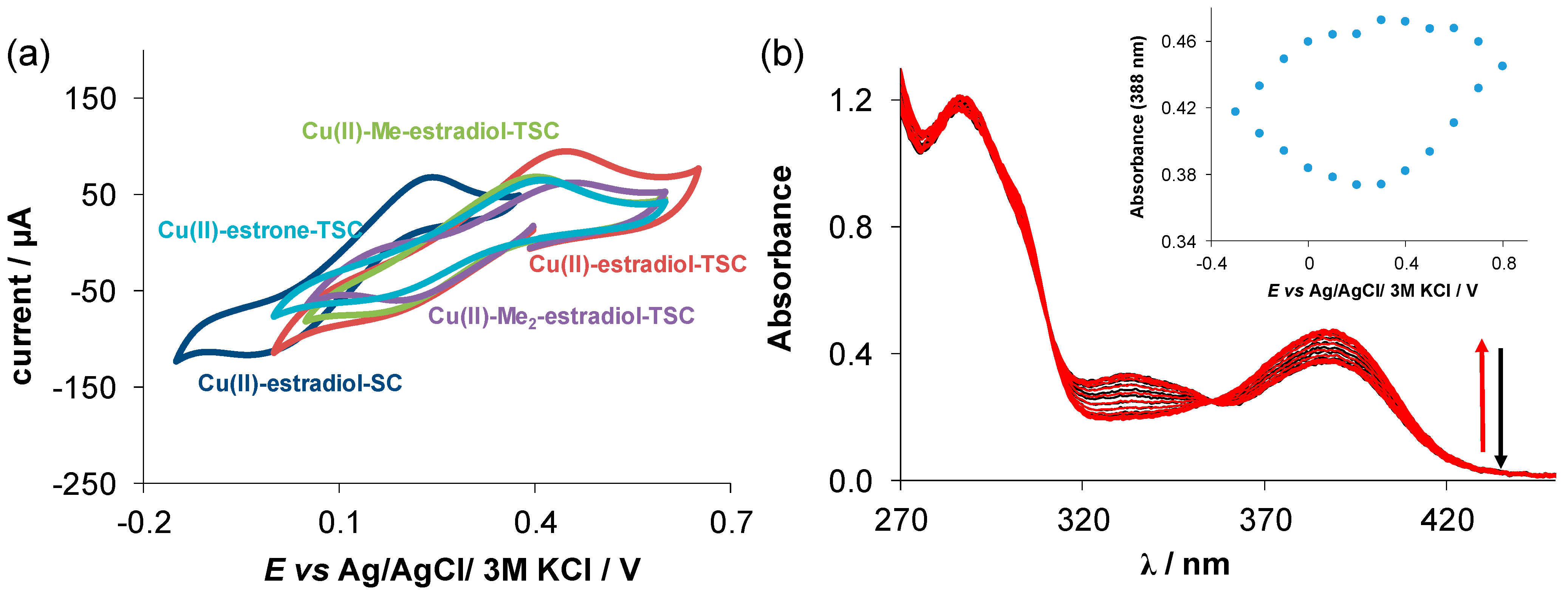
(a) Cyclic voltammograms of the Cu(II)-(T)SC (1:1) chemical systems in 90% (v/v) DMSO/H2O at 10 mV/s scan rate (cligand =1 mM, cCu(II) = 1 mM, pH = 7.40 (10 mM HEPES), T = 25 °C, I = 0.1 M (nBu4NPF6)) E’ vs. Ag/AgCl/3M KCl (V): Cu(II)–estradiol-SC = +0.10; Cu(II)-estrone-TSC = +0.28; Cu(II)-estradiol-TSC = +0.31; Cu(II)Me–estradiol-TSC = +0.29; Cu(II)-Me2-estradiol-TSC = +0.33; (b) UV–vis absorption spectra recorded for the Cu(II)-estradiol-SC system at the various potential values using the spectroelectrochemical cell. Inserted figure shows the absorbance values at 388 nm plotted against the potential. (cCu(II) = 100 µM; ℓ = 1 cm; I = 0.1 M (NBu4PF6); T = 25.0 °C, scan rate: 10 mV/s).
Figure 6.
(a) Cyclic voltammograms of the Cu(II)-(T)SC (1:1) chemical systems in 90% (v/v) DMSO/H2O at 10 mV/s scan rate (cligand =1 mM, cCu(II) = 1 mM, pH = 7.40 (10 mM HEPES), T = 25 °C, I = 0.1 M (nBu4NPF6)) E’ vs. Ag/AgCl/3M KCl (V): Cu(II)–estradiol-SC = +0.10; Cu(II)-estrone-TSC = +0.28; Cu(II)-estradiol-TSC = +0.31; Cu(II)Me–estradiol-TSC = +0.29; Cu(II)-Me2-estradiol-TSC = +0.33; (b) UV–vis absorption spectra recorded for the Cu(II)-estradiol-SC system at the various potential values using the spectroelectrochemical cell. Inserted figure shows the absorbance values at 388 nm plotted against the potential. (cCu(II) = 100 µM; ℓ = 1 cm; I = 0.1 M (NBu4PF6); T = 25.0 °C, scan rate: 10 mV/s).
Figure 7.
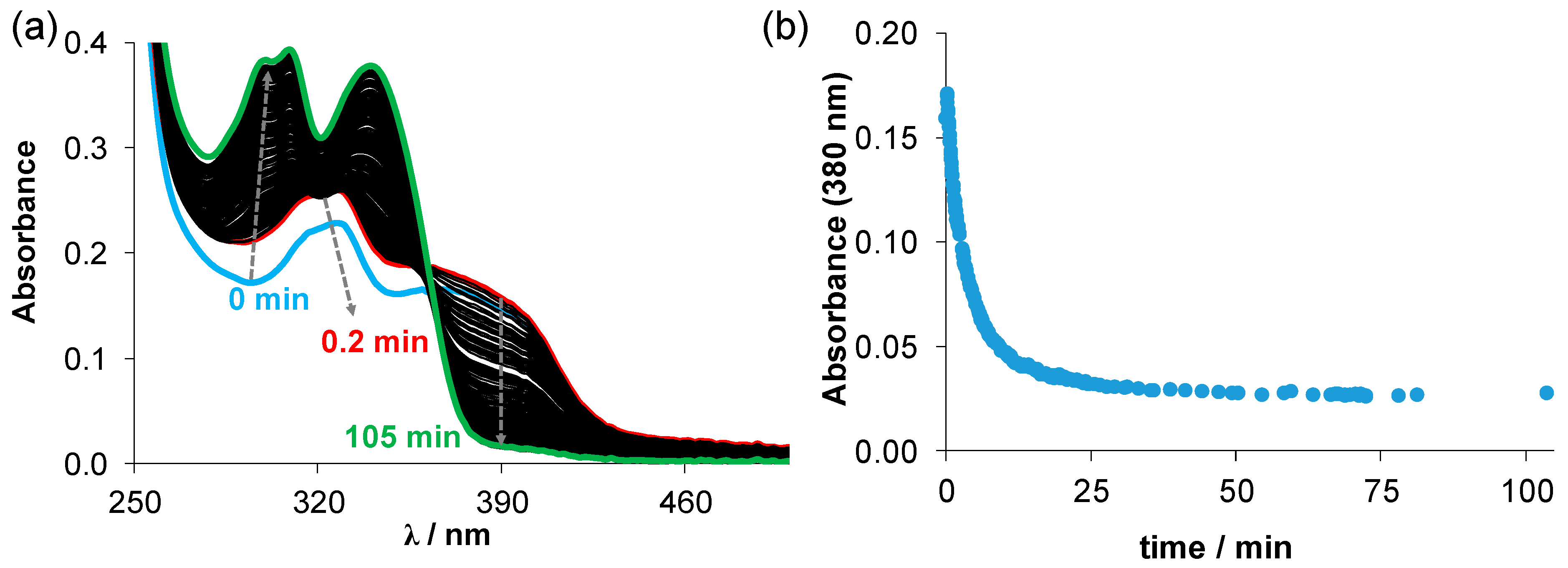
(a) Time-dependent changes of the UV–vis spectra recorded for the Cu-Me-estradiol-TSC complex (25 µM) in the presence of 50 equiv. GSH (1.25 mM) at pH 7.4 (10 mM HEPES) in 30% (v/v) DMSO/H2O under anaerobic conditions. (b) Absorbance values at 380 nm plotted against the time. (T = 25 °C; I = 0.1 M (KCl)).
Figure 7.
(a) Time-dependent changes of the UV–vis spectra recorded for the Cu-Me-estradiol-TSC complex (25 µM) in the presence of 50 equiv. GSH (1.25 mM) at pH 7.4 (10 mM HEPES) in 30% (v/v) DMSO/H2O under anaerobic conditions. (b) Absorbance values at 380 nm plotted against the time. (T = 25 °C; I = 0.1 M (KCl)).
Figure 8.
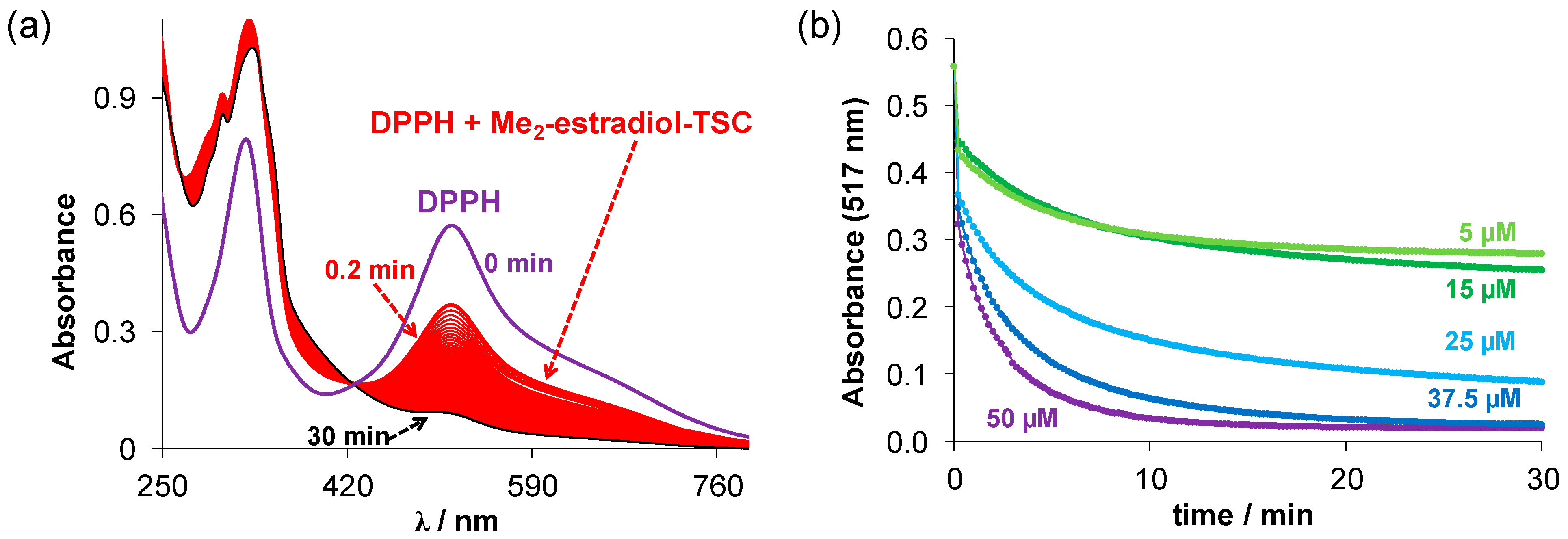
(a) Time-dependent changes of the UV–vis spectra of the DPPH• (c = 50 μM) in the presence of Me2-estradiol-TSC (50 µM) in ethanol. (T = 25 °C); (b) absorbance changes of DPPH at 517 nm upon reaction with Me2-estradiol-TSC at different concentrations in ethanol plotted against time. (cDPPH = 50 µM; cligand = 50, 37.5; 25; 15; 5 µM; ℓ = 1 cm; T = 25 °C).
Figure 8.
(a) Time-dependent changes of the UV–vis spectra of the DPPH• (c = 50 μM) in the presence of Me2-estradiol-TSC (50 µM) in ethanol. (T = 25 °C); (b) absorbance changes of DPPH at 517 nm upon reaction with Me2-estradiol-TSC at different concentrations in ethanol plotted against time. (cDPPH = 50 µM; cligand = 50, 37.5; 25; 15; 5 µM; ℓ = 1 cm; T = 25 °C).
Figure 9.
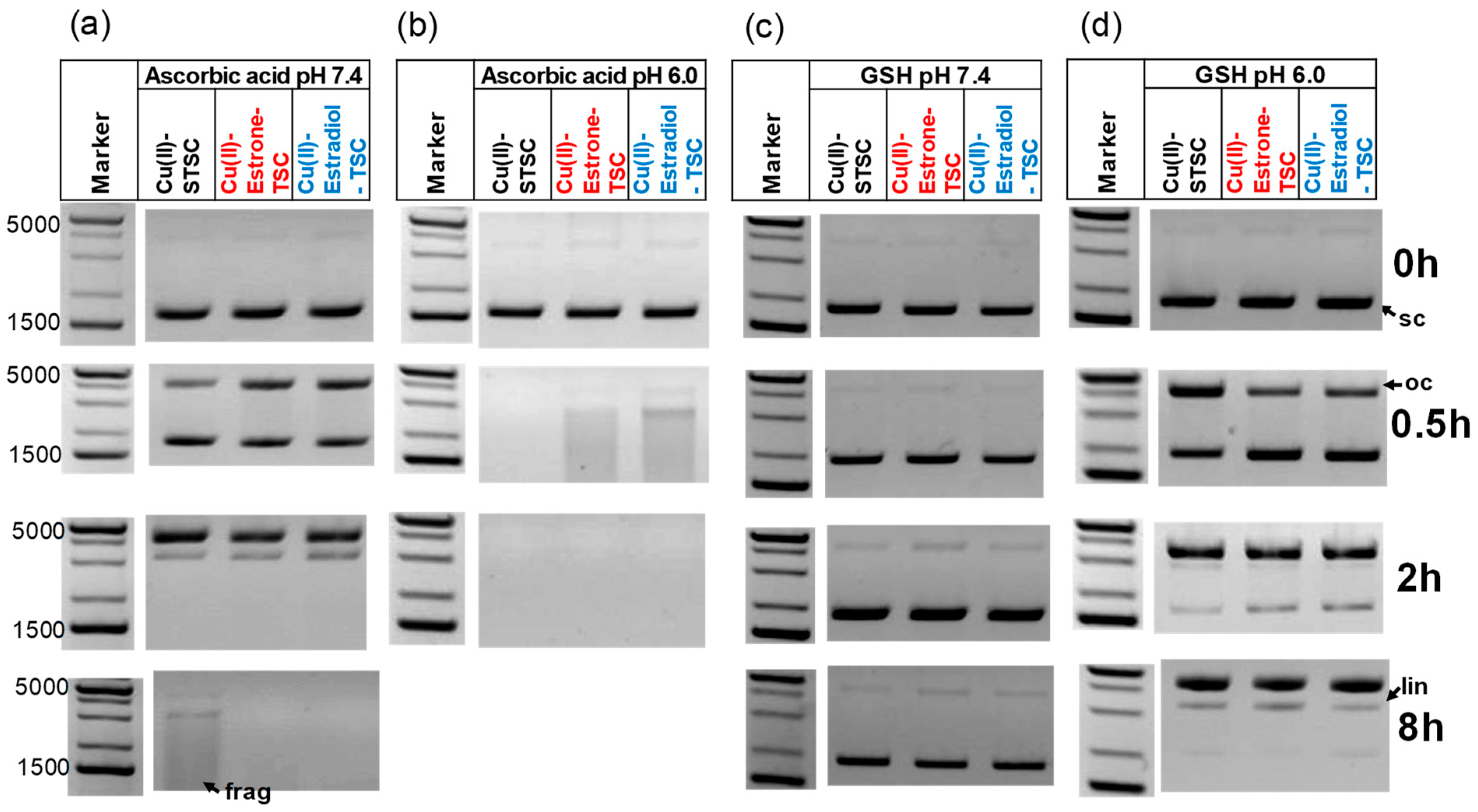
Representative DNA cleavage activity of the Cu(II)-TSC complexes analyzed by 1% (w/v) agarose gel electrophoresis in the presence of 1 mM ascorbic acid (a) at pH 7.4 and (b) at pH 6; or in the presence of 1 mM GSH (c) at pH 7.4 and (d) at pH 6.0. The reaction mixtures contained 100 ng pUC119ΔH+N-N6 plasmid DNA. (cL = 20 μM; cCu(II) = 18 μM; (10 mM HEPES); incubation time at 37 °C: 0, 0.5, 2 and 8 h). Abbreviations indicate various forms of the plasmid DNA: oc—open circular, lin—linear, sc—supercoiled frag—fragmented. 1 kb Plus DNA. Ladder was used as a marker.
Figure 9.
Representative DNA cleavage activity of the Cu(II)-TSC complexes analyzed by 1% (w/v) agarose gel electrophoresis in the presence of 1 mM ascorbic acid (a) at pH 7.4 and (b) at pH 6; or in the presence of 1 mM GSH (c) at pH 7.4 and (d) at pH 6.0. The reaction mixtures contained 100 ng pUC119ΔH+N-N6 plasmid DNA. (cL = 20 μM; cCu(II) = 18 μM; (10 mM HEPES); incubation time at 37 °C: 0, 0.5, 2 and 8 h). Abbreviations indicate various forms of the plasmid DNA: oc—open circular, lin—linear, sc—supercoiled frag—fragmented. 1 kb Plus DNA. Ladder was used as a marker.
Figure 10.
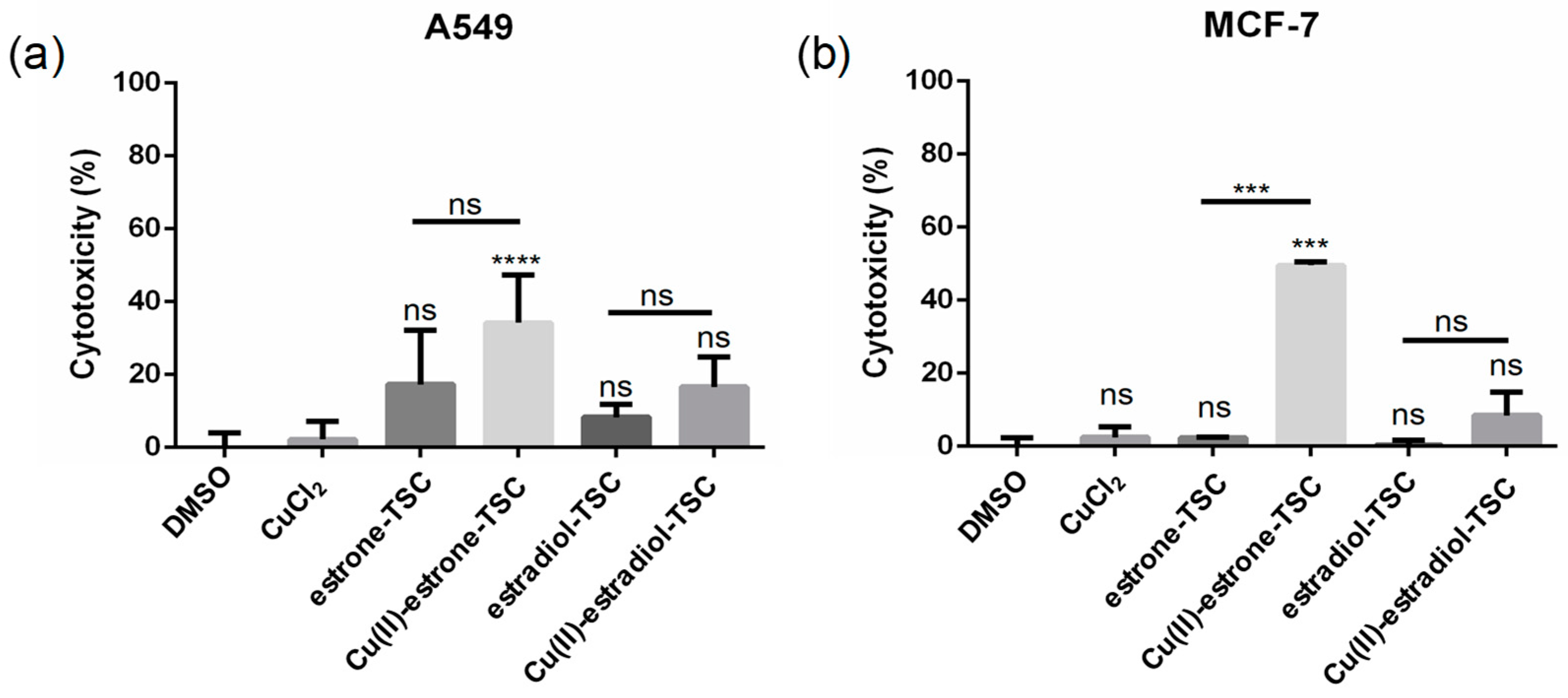
LDH assay to measure the reduction in plasma membrane integrity: cytotoxicity (%) obtained on (a) A549 and (b) MCF-7 cells upon the treatment with estrone-TSC and estradiol-TSC and their Cu(II) complexes (20 μM) in addition to the DMSO blank (90% DMSO and 10% phosphate buffered saline (PBS)) and CuCl2 (20 μM), as indicated in the figure using 24 h incubation time. (ns: not-significant; ***: p < 0.005; ****: p < 0.001).
Figure 10.
LDH assay to measure the reduction in plasma membrane integrity: cytotoxicity (%) obtained on (a) A549 and (b) MCF-7 cells upon the treatment with estrone-TSC and estradiol-TSC and their Cu(II) complexes (20 μM) in addition to the DMSO blank (90% DMSO and 10% phosphate buffered saline (PBS)) and CuCl2 (20 μM), as indicated in the figure using 24 h incubation time. (ns: not-significant; ***: p < 0.005; ****: p < 0.001).
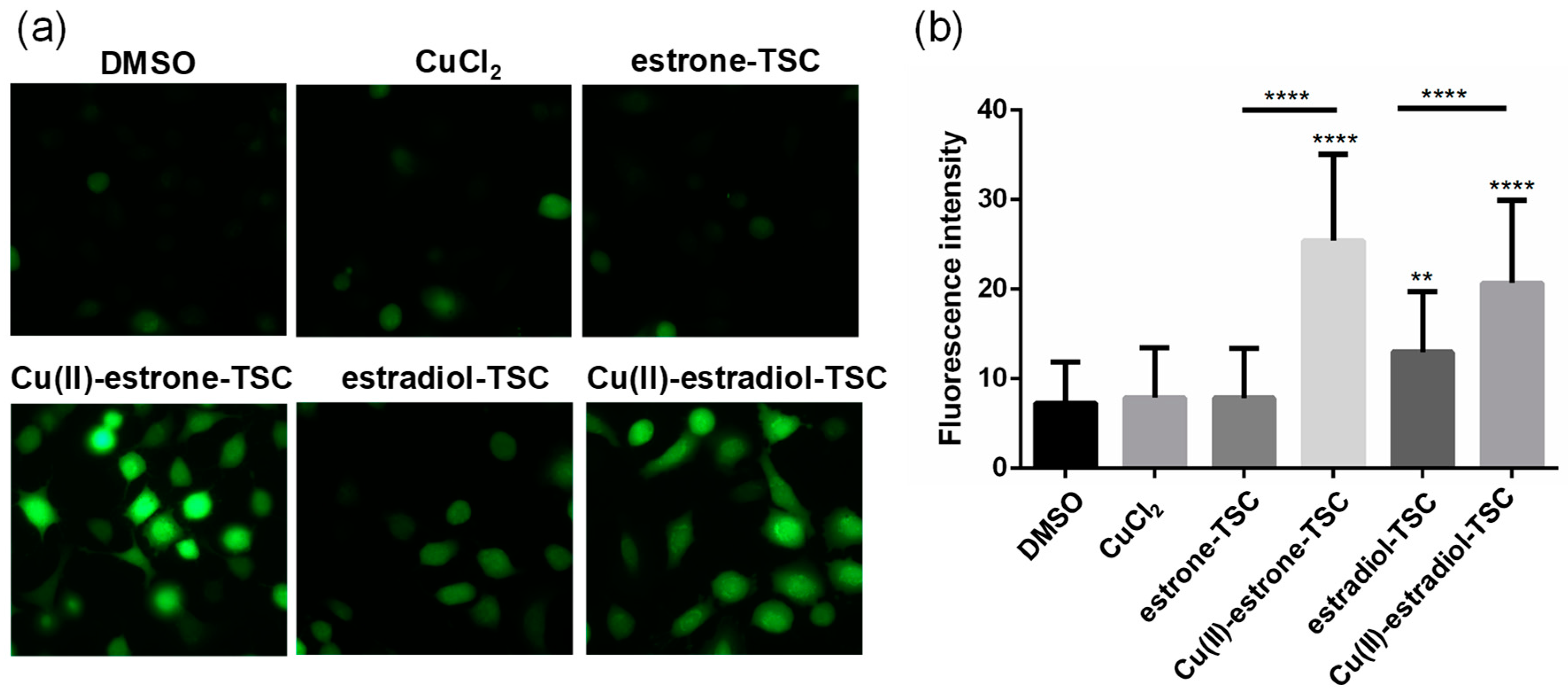
Figure 11.
(a) Representative fluorescence microscopic images show the DCFDA staining to determine ROS levels in A549 cells upon the treatment with estrone-TSC and estradiol-TSC and their Cu(II) complexes (20 μM) in addition to the DMSO blank (90% DMSO and 10% PBS) and CuCl2 (20 μM) as indicated in the figure using 6 h incubation time. (b) Level of ROS generation measured by DCFDA staining on A549 cells. (**: p < 0.01; ****: p < 0.001).
Figure 11.
(a) Representative fluorescence microscopic images show the DCFDA staining to determine ROS levels in A549 cells upon the treatment with estrone-TSC and estradiol-TSC and their Cu(II) complexes (20 μM) in addition to the DMSO blank (90% DMSO and 10% PBS) and CuCl2 (20 μM) as indicated in the figure using 6 h incubation time. (b) Level of ROS generation measured by DCFDA staining on A549 cells. (**: p < 0.01; ****: p < 0.001).
Table 1.
pKa values of the studied compounds determined by UV–vis spectrophotometric titrations in 30% (v/v) DMSO/H2O (T = 25 °C, I = 0.10 M (KCl)).
Table 1.
pKa values of the studied compounds determined by UV–vis spectrophotometric titrations in 30% (v/v) DMSO/H2O (T = 25 °C, I = 0.10 M (KCl)).
| | pKa |
|---|
| estradiol-SC | 10.02 ± 0.03 |
| estradiol-TSC | 9.23 ± 0.03 |
| Me-estradiol-TSC | 9.28 ± 0.03 |
| Me2-estradiol-TSC | 9.20 ± 0.03 |
| estrone-SC | 9.50 1 |
| estrone-TSC | 9.00 2 |
| Me-estrone-TSC | 9.21 3 |
| Me2-estrone-TSC | 9.02 ± 0.03 4 |
Table 2.
Overall stability constants (β), pKa of the Cu(II) complexes estradiol-(T)SCs and estrone-(T)SC for comparison, determined by UV–vis titrations, and calculated pCu values at pH 5.0, 6.0, 7.4 using cCu(II) = 20 µM, cligand = 20 µM. Observed rate constants (kobs) obtained for the direct redox reaction of the Cu(II) complexes (25 µM) with GSH (1.25 mM) at pH 7.4 under anaerobic conditions. (30% (v/v) DMSO/H2O; T = 25.0 °C; I = 0.1 M (KCl)).
Table 2.
Overall stability constants (β), pKa of the Cu(II) complexes estradiol-(T)SCs and estrone-(T)SC for comparison, determined by UV–vis titrations, and calculated pCu values at pH 5.0, 6.0, 7.4 using cCu(II) = 20 µM, cligand = 20 µM. Observed rate constants (kobs) obtained for the direct redox reaction of the Cu(II) complexes (25 µM) with GSH (1.25 mM) at pH 7.4 under anaerobic conditions. (30% (v/v) DMSO/H2O; T = 25.0 °C; I = 0.1 M (KCl)).
| | logβ [CuL]+ | logβ [CuLH−1] | pKa [CuL]+ | pCu5.0 | pCu6.0 5 | pCu7.4 6 | kobs (min−1) |
|---|
| estradiol-SC | 11.63 ± 0.03 | 5.16 ± 0.03 | 6.48 | 5.7 | 6.3 | 7.8 | fast reaction |
| estradiol-TSC | 13.79 ± 0.03 | 9.84 ± 0.03 | 3.95 | 7.8 | 9.3 | 12.1 | 0.07 ± 0.03 |
| Me-estradiol-TSC | 13.68 ± 0.03 | 9.59 ± 0.03 | 4.08 | 7.6 | 9.1 | 11.8 | 0.22 ± 0.02 |
| Me2-estradiol-TSC | 13.96 ± 0.03 | 10.11 ± 0.03 | 3.85 | 8.0 | 9.6 | 12.4 | 0.31 ± 0.06 |
| estrone-SC | 11.26 1 | 4.82 1 | 6.44 1 | 5.8 | 6.3 | 7.5 1 | fast reaction 1 |
| estrone-TSC | 13.18 2 | 9.44 2 | 3.74 2 | 7.7 | 9.2 | 11.9 2 | 0.13 3 |
| Me-estrone-TSC | 13.64 3 | 9.66 3 | 3.98 3 | 7.7 | 9.2 | 11.9 3 | 0.05 4 |
| Me2-estrone-TSC | 13.36 ± 0.08 4 | 9.44 ± 0.07 4 | 3.92 4 | 8.0 | 9.7 | 12.5 4 | 0.05 4 |
Table 3.
Isotropic EPR parameters obtained by the simulation of room temperature EPR spectra recorded in DMSO of Cu(II) complexes formed with estradiol-(T)SCs. The unit of the coupling values is 10−4 cm−1. The experimental errors were ±0.001 for g0 and ±1 G for couplings and linewidth parameters.
Table 3.
Isotropic EPR parameters obtained by the simulation of room temperature EPR spectra recorded in DMSO of Cu(II) complexes formed with estradiol-(T)SCs. The unit of the coupling values is 10−4 cm−1. The experimental errors were ±0.001 for g0 and ±1 G for couplings and linewidth parameters.
| Complexes | Components | g0 | A0 | a0N | α (G) | β (G) | γ (G) | Component Ratio |
|---|
| Cu(II)-estradiol-SC | [CuLH−1] | 2.099 | 56.6 | 15.4 | 36.0 | −20.0 | 3.7 | 50% |
| [CuL]+ | 2.118 | 48.0 | 14.3 | 54.0 | −25.0 | 1.8 | 50% |
| Cu(II)-estradiol-TSC | [CuLH−1] | 2.090 | 70.2 | 17.5 | 35.7 | −21.3 | 3.0 | 100% |
| Cu(II)-Me-estradiol-TSC | [CuLH−1] | 2.088 | 68.7 | 18.0 | 36.9 | −23.9 | 5.4 | 100% |
| Cu(II)-Me2-estradiol-TSC | [CuLH−1] | 2.086 | 67.1 | 17.5 | 39.8 | −22.3 | 3.2 | 82% |
| [CuL]+ | 2.102 | 56.3 | 17.0 | 35.2 | −21.5 | 3.5 | 18% |
Table 4.
Anisotropic EPR parameters obtained for the Cu(II) complexes of estradiol-(T)SC ligands by the simulation of frozen solution (77 K) EPR spectra recorded in 20% (v/v) MeOH/DMSO. The unit of the coupling values is 10−4 cm−1 unit. The experimental errors were ±0.002 for gx and gy, ±0.001 for gz, ±2 G for Ax and Ay, and ±1 G for Az and nitrogen couplings. Isotropic values of the g-tensor are calculated via equation: g0 = (gx + gy + gz)/3.
Table 4.
Anisotropic EPR parameters obtained for the Cu(II) complexes of estradiol-(T)SC ligands by the simulation of frozen solution (77 K) EPR spectra recorded in 20% (v/v) MeOH/DMSO. The unit of the coupling values is 10−4 cm−1 unit. The experimental errors were ±0.002 for gx and gy, ±0.001 for gz, ±2 G for Ax and Ay, and ±1 G for Az and nitrogen couplings. Isotropic values of the g-tensor are calculated via equation: g0 = (gx + gy + gz)/3.
| | Estradiol-SC | Estradiol-TSC | Me-Estradiol-TSC | Me2-Estradiol-TSC | Oligomers 1 |
|---|
| gx | 2.054 | 2.030 | 2.028 | 2.030 | 2.088 |
| gy | 2.063 | 2.056 | 2.054 | 2.056 | – |
| gz | 2.299 | 2.209 | 2.206 | 2.222 | – |
| Ax | 19.22 | 23.7 | 25.7 | 24.2 | 44 |
| Ay | 4.8 | 17.8 | 17.9 | 15.7 | – |
| Az | 166.4 | 180.0 | 178.1 | 174.2 | – |
| axN | 12.1 | 14.6 | 14.4 | 4.4 | – |
| ayN | 15.6 | 10.1 | 9.9 | 9.9 | – |
| azN | 12.9 | 8.6 | 8.4 | 14.4 | – |
| g0,calc | 2.139 | 2.098 | 2.096 | 2.103 | – |
Table 5.
DPPH free radical scavenging activity of estradiol-(T)SC, estrone-TSC, STSC, estradiol and Trolox expressed as IC50 and Trolox equivalent antioxidant capacity (TEAC) values, and % scavenging effect at 1:1 DPPH-to-compound ratio after 30 min. (T = 25 °C; in ethanol, cDPPH = 50 µM; ccompound = 5−50 µM).
Table 5.
DPPH free radical scavenging activity of estradiol-(T)SC, estrone-TSC, STSC, estradiol and Trolox expressed as IC50 and Trolox equivalent antioxidant capacity (TEAC) values, and % scavenging effect at 1:1 DPPH-to-compound ratio after 30 min. (T = 25 °C; in ethanol, cDPPH = 50 µM; ccompound = 5−50 µM).
| | IC50 (µM) | % Scavenging Effect 1 | TEAC |
|---|
| estradiol-SC | >50 2 | 6.5 | - |
| estradiol-TSC | 14.6 | 72.0 | 0.8 |
| Me-estradiol-TSC | 15.4 | 31.0 | 0.8 |
| Me2-estradiol-TSC | 13.6 | 100 | 0.9 |
| estrone-TSC | 17.8 | 49.8 | 0.7 |
| STSC | 19.0 | 23.4 | 0.6 |
| estradiol | >50 2 | 0.5 | - |
| estradiol-SC+1 equiv. Cu(II) | >50 2 | 8.5 | - |
| estradiol-TSC+1 equiv. Cu(II) | 15.7 | 90.8 | 0.8 |
| Me-estradiol-TSC+1 equiv. Cu(II) | 20.3 | 33.6 | 0.6 |
| Me2-estradiol-TSC+1 equiv. Cu(II) | 16.4 | 40.4 | 0.7 |
| estrone-TSC+1 equiv. Cu(II) | 34.8 | 11.1 | 0.3 |
| STSC+1 equiv. Cu(II) | >50 2 | 29.4 | - |
| Trolox | 12 | 100 | 1 |
Table 6.
MIC values determined for the ligands in the absence and presence of 1 equiv. Cu(II) on Gram-positive bacteria strains.
Table 6.
MIC values determined for the ligands in the absence and presence of 1 equiv. Cu(II) on Gram-positive bacteria strains.
| | MIC (µM) |
|---|
| | E. faecalis ATCC 29212 | S. aureus ATCC 25928 |
|---|
| estradiol-SC | >100 | 50 |
| estradiol-TSC | >100 | 25 |
| Me-estradiol-TSC | >100 | >100 |
| Me2-estradiol-TSC | >100 | >100 |
| estrone-TSC | >100 | >100 |
| estradiol-SC+1 equiv. Cu(II) | >100 | 50 |
| estradiol-TSC+1 equiv. Cu(II) | 12.5 | 3.125 |
| Me-estradiol-TSC+1 equiv. Cu(II) | 50 | 6.25 |
| Me2-estradiol-TSC+1 equiv. Cu(II) | 12.5 | 0.78 |
| estrone-TSC+1 equiv. Cu(II) | 12.5 | 6.25 |
| CuCl2 | >100 | >100 |
Table 7.
IC50 values (µM) of the complexes and the ligands determined on various cancer cell lines. (24 h incubation time).
Table 7.
IC50 values (µM) of the complexes and the ligands determined on various cancer cell lines. (24 h incubation time).
| | IC50 Values (µM) |
|---|
| | MCF-7 | MCF-7 KCR | DU-145 | A549 |
|---|
| DMSO | >20 | >20 | >20 | >20 |
| CuCl2 | >20 | >20 | >20 | >20 |
| estrone-TSC | >20 | >20 | >20 | 16.71 ± 1.03 |
| Cu(II)-estrone-TSC | 4.28 ± 1.06 | 12.75 ± 1.10 | 11.37 ± 1.05 | 4.03 ± 1.01 |
| estradiol-SC | >20 | >20 | >20 | >20 |
| Cu(II)-estradiol-SC | >20 | >20 | >20 | >20 |
| estradiol-TSC | >20 | >20 | >20 | 20.54 ± 1.07 |
| Cu(II)-estradiol-TSC | 8.40 ± 1.11 | 12.95 ± 1.09 | >20 | 10.03 ± 1.04 |
| Me-estradiol-TSC | >20 | >20 | >20 | 15.79 ± 1.03 |
| Cu(II)-Me-estradiol-TSC | 16.74 ± 1.66 | >20 | 15.84 ± 1.08 | 3.71 ± 1.02 |
| Me2-estradiol-TSC | >20 | >20 | >20 | >20 |
| Cu(II)-Me2-estradiol-TSC | 10.05 ± 1.36 | >20 | 7.85 ± 1.08 | 3.44 ± 1.02 |