Gold Nanoparticles as a Biosensor for Cancer Biomarker Determination
Abstract
1. Introduction
2. Synthesis of Gold Nanoparticles
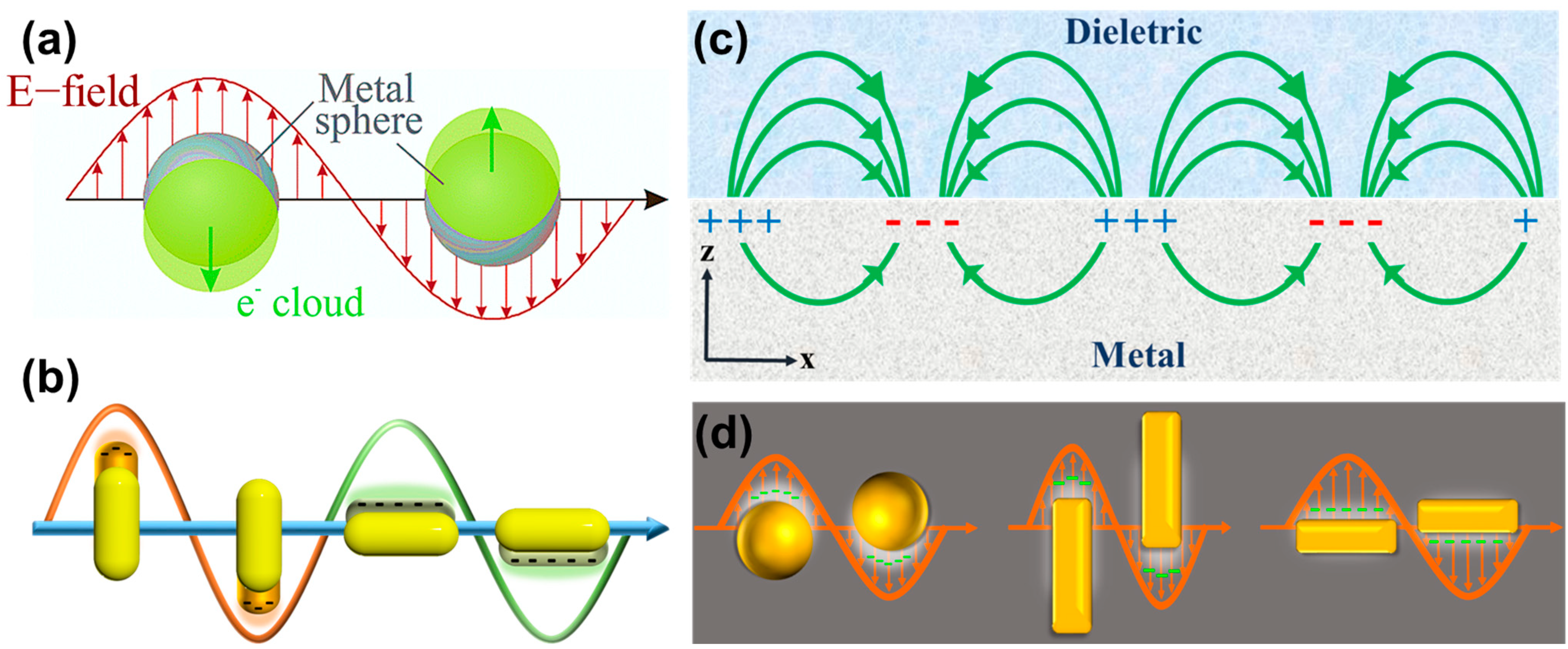
2.1. Surfactant-Preferential-Binding-Directed Growth
2.2. Electric-Field-Directed Growth Mechanism
3. Biological Characteristics of AuNPs
3.1. Localized Surface Plasmon Resonance (LSPR)
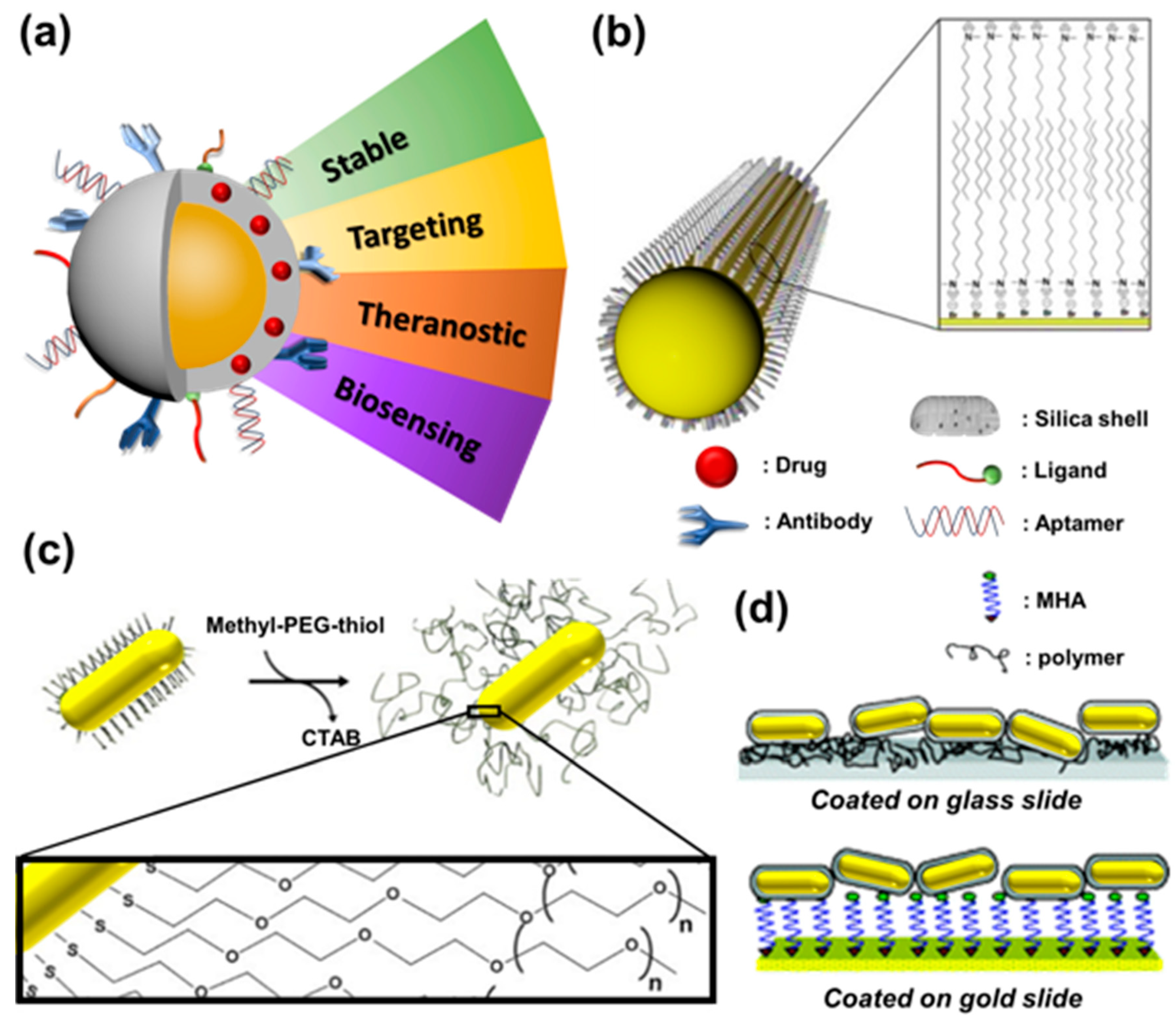
3.2. Surface Modification of Gold Nanoparticles
3.2.1. Ligand Exchange
3.2.2. Electrostatic Adsorption
3.2.3. Electrostatic Adsorption
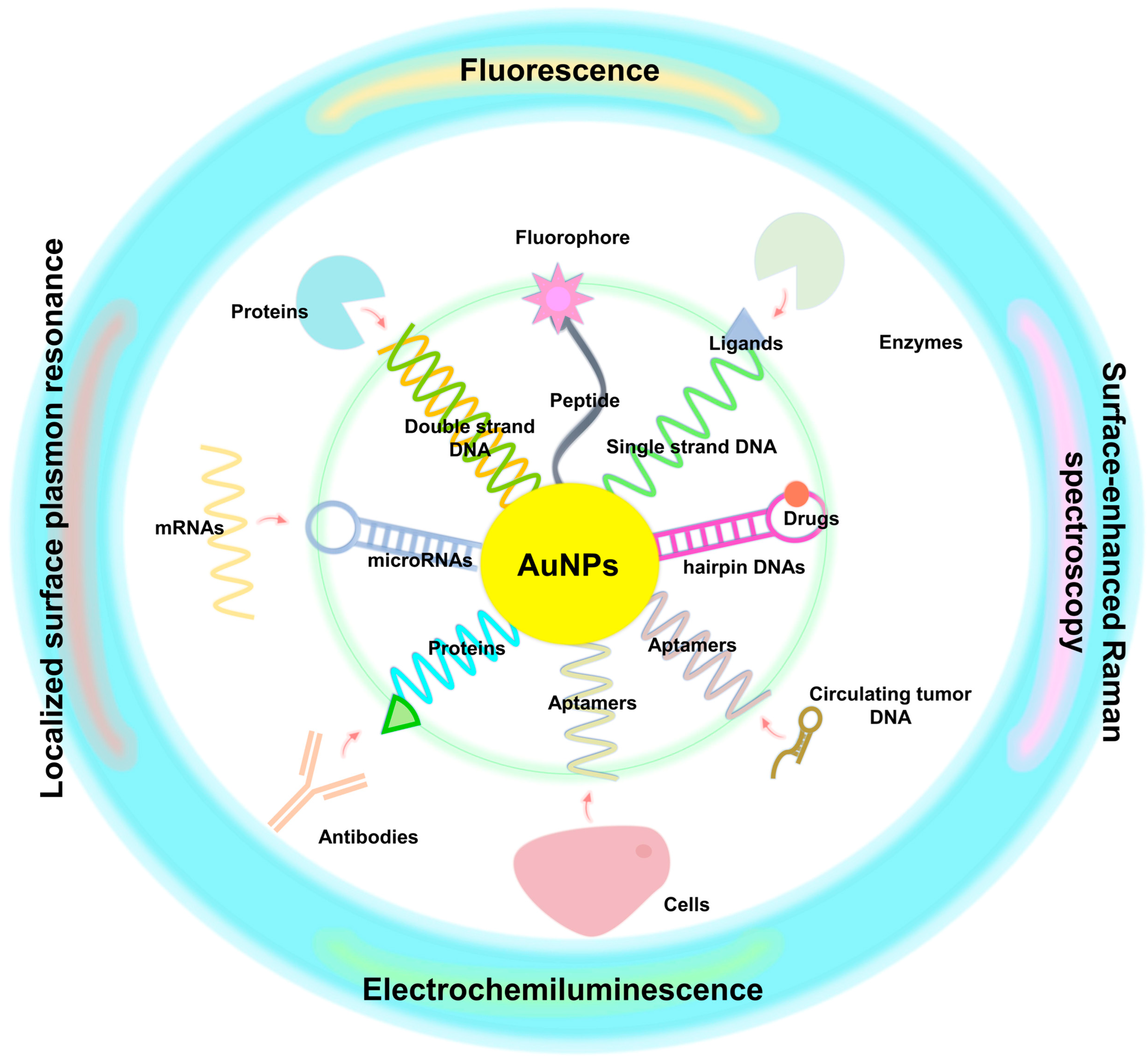
3.3. Biosensing Assays
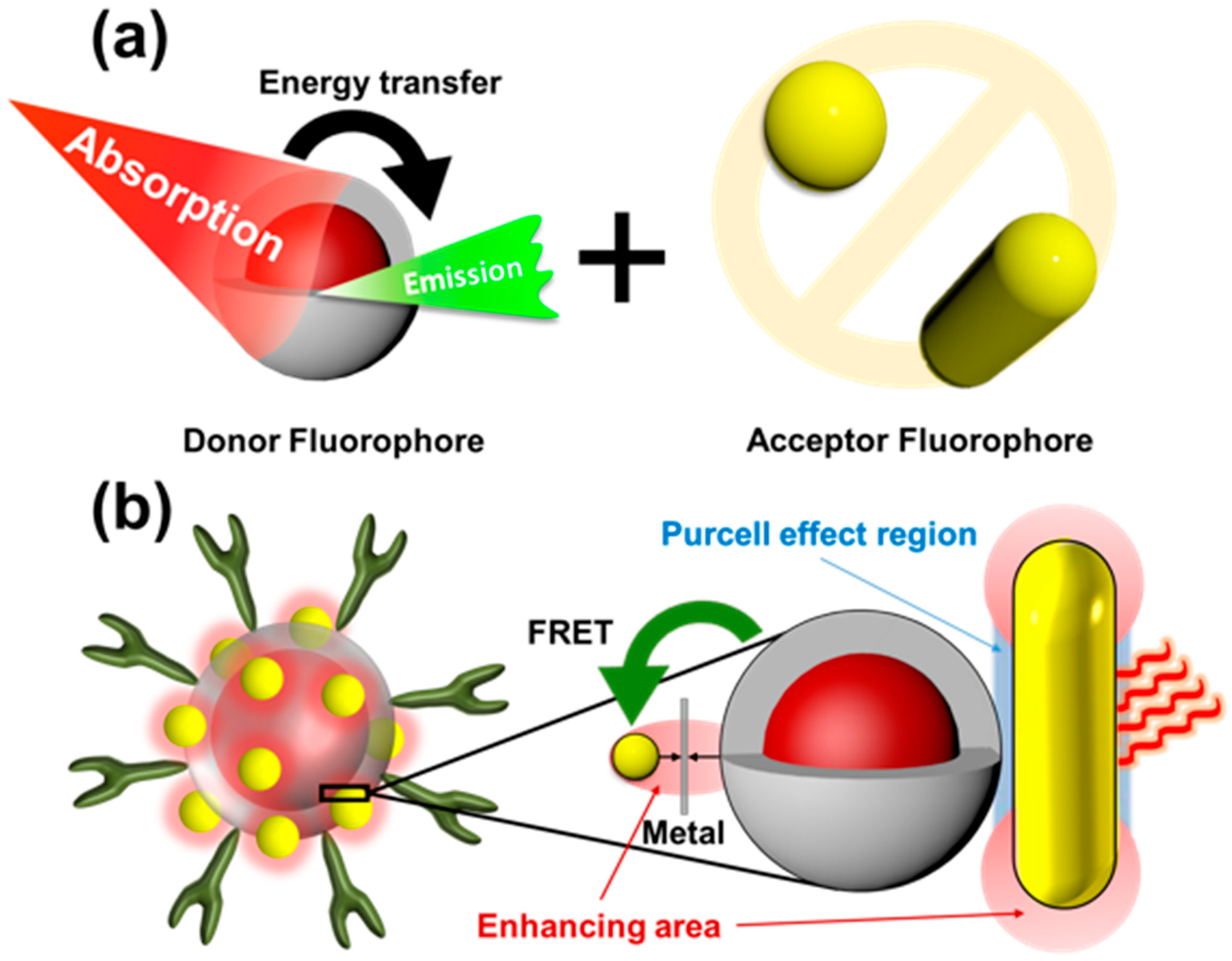
3.3.1. Förster Resonance Energy Transfer (FRET)
3.3.2. LSPR Electric Field Enhancement
3.3.3. Fluorescent and Colorimetric Assay
4. Clinical Biomarkers for Early Cancer Detection
4.1. DNA
4.2. RNA/miRNA
4.3. Protein
5. Biosensing of Cancer Biomarkers
5.1. Optical Biosensing
5.2. Enzyme-Dependent Biosensing
6. Clinical Applications of Cancer Based AuNP Biomarkers
6.1. Single-Point Mutations or Single-Nucleotide Polymorphism (SNP)
6.2. Exon or Gene Copy-Number Changes Detection
6.3. Protein Structural Modifications Detection
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daniel, M.C.; Astruc, D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar] [CrossRef]
- Baranova, N.N.; Zotov, A.V.; Bannykh, L.N.; Darina, T.G.; Savelev, B.V. Experimental-Study of the Solubility of Gold in Water under 450-Degrees-C and 500-Atm in Relation to Redox Conditions. Geokhimiya 1983, 8, 1133–1138. [Google Scholar]
- Nerle, U.; Hodlur, R.M.; Rabinal, M.K. A sharp and visible range plasmonic in heavily doped metal oxide films. Mater. Res. Express 2014, 1, 015910. [Google Scholar] [CrossRef]
- Sun, M.Z.; Xu, L.G.; Ma, W.; Wu, X.L.; Kuang, H.; Wang, L.B.; Xu, C.L. Hierarchical Plasmonic Nanorods and Upconversion Core-Satellite Nanoassemblies for Multimodal Imaging-Guided Combination Phototherapy. Adv. Mater. 2016, 28, 898–904. [Google Scholar] [CrossRef]
- Martin, C.R. Nanomaterials: A Membrane-Based Synthetic Approach. Science 1994, 266, 1961–1966. [Google Scholar] [CrossRef]
- Billot, L.; de la Chapelle, M.L.; Grimault, A.-S.; Vial, A.; Barchiesi, D.; Bijeon, J.-L.; Adam, P.-M.; Royer, P. Surface enhanced Raman scattering on gold nanowire arrays: Evidence of strong multipolar surface plasmon resonance enhancement. Chem. Phys. Lett. 2006, 422, 303–307. [Google Scholar] [CrossRef]
- Taub, N.; Krichevski, A.O.; Markovich, G. Growth of Gold Nanorods on Surfaces. J. Phys. Chem. B 2003, 107, 11579–11582. [Google Scholar] [CrossRef]
- Reetz, M.T.; Helbig, W. Size-Selective Synthesis of Nanostructured Transition Metal Clusters. J. Am. Chem. Soc. 1994, 116, 7401–7402. [Google Scholar] [CrossRef]
- Jana, N.R. Gram-Scale Synthesis of Soluble, Near-Monodisperse Gold Nanorods and Other Anisotropic Nanoparticles. Small 2005, 1, 875–882. [Google Scholar] [CrossRef]
- Jana, N.R.; Gearheart, L.; Murphy, C.J. Evidence for Seed-Mediated Nucleation in the Chemical Reduction of Gold Salts to Gold Nanoparticles. Chem. Mater. 2001, 13, 2313–2322. [Google Scholar] [CrossRef]
- Wiesner, J.; Wokaun, A. Anisometric Gold Colloids—Preparation, Characterization, and Optical-Properties. Chem. Phys. Lett. 1989, 157, 569–575. [Google Scholar] [CrossRef]
- Nikoobakht, B.; El-Sayed, M.A. Preparation and Growth Mechanism of Gold Nanorods (NRs) Using Seed-Mediated Growth Method. Chem. Mater. 2003, 15, 1957–1962. [Google Scholar] [CrossRef]
- Johnson, C.J.; Dujardin, E.; Davis, S.A.; Murphy, C.J.; Mann, S. Growth and form of gold nanorods prepared by seed-mediated, surfactant-directed synthesis. J. Mater. Chem. 2002, 12, 1765–1770. [Google Scholar] [CrossRef]
- Murphy, C.J.; San, T.K.; Gole, A.M.; Orendorff, C.J.; Gao, J.X.; Gou, L.; Hunyadi, S.E.; Li, T. Anisotropic metal nanoparticles: Synthesis, assembly, and optical applications. J. Phys. Chem. B 2005, 109, 13857–13870. [Google Scholar] [CrossRef]
- Pérez-Juste, J.; Liz-Marzán, L.M.; Carnie, S.; Chan, D.Y.C.; Mulvaney, P. Electric-Field-Directed Growth of Gold Nanorods in Aqueous Surfactant Solutions. Adv. Funct. Mater. 2004, 14, 571–579. [Google Scholar] [CrossRef]
- Alkilany, A.M.; Thompson, L.B.; Boulos, S.P.; Sisco, P.N.; Murphy, C.J. Gold nanorods: Their potential for photothermal therapeutics and drug delivery, tempered by the complexity of their biological interactions. Adv. Drug Deliv. Rev. 2012, 64, 190–199. [Google Scholar] [CrossRef]
- Wood, R.W. On a Remarkable Case of Uneven Distribution of Light in a Diffraction Grating Spectrum. PPSL 1902, 18, 269. [Google Scholar] [CrossRef]
- Fano, U. The Theory of Anomalous Diffraction Gratings and of Quasi-Stationary Waves on Metallic Surfaces (Sommerfeld’s Waves). J. Opt. Soc. Am. 1941, 31, 213–222. [Google Scholar] [CrossRef]
- Kelly, K.L.; Coronado, E.; Zhao, L.L.; Schatz, G.C. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. J. Phys. Chem. B 2003, 107, 668–677. [Google Scholar] [CrossRef]
- Weissleder, R. A clearer vision for in vivo imaging. Nat. Biotechnol. 2001, 19, 316–317. [Google Scholar] [CrossRef]
- Jain, P.K.; Huang, X.H.; El-Sayed, I.H.; El-Sayed, M.A. Noble Metals on the Nanoscale: Optical and Photothermal Properties and Some Applications in Imaging, Sensing, Biology, and Medicine. Accounts Chem. Res. 2008, 41, 1578–1586. [Google Scholar] [CrossRef]
- Ye, X.; Jin, L.; Caglayan, H.; Chen, J.; Xing, G.; Zheng, C.; Doan-Nguyen, V.; Kang, Y.; Engheta, N.; Kagan, C.R.; et al. Improved Size-Tunable Synthesis of Monodisperse Gold Nanorods through the Use of Aromatic Additives. ACS Nano 2012, 6, 2804–2817. [Google Scholar] [CrossRef]
- Wijaya, A.; Schaffer, S.B.; Pallares, I.G.; Hamad-Schifferli, K. Selective Release of Multiple DNA Oligonucleotides from Gold Nanorods. ACS Nano 2008, 3, 80–86. [Google Scholar] [CrossRef]
- Chen, C.-C.; Lin, Y.-P.; Wang, C.-W.; Tzeng, H.-C.; Wu, C.-H.; Chen, Y.-C.; Chen, C.-P.; Chen, L.-C.; Wu, Y.-C. DNA−Gold Nanorod Conjugates for Remote Control of Localized Gene Expression by near Infrared Irradiation. J. Am. Chem. Soc. 2006, 128, 3709–3715. [Google Scholar] [CrossRef]
- Lee, S.E.; Liu, G.L.; Kim, F.; Lee, L.P. Remote Optical Switch for Localized and Selective Control of Gene Interference. Nano Lett. 2009, 9, 562–570. [Google Scholar] [CrossRef]
- Dujardin, E.; Mann, S.; Hsin, L.-B.; Wang, C.R.C. DNA-driven self-assembly of gold nanorods. Chem. Commun. 2001, 1264–1265. [Google Scholar] [CrossRef]
- Huff, T.B.; Tong, L.; Zhao, Y.; Hansen, M.N.; Cheng, J.-X.; Wei, A. Hyperthermic effects of gold nanorods on tumor cells. Nanomedicine 2007, 2, 125–132. [Google Scholar] [CrossRef]
- von Maltzahn, G.; Park, J.-H.; Agrawal, A.; Bandaru, N.K.; Das, S.K.; Sailor, M.J.; Bhatia, S.N. Computationally Guided Photothermal Tumor Therapy Using Long-Circulating Gold Nanorod Antennas. Cancer Res 2009, 69, 3892–3900. [Google Scholar] [CrossRef]
- Yu, C.; Varghese, L.; Irudayaraj, J. Surface Modification of Cetyltrimethylammonium Bromide-Capped Gold Nanorods to Make Molecular Probes. Langmuir 2007, 23, 9114–9119. [Google Scholar] [CrossRef]
- Pissuwan, D.; Valenzuela, S.M.; Killingsworth, M.C.; Xu, X.; Cortie, M.B. Targeted destruction of murine macrophage cells with bioconjugated gold nanorods. J. Nanoparticle Res. 2007, 9, 1109–1124. [Google Scholar] [CrossRef]
- Hauck, T.S.; Ghazani, A.A.; Chan, W.C.W. Assessing the Effect of Surface Chemistry on Gold Nanorod Uptake, Toxicity, and Gene Expression in Mammalian Cells. Small 2008, 4, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Gole, A.; Murphy, C. Polyelectrolyte-Coated Gold Nanorods: Synthesis, Characterization and Immobilization. Chem. Mater. 2005, 17, 1325–1330. [Google Scholar] [CrossRef]
- Alkilany, A.M.; Thompson, L.B.; Murphy, C.J. Polyelectrolyte Coating Provides a Facile Route to Suspend Gold Nanorods in Polar Organic Solvents and Hydrophobic Polymers. ACS Appl. Mater. Interfaces 2010, 2, 3417–3421. [Google Scholar] [CrossRef]
- Sendroiu, I.E.; Warner, M.E.; Corn, R.M. Fabrication of Silica-Coated Gold Nanorods Functionalized with DNA for Enhanced Surface Plasmon Resonance Imaging Biosensing Applications. Langmuir 2009, 25, 11282–11284. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Wang, L.M.; Wang, J.; Jiang, X.M.; Li, X.H.; Hu, Z.J.; Ji, Y.H.; Wu, X.C.; Chen, C.Y. Mesoporous Silica-Coated Gold Nanorods as a Light-Mediated Multifunctional Theranostic Platform for Cancer Treatment. Adv. Mater. 2012, 24, 1418–1423. [Google Scholar] [CrossRef]
- Malsch, I. Nanotechnology in Europe: Scientific trends and organizational dynamics. Nanotechnology 1999, 10, 1–7. [Google Scholar] [CrossRef]
- Ojea-Jiménez, I.; Tort, O.; Lorenzo, J.; Puntes, V. Engineered nonviral nanocarriers for intracellular gene delivery applications. Biomed. Mater. 2012, 7, 54106. [Google Scholar] [CrossRef] [PubMed]
- Roduner, E. Size matters: Why nanomaterials are different. Chem. Soc. Rev. 2006, 35, 583–592. [Google Scholar] [CrossRef]
- Li, Y.; Boone, E.; El-Sayed, M.A. Size Effects of PVP−Pd Nanoparticles on the Catalytic Suzuki Reactions in Aqueous Solution. Langmuir 2002, 18, 4921–4925. [Google Scholar] [CrossRef]
- Zhan, Q.; Zhang, X.; Zhao, Y.; Liu, J.; He, S. Tens of thousands-fold upconversion luminescence enhancement induced by a single gold nanorod. Laser Photon- Rev. 2015, 9, 479–487. [Google Scholar] [CrossRef]
- Dadmehr, M.; Shahi, S.C.; Malekkiani, M.; Korouzhdehi, B.; Tavassoli, A. A stem-loop like aptasensor for sensitive detection of aflatoxin based on graphene oxide/AuNPs nanocomposite platform. Food Chem. 2023, 402, 134212. [Google Scholar] [CrossRef]
- Hu, A.; Chen, G.; Yang, T.; Ma, C.; Li, L.; Gao, H.; Gu, J.; Zhu, C.; Wu, Y.; Li, X.; et al. A fluorescent probe based on FRET effect between carbon nanodots and gold nanoparticles for sensitive detection of thiourea. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2022, 281, 121582. [Google Scholar] [CrossRef]
- Wu, D.M.; García-Etxarri, A.; Salleo, A.; Dionne, J.A. Plasmon-Enhanced Upconversion. J. Phys. Chem. Lett. 2014, 5, 4020–4031. [Google Scholar] [CrossRef]
- Li, Y.; Wen, T.; Zhao, R.; Liu, X.; Ji, T.; Wang, H.; Shi, X.; Shi, J.; Wei, J.; Zhao, Y.; et al. Localized Electric Field of Plasmonic Nanoplatform Enhanced Photodynamic Tumor Therapy. ACS Nano 2014, 8, 11529–11542. [Google Scholar] [CrossRef]
- Huang, K.-W.; Yu, C.-J.; Tseng, W.-L. Sensitivity enhancement in the colorimetric detection of lead(II) ion using gallic acid–capped gold nanoparticles: Improving size distribution and minimizing interparticle repulsion. Biosens. Bioelectron. 2010, 25, 984–989. [Google Scholar] [CrossRef]
- Wang, C.-I.; Huang, C.-C.; Lin, Y.-W.; Chen, W.-T.; Chang, H.-T. Catalytic gold nanoparticles for fluorescent detection of mercury(II) and lead(II) ions. Anal. Chim. Acta 2012, 745, 124–130. [Google Scholar] [CrossRef]
- Li, Y.; Tang, L.; Zhu, C.; Liu, X.; Wang, X.; Liu, Y. Fluorescent and colorimetric assay for determination of Cu(II) and Hg(II) using AuNPs reduced and wrapped by carbon dots. Mikrochim. Acta 2021, 189, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dadmehr, M.; Mortezaei, M.; Korouzhdehi, B. Dual mode fluorometric and colorimetric detection of matrix metalloproteinase MMP-9 as a cancer biomarker based on AuNPs@gelatin/ AuNCs nanocomposite. Biosens. Bioelectron. 2023, 220. [Google Scholar] [CrossRef]
- Shahi, S.C.; Dadmehr, M.; Korouzhdehi, B.; Tavassoli, A. A novel colorimetric biosensor for sensitive detection of aflatoxin mediated by bacterial enzymatic reaction in saffron samples. Nanotechnology 2021, 32, 505503. [Google Scholar] [CrossRef]
- Strasser, B.J. A world in one dimension: Linus Pauling, Francis Crick and the central dogma of molecular biology. Hist. Philos. Life Sci. 2006, 28, 491–512. [Google Scholar]
- Zhou, V.W.; Goren, A.; Bernstein, B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2010, 12, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Tsankova, N.; Renthal, W.; Kumar, A.; Nestler, E.J. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007, 8, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Laird, P.W. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef]
- Olkhov-Mitsel, E.; Bapat, B. Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med. 2012, 1, 237–260. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, C.-Y. Single base extension reaction-based surface enhanced Raman spectroscopy for DNA methylation assay. Biosens. Bioelectron. 2012, 31, 451–457. [Google Scholar] [CrossRef]
- Liu, P.; Wang, D.; Zhou, Y.; Wang, H.; Yin, H.; Ai, S. DNA methyltransferase detection based on digestion triggering the combination of poly adenine DNA with gold nanoparticles. Biosens. Bioelectron. 2016, 80, 74–78. [Google Scholar] [CrossRef]
- Guthula, L.S.; Yeh, K.-T.; Huang, W.-L.; Chen, C.-H.; Chen, Y.-L.; Huang, C.-J.; Chau, L.-K.; Chan, M.W.; Lin, S.-H. Quantitative and amplification-free detection of SOCS-1 CpG methylation percentage analyses in gastric cancer by fiber optic nanoplasmonic biosensor. Biosens. Bioelectron. 2022, 214, 114540. [Google Scholar] [CrossRef]
- Rasheed, P.A.; Sandhyarani, N. Femtomolar level detection of BRCA1 gene using a gold nanoparticle labeled sandwich type DNA sensor. Colloids Surfaces B: Biointerfaces 2014, 117, 7–13. [Google Scholar] [CrossRef]
- Feng, D.; Su, J.; He, G.; Xu, Y.; Wang, C.; Zheng, M.; Qian, Q.; Mi, X. Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles. Biosensors 2020, 10, 78. [Google Scholar] [CrossRef]
- Bai, Y.; Li, H.; Xu, J.; Huang, Y.; Zhang, X.; Weng, J.; Li, Z.; Sun, L. Ultrasensitive colorimetric biosensor for BRCA1 mutation based on multiple signal amplification strategy. Biosens. Bioelectron. 2020, 166, 112424. [Google Scholar] [CrossRef]
- Liang, Z.; Nie, Y.; Zhang, X.; Wang, P.; Ma, Q. Multiplex Electrochemiluminescence Polarization Assay Based on the Surface Plasmon Coupling Effect of Au NPs and Ag@Au NPs. Anal. Chem. 2021, 93, 7491–7498. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, V.C.; Soares, J.C.; Soares, A.C.; Braz, D.C.; Melendez, M.E.; Ribas, L.C.; Scabini, L.F.; Bruno, O.M.; Carvalho, A.L.; Reis, R.M.; et al. Electrochemical and optical detection and machine learning applied to images of genosensors for diagnosis of prostate cancer with the biomarker PCA3. Talanta 2020, 222, 121444. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef]
- Ciesiolka, J.; Yarus, M. Small RNA-divalent domains. RNA 1996, 2, 785–793. [Google Scholar]
- Hofmann, H.P.; Limmer, S.; Hornung, V.; Sprinzl, M. Ni2+-binding RNA motifs with an asymmetric purine-rich internal loop and a G-A base pair. RNA 1997, 3, 1289–1300. [Google Scholar]
- Rajendran, M.; Ellington, A.D. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal. Bioanal. Chem. 2007, 390, 1067–1075. [Google Scholar] [CrossRef]
- Ding, C.; Wei, S.; Liu, H. Electrochemiluminescent Determination of Cancer Cells Based on Aptamers, Nanoparticles, and Magnetic Beads. Chem. – A Eur. J. 2012, 18, 7263–7268. [Google Scholar] [CrossRef]
- Liu, H.; Xu, S.; He, Z.; Deng, A.; Zhu, J.-J. Supersandwich Cytosensor for Selective and Ultrasensitive Detection of Cancer Cells Using Aptamer-DNA Concatamer-Quantum Dots Probes. Anal. Chem. 2013, 85, 3385–3392. [Google Scholar] [CrossRef]
- Wang, X.; Shu, G.; Gao, C.; Yang, Y.; Xu, Q.; Tang, M. Electrochemical biosensor based on functional composite nanofibers for detection of K-ras gene via multiple signal amplification strategy. Anal. Biochem. 2014, 466, 51–58. [Google Scholar] [CrossRef]
- Bao, C.; Conde, J.; Curtin, J.; Artzi, N.; Tian, F.; Cui, D. Bioresponsive antisense DNA gold nanobeacons as a hybrid in vivo theranostics platform for the inhibition of cancer cells and metastasis. Sci. Rep. 2015, 5, 12297. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, T.; Tan, W.; Fan, Z.H. Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. ACS Nano 2013, 7, 7067–7076. [Google Scholar] [CrossRef] [PubMed]
- Borghei, Y.-S.; Hosseini, M.; Dadmehr, M.; Hosseinkhani, S.; Ganjali, M.R.; Sheikhnejad, R. Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal. Chim. Acta 2016, 904, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Guo, Z.; Cao, Y.; Zhang, W.; Chen, Y. A dual biomarker detection platform for quantitating circulating tumor DNA (ctDNA). Nanotheranostics 2018, 2, 12–20. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Zhong, H.; Li, J.; Ding, C. Near-Infrared Light-Activated Pt@Au Nanorings-Based Probe for Fluorescence Imaging and Targeted Photothermal Therapy of Cancer Cells. ACS Appl. Bio Mater. 2019, 2, 5012–5020. [Google Scholar] [CrossRef]
- Wang, C.; Wang, W.; Xu, Y.; Zhao, X.; Li, S.; Qian, Q.; Mi, X. Tetrahedral DNA Framework-Programmed Electrochemical Biosenors with Gold Nanoparticles for Ultrasensitive Cell-Free DNA Detection. Nanomaterials 2022, 12, 666. [Google Scholar] [CrossRef]
- Avelino, K.Y.; Oliveira, L.S.; Lucena-Silva, N.; de Melo, C.P.; de Andrade, C.A.S.; Oliveira, M.D. Metal-polymer hybrid nanomaterial for impedimetric detection of human papillomavirus in cervical specimens. J. Pharm. Biomed. Anal. 2020, 185, 113249. [Google Scholar] [CrossRef]
- Ilbeigi, S.; Vais, R.D.; Sattarahmady, N. Photo-genosensor for Trichomonas vaginalis based on gold nanoparticles-genomic DNA. Photodiagnosis Photodyn. Ther. 2021, 34, 102290. [Google Scholar] [CrossRef]
- Zhu, D.; Zhao, D.; Huang, J.; Zhu, Y.; Chao, J.; Su, S.; Li, J.; Wang, L.; Shi, J.; Zuo, X.; et al. Poly-adenine-mediated fluorescent spherical nucleic acid probes for live-cell imaging of endogenous tumor-related mRNA. Nanomedicine: Nanotechnology, Biol. Med. 2018, 14, 1797–1807. [Google Scholar] [CrossRef]
- Shawky, S.M.; Awad, A.M.; Abugable, A.A.; El-Khamisy, S.F. Gold nanoparticles—An optical biosensor for RNA quantification for cancer and neurologic disorders diagnosis. Int. J. Nanomed. 2018, 13, 8137–8151. [Google Scholar] [CrossRef]
- Li, H.; Warden, A.R.; Su, W.; He, J.; Zhi, X.; Wang, K.; Zhu, L.; Shen, G.; Ding, X. Highly sensitive and portable mRNA detection platform for early cancer detection. J. Nanobiotechnology 2021, 19, 1–10. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, L.; Wang, G.; Feng, Q.; Liu, L. Label-free and sensitive strategy for microRNAs detection based on the formation of boronate ester bonds and the dual-amplification of gold nanoparticles. Biosens. Bioelectron. 2013, 47, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xia, N.; Liu, H.; Kang, X.; Liu, X.; Xue, C.; He, X. Highly sensitive and label-free electrochemical detection of microRNAs based on triple signal amplification of multifunctional gold nanoparticles, enzymes and redox-cycling reaction. Biosens. Bioelectron. 2014, 53, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Borghei, Y.-S.; Hosseini, M. A New Eye Dual-readout Method for MiRNA Detection based on Dissolution of Gold nanoparticles via LSPR by CdTe QDs Photoinduction. Sci. Rep. 2019, 9, 5453. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; He, G.; Wang, C.; Li, S.; Zhao, X.; Xu, Y.; Mi, X. Poly-adenine-mediated spherical nucleic acid probes for live cell fluorescence imaging of tumor-related microRNAs. Mol. Biol. Rep. 2022, 49, 3705–3712. [Google Scholar] [CrossRef]
- Pothipor, C.; Aroonyadet, N.; Bamrungsap, S.; Jakmunee, J.; Ounnunkad, K. A highly sensitive electrochemical microRNA-21 biosensor based on intercalating methylene blue signal amplification and a highly dispersed gold nanoparticles/graphene/polypyrrole composite. Analyst 2021, 146, 2679–2688. [Google Scholar] [CrossRef]
- Zhao, J.; He, C.; Wu, W.; Yang, H.; Dong, J.; Wen, L.; Hu, Z.; Yang, M.; Hou, C.; Huo, D. MXene-MoS2 heterostructure collaborated with catalyzed hairpin assembly for label-free electrochemical detection of microRNA-21. Talanta 2021, 237, 122927. [Google Scholar] [CrossRef]
- Liu, S.; Su, W.; Li, Z.; Ding, X. Electrochemical detection of lung cancer specific microRNAs using 3D DNA origami nanostructures. Biosens. Bioelectron. 2015, 71, 57–61. [Google Scholar] [CrossRef]
- Shen, D.; Hu, W.; He, Q.; Yang, H.; Cui, X.; Zhao, S. A highly sensitive electrochemical biosensor for microRNA122 detection based on a target-induced DNA nanostructure. Anal. Methods 2021, 13, 2823–2829. [Google Scholar] [CrossRef]
- Lu, H.; Hailin, T.; Yi, X.; Wang, J. Three-Dimensional DNA Nanomachine Combined with Toehold-Mediated Strand Displacement Reaction for Sensitive Electrochemical Detection of MiRNA. Langmuir 2020, 36, 10708–10714. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, X.; Li, P.; Lin, X.; Wang, J.; Hu, Z.; Zhang, P.; Chen, D.; Cai, H.; Niessner, R.; et al. Ultrasensitive and Simultaneous SERS Detection of Multiplex MicroRNA Using Fractal Gold Nanotags for Early Diagnosis and Prognosis of Hepatocellular Carcinoma. Anal. Chem. 2021, 93, 8799–8809. [Google Scholar] [CrossRef]
- Kim, W.H.; Lee, J.U.; Jeon, M.J.; Park, K.H.; Sim, S.J. Three-dimensional hierarchical plasmonic nano-architecture based label-free surface-enhanced Raman spectroscopy detection of urinary exosomal miRNA for clinical diagnosis of prostate cancer. Biosens. Bioelectron. 2022, 205, 114116. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Ye, S.; Li, R.; Li, H. A One–Two–Three Multifunctional System for Enhanced Imaging and Detection of Intracellular MicroRNA and Chemogene Therapy. ACS Appl. Mater. Interfaces 2021, 13, 27825–27835. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, J.; Li, T.; Shen, R.; Li, G.; Ling, L. Strand displacement amplification-coupled dynamic light scattering method to detect urinary telomerase for non-invasive detection of bladder cancer. Biosens. Bioelectron. 2019, 131, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Pu, F.; Ren, J.; Qu, X. Primer-Modified G-Quadruplex-Au Nanoparticles for Colorimetric Assay of Human Telomerase Activity and Initial Screening of Telomerase Inhibitors. Methods Mol. Biol. 2019, 2035, 347–356. [Google Scholar] [CrossRef]
- Liu, L.; Chang, Y.; Ji, X.; Chen, J.; Zhang, M.; Yang, S. Surface-tethered electrochemical biosensor for telomerase detection by integration of homogeneous extension and hybridization reactions. Talanta 2023, 253, 123597. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, D.; Lu, Y.; Liu, S.; Zhang, J.; Pu, Y.; Wei, W. Fluorescence imaging of FEN1 activity in living cells based on controlled-release of fluorescence probe from mesoporous silica nanoparticles. Biosens. Bioelectron. 2022, 214, 114529. [Google Scholar] [CrossRef]
- Aayanifard, Z.; Alebrahim, T.; Pourmadadi, M.; Yazdian, F.; Dinani, H.S.; Rashedi, H.; Omidi, M. Ultra pH-sensitive detection of total and free prostate-specific antigen using electrochemical aptasensor based on reduced graphene oxide/gold nanoparticles emphasis on TiO2/carbon quantum dots as a redox probe. Eng. Life Sci. 2021, 21, 739–752. [Google Scholar] [CrossRef]
- Wei, B.; Mao, K.; Liu, N.; Zhang, M.; Yang, Z. Graphene nanocomposites modified electrochemical aptamer sensor for rapid and highly sensitive detection of prostate specific antigen. Biosens. Bioelectron. 2018, 121, 41–46. [Google Scholar] [CrossRef]
- Poturnayová, A.; Dzubinová, L.; Buríková, M.; Bízik, J.; Hianik, T. Detection of Breast Cancer Cells Using Acoustics Aptasensor Specific to HER2 Receptors. Biosensors 2019, 9, 72. [Google Scholar] [CrossRef]
- Chen, W.; Li, Z.; Wu, T.; Li, J.; Li, X.; Liu, L.; Bai, H.; Ding, S.; Li, X.; Yu, X. Surface plasmon resonance biosensor for exosome detection based on reformative tyramine signal amplification activated by molecular aptamer beacon. J. Nanobiotechnology 2021, 19, 1–10. [Google Scholar] [CrossRef]
- Gundagatti, S.; Srivastava, S. Development of Electrochemical Biosensor for miR204-Based Cancer Diagnosis. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Ding, L.; Gong, P.; Huang, J. A colorimetric detection of microRNA-148a in gastric cancer by gold nanoparticle–RNA conjugates. Nanotechnology 2019, 31, 095501. [Google Scholar] [CrossRef] [PubMed]
- Miti, A.; Thamm, S.; Müller, P.; Csáki, A.; Fritzsche, W.; Zuccheri, G. A miRNA biosensor based on localized surface plasmon resonance enhanced by surface-bound hybridization chain reaction. Biosens. Bioelectron. 2020, 167, 112465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Song, S.T.; Huang, S.; Yang, L.; Min, Q.H.; Wu, X.C.; Lu, F.; Zhu, J.J. Lighting Up MicroRNA in Living Cells by the Disassembly of Lock-Like DNA-Programmed UCNPs-AuNPs through the Target Cycling Amplification Strategy. Small 2018, 14, 1802292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, K.; Wei, W.; Liu, Y.; Liu, S. Multifunctional Plasmonic Core-Satellites Nanoprobe for Cancer Diagnosis and Therapy Based on a Cascade Reaction Induced by MicroRNA. Anal. Chem. 2021, 93, 9521–9530. [Google Scholar] [CrossRef]
- Zhang, Y.; Chai, Y.; Wang, H.; Yuan, R. Target-Induced 3D DNA Network Structure as a Novel Signal Amplifier for Ultrasensitive Electrochemiluminescence Detection of MicroRNAs. Anal. Chem. 2019, 91, 14368–14374. [Google Scholar] [CrossRef]
- Cui, A.; Zhang, J.; Bai, W.; Sun, H.; Bao, L.; Ma, F.; Li, Y. Signal-on electrogenerated chemiluminescence biosensor for ultrasensitive detection of microRNA-21 based on isothermal strand-displacement polymerase reaction and bridge DNA-gold nanoparticles. Biosens. Bioelectron. 2019, 144, 111664. [Google Scholar] [CrossRef]
- Li, J.; Huang, J.; Yang, X.; Yang, Y.; Quan, K.; Xie, N.; Wu, Y.; Ma, C.; Wang, K. Two-Color-Based Nanoflares for Multiplexed MicroRNAs Imaging in Live Cells. Nanotheranostics 2018, 2, 96–105. [Google Scholar] [CrossRef]
- Qi, G.; Yi, X.; Wang, M.; Sun, D.; Zhu, H. SERS and fluorescence dual-channel microfluidic droplet platform for exploring telomerase activity at single-cell level. Anal. 2022, 147, 5062–5067. [Google Scholar] [CrossRef]
- Mahani, M.; Taheri, M.; Divsar, F.; Khakbaz, F.; Nomani, A.; Ju, H. Label-free triplex DNA-based biosensing of transcription factor using fluorescence resonance energy transfer between N-doped carbon dot and gold nanoparticle. Anal. Chim. Acta 2021, 1181, 338919. [Google Scholar] [CrossRef]
- Huang, J.-Y.; Lin, H.-T.; Chen, T.-H.; Chen, C.-A.; Chang, H.-T.; Chen, C.-F. Signal Amplified Gold Nanoparticles for Cancer Diagnosis on Paper-Based Analytical Devices. ACS Sens. 2018, 3, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Bo, B.; Zhang, T.; Jiang, Y.; Cui, H.; Miao, P. Triple Signal Amplification Strategy for Ultrasensitive Determination of miRNA Based on Duplex Specific Nuclease and Bridge DNA–Gold Nanoparticles. Anal. Chem. 2018, 90, 2395–2400. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Shangguan, J.; Guo, Q.; Ma, W.; Wang, H.; Jia, R.; Ye, Z.; He, X.; Wang, K. Colorimetric and fluorescent dual-mode detection of microRNA based on duplex-specific nuclease assisted gold nanoparticle amplification. Anal. 2019, 144, 4917–4924. [Google Scholar] [CrossRef]
- Sun, Z.; Li, J.; Yang, Y.; Tong, Y.; Li, H.; Wang, C.; Du, L.; Jiang, Y. Ratiometric Fluorescent Biosensor Based on Self-Assembled Fluorescent Gold Nanoparticles and Duplex-Specific Nuclease-Assisted Signal Amplification for Sensitive Detection of Exosomal miRNA. Bioconjugate Chem. 2022, 33, 1698–1706. [Google Scholar] [CrossRef] [PubMed]
- Ki, J.S.; Lee, H.Y.; Son, H.Y.; Huh, Y.-M.; Haam, S. Sensitive Plasmonic Detection of miR-10b in Biological Samples Using Enzyme-Assisted Target Recycling and Developed LSPR Probe. ACS Appl. Mater. Interfaces 2019, 11, 18923–18929. [Google Scholar] [CrossRef]
- Ma, D.; Huang, C.; Zheng, J.; Tang, J.; Li, J.; Yang, J.; Yang, R. Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 2018, 101, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Yang, L.; Lu, F.; Wu, X.C.; Zhu, J.J. A Universal Upconversion Sensing Platform for the Sensitive Detection of Tumour-Related ncRNA through an Exo III-Assisted Cycling Amplification Strategy. Small 2018, 14, 1703858. [Google Scholar] [CrossRef]
- Wang, W.; Liu, J.; Wu, L.-A.; Ko, C.-N.; Wang, X.; Lin, C.; Liu, J.; Ling, L.; Wang, J. Nicking enzyme-free strand displacement amplification-assisted CRISPR-Cas-based colorimetric detection of prostate-specific antigen in serum samples. Anal. Chim. Acta 2022, 1195, 339479. [Google Scholar] [CrossRef]
- Li, Y.; Wark, A.W.; Lee, H.J.; Corn, R.M. Single-Nucleotide Polymorphism Genotyping by Nanoparticle-Enhanced Surface Plasmon Resonance Imaging Measurements of Surface Ligation Reactions. Anal. Chem. 2006, 78, 3158–3164. [Google Scholar] [CrossRef]
- Gao, J.; Ma, L.; Lei, Z.; Wang, Z. Multiple detection of single nucleotide polymorphism by microarray-based resonance light scattering assay with enlarged gold nanoparticle probes. Analyst 2016, 141, 1772–1778. [Google Scholar] [CrossRef]
- Lyu, N.; Rajendran, V.K.; Li, J.; Engel, A.; Molloy, M.P.; Wang, Y. Highly specific detection of KRAS single nucleotide polymorphism by asymmetric PCR/SERS assay. Analyst 2021, 146, 5714–5721. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kang, J.; Baek, I.; You, J.; Jang, K.; Na, S. Highly sensitive and selective detection of single-nucleotide polymorphisms using gold nanoparticle MutS enzymes and a micro cantilever resonator. Talanta 2019, 205, 120154. [Google Scholar] [CrossRef] [PubMed]
- Kalligosfyri, P.; Nikou, S.; Bravou, V.; Kalogianni, D.P. Liquid biopsy genotyping by a simple lateral flow strip assay with visual detection. Anal. Chim. Acta 2021, 1163, 338470. [Google Scholar] [CrossRef] [PubMed]
- Kalligosfyri, P.M.; Nikou, S.; Karteri, S.; Kalofonos, H.P.; Bravou, V.; Kalogianni, D.P. Rapid Multiplex Strip Test for the Detection of Circulating Tumor DNA Mutations for Liquid Biopsy Applications. Biosensors 2022, 12, 97. [Google Scholar] [CrossRef]
- Wang, Y.; Kong, S.L.; Di Su, X. A centrifugation-assisted visual detection of SNP in circulating tumor DNA using gold nanoparticles coupled with isothermal amplification. RSC Adv. 2020, 10, 1476–1483. [Google Scholar] [CrossRef]
- Lee, H.; Kang, T.; Yoon, K.-A.; Lee, S.Y.; Joo, S.-W.; Lee, K. Colorimetric detection of mutations in epidermal growth factor receptor using gold nanoparticle aggregation. Biosens. Bioelectron. 2010, 25, 1669–1674. [Google Scholar] [CrossRef]
- You, J.; Park, C.; Jang, K.; Park, J.; Na, S. Novel Detection Method for Circulating EGFR Tumor DNA Using Gravitationally Condensed Gold Nanoparticles and Catalytic Walker DNA. Materials 2022, 15, 3301. [Google Scholar] [CrossRef]
- New, S.Y.; Aung, K.M.M.; Lim, G.L.; Hong, S.; Tan, S.K.; Lu, Y.; Cheung, E.; Su, X. Fast Screening of Ligand-Protein Interactions based on Ligand-Induced Protein Stabilization of Gold Nanoparticles. Anal. Chem. 2014, 86, 2361–2370. [Google Scholar] [CrossRef]
- Liu, Z.; Liang, Y.; Cao, W.; Gao, W.; Tang, B. Proximity-Induced Hybridization Chain Reaction-Based Photoacoustic Imaging System for Amplified Visualization Protein-Specific Glycosylation in Mice. Anal. Chem. 2021, 93, 8915–8922. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.-H.; Chan, M.-H.; Chang, Y.-C.; Hsiao, M. Gold Nanoparticles as a Biosensor for Cancer Biomarker Determination. Molecules 2023, 28, 364. https://doi.org/10.3390/molecules28010364
Li C-H, Chan M-H, Chang Y-C, Hsiao M. Gold Nanoparticles as a Biosensor for Cancer Biomarker Determination. Molecules. 2023; 28(1):364. https://doi.org/10.3390/molecules28010364
Chicago/Turabian StyleLi, Chien-Hsiu, Ming-Hsien Chan, Yu-Chan Chang, and Michael Hsiao. 2023. "Gold Nanoparticles as a Biosensor for Cancer Biomarker Determination" Molecules 28, no. 1: 364. https://doi.org/10.3390/molecules28010364
APA StyleLi, C.-H., Chan, M.-H., Chang, Y.-C., & Hsiao, M. (2023). Gold Nanoparticles as a Biosensor for Cancer Biomarker Determination. Molecules, 28(1), 364. https://doi.org/10.3390/molecules28010364






