Molecular Mechanisms of Aggregation of Canine SOD1 E40K Amyloidogenic Mutant Protein
Abstract
1. Introduction
2. Results
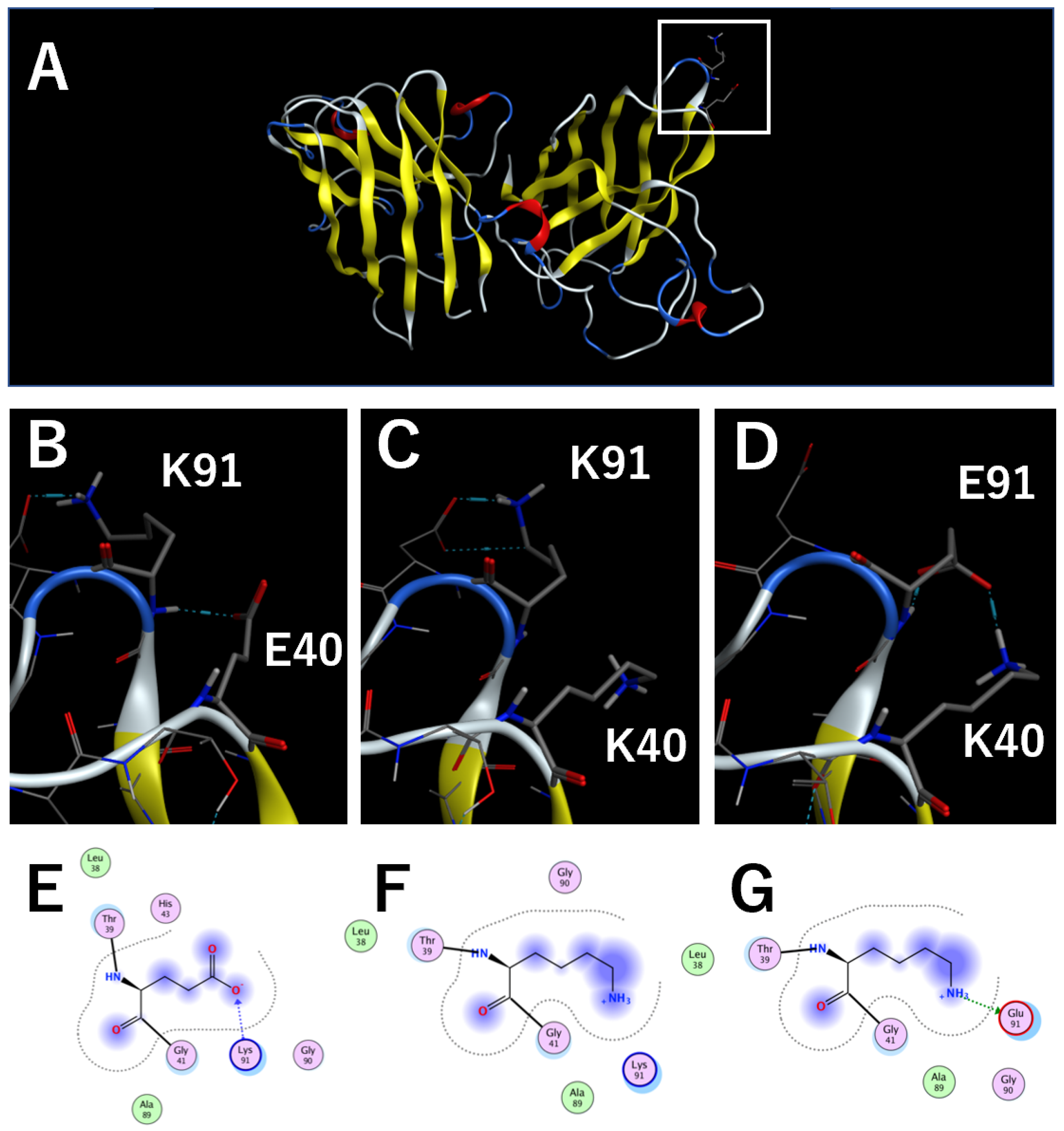
2.1. Structural Analysis of cSOD1 Proteins
2.2. DSF Evaluation of Structural Stability of cSOD1 WT and Mutants
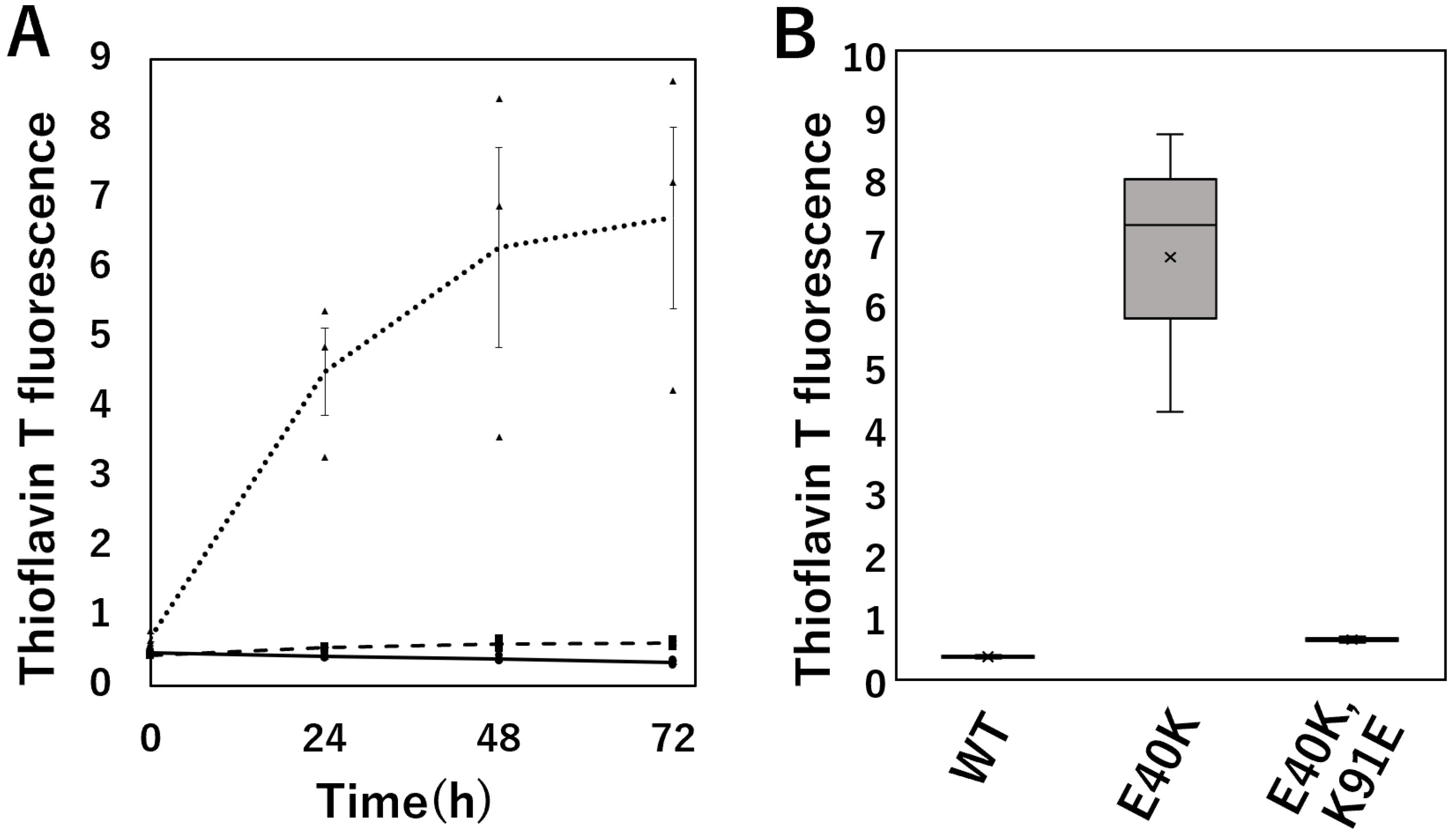
2.3. Thioflavin T (ThT) Fluorescence Assay Evaluation of Amyloid Fibril Formation of cSOD1 WT and Mutants
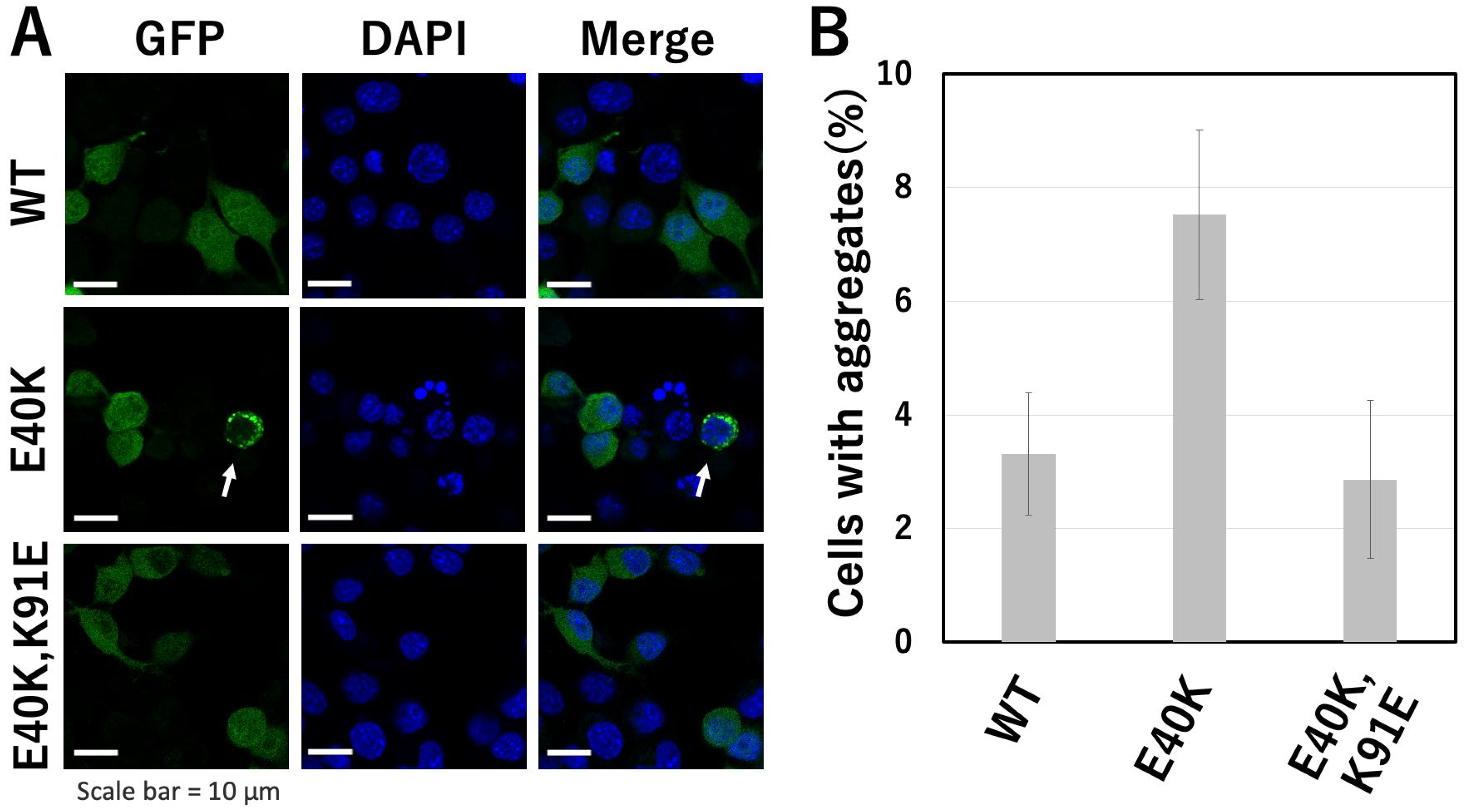
2.4. Evaluation of Aggregate Formation of GFP-Tagged cSOD1(WT) and Mutants in Transfectants
3. Discussion
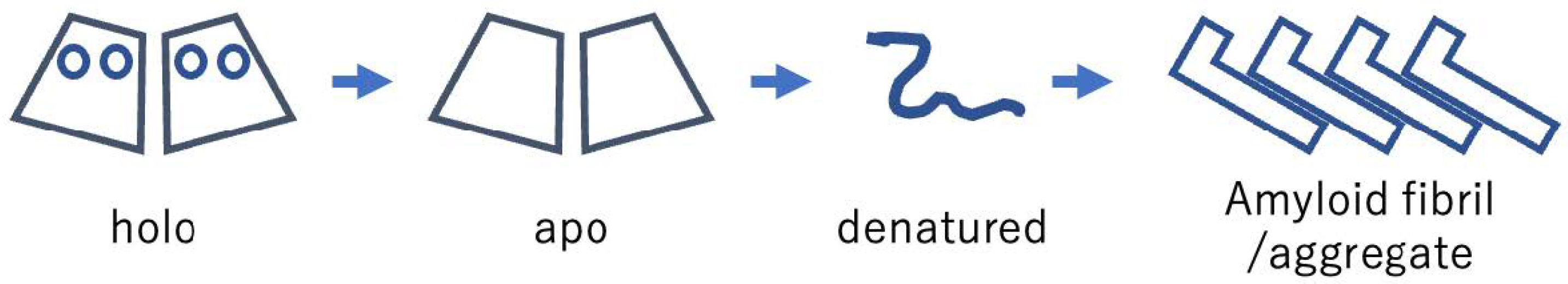
3.1. Denaturation and Aggregation of cSOD1
3.2. The E40K Mutant Is Less Stable and Displays Higher Amyloidogenic Propensity In Vitro and in Cells
3.3. The E40K and K91E Double Mutant Recover Stability and Lessen Amyloidogenic Propensity In Vitro and in Cells
3.4. Importance of the Interaction between Amino Acid Residues E40 and K91 in cSOD1
3.5. Molecular Evolution and Equine SOD1
4. Materials and Methods
4.1. Computational Analysis of cSOD1 Structures
4.2. Preparation of Recombinant Proteins
4.3. DSF
4.4. ThT Fluorescence Assay to Monitor Amyloid Formation
4.5. Aggregate Formation in cSOD1 Transfectants
4.6. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiernan, M.C.; Vucic, S.; Cheah, B.C.; Turner, M.R.; Eisen, A.; Hardiman, O.; Burrell, J.R.; Zoing, M.C. Amyotrophic Lateral Sclerosis. Lancet 2011, 377, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.J.; Talbot, K. Transgenics, Toxicity and Therapeutics in Rodent Models of Mutant SOD1-Mediated Familial ALS. Prog. Neurobiol. 2008, 85, 94–134. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.-X.; et al. Mutations in Cu/Zn Superoxide Dismutase Gene Are Associated with Familial Amyotrophic Lateral Sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Awano, T.; Johnson, G.S.; Wade, C.M.; Katz, M.L.; Johnson, G.C.; Taylor, J.F.; Perloski, M.; Biagi, T.; Baranowska, I.; Long, S.; et al. Genome-Wide Association Analysis Reveals a SOD1 Mutation in Canine Degenerative Myelopathy That Resembles Amyotrophic Lateral Sclerosis. Proc. Natl. Acad. Sci. USA 2009, 106, 2794–2799. [Google Scholar] [CrossRef] [PubMed]
- Nakamae, S.; Kobatake, Y.; Suzuki, R.; Tsukui, T.; Kato, S.; Yamato, O.; Sakai, H.; Urushitani, M.; Maeda, S.; Kamishina, H. Accumulation and Aggregate Formation of Mutant Superoxide Dismutase 1 in Canine Degenerative Myelopathy. Neuroscience 2015, 303, 229–240. [Google Scholar] [CrossRef]
- Oyake, K.; Kobatake, Y.; Shibata, S.; Sakai, H.; Saito, M.; Yamato, O.; Kushida, K.; Maeda, S.; Kamishina, H. Changes in Respiratory Function in Pembroke Welsh Corgi Dogs with Degenerative Myelopathy. J. Vet. Med. Sci. 2016, 78, 1323–1327. [Google Scholar] [CrossRef]
- Coates, J.R.; March, P.A.; Oglesbee, M.; Ruaux, C.G.; Olby, N.J.; Berghaus, R.D.; O’Brien, D.P.; Keating, J.H.; Johnson, G.S.; Williams, D.A. Clinical Characterization of a Familial Degenerative Myelopathy in Pembroke Welsh Corgi Dogs. J. Vet. Intern. Med. 2007, 21, 1323–1331. [Google Scholar] [CrossRef]
- Kobatake, Y.; Nakata, K.; Sakai, H.; Sasaki, J.; Yamato, O.; Takashima, S.; Nishii, N.; Maeda, S.; Islam, M.S.; Kamishina, H. The Long-Term Clinical Course of Canine Degenerative Myelopathy and Therapeutic Potential of Curcumin. Vet. Sci. 2021, 8, 192. [Google Scholar] [CrossRef]
- Nardone, R.; Höller, Y.; Taylor, A.C.; Lochner, P.; Tezzon, F.; Golaszewski, S.; Brigo, F.; Trinka, E. Canine Degenerative Myelopathy: A Model of Human Amyotrophic Lateral Sclerosis. Zoology 2016, 119, 64–73. [Google Scholar] [CrossRef]
- Coates, J.R.; Wininger, F.A. Canine Degenerative Myelopathy. Vet. Clin. North Am. Small Anim. Pract. 2010, 40, 929–950. [Google Scholar] [CrossRef]
- Dion, P.A.; Daoud, H.; Rouleau, G.A. Genetics of Motor Neuron Disorders: New Insights into Pathogenic Mechanisms. Nat. Rev. Genet. 2009, 10, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Hayward, C.; Brock, D.J.; Minns, R.A.; Swingler, R.J. Homozygosity for Asn86Ser Mutation in the CuZn-Superoxide Dismutase Gene Produces a Severe Clinical Phenotype in a Juvenile Onset Case of Familial Amyotrophic Lateral Sclerosis. J. Med. Genet. 1998, 35, 174. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Je, G.; Pacut, P.; Keyhanian, K.; Gao, J.; Ghasemi, M. Gene Therapy in Amyotrophic Lateral Sclerosis. Cells 2022, 11, 2066. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.M.; Al-Chalabi, A. Clinical Genetics of Amyotrophic Lateral Sclerosis: What Do We Really Know? Nat. Rev. Neurol. 2011, 7, 603–615. [Google Scholar] [CrossRef]
- Zeng, R.; Coates, J.R.; Johnson, G.C.; Hansen, L.; Awano, T.; Kolicheski, A.; Ivansson, E.; Perloski, M.; Lindblad-Toh, K.; O’Brien, D.P.; et al. Breed Distribution of SOD1 Alleles Previously Associated with Canine Degenerative Myelopathy. J. Vet. Intern. Med. 2014, 28, 515–521. [Google Scholar] [CrossRef]
- Wininger, F.A.; Zeng, R.; Johnson, G.S.; Katz, M.L.; Johnson, G.C.; Bush, W.W.; Jarboe, J.M.; Coates, J.R. Degenerative Myelopathy in a Bernese Mountain Dog with a Novel SOD1 Missense Mutation. J. Vet. Intern. Med. 2011, 25, 1166–1170. [Google Scholar] [CrossRef]
- McCord, J.M.; Fridovich, I. Superoxide Dismutase: An Enzymic Function for Erythrocuprein (Hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef]
- Fridovich, I. Superoxide Radical and the Bactericidal Action of Phagocytes. New Engl. J. Med. 1974, 290, 624–625. [Google Scholar] [CrossRef]
- Parge, H.E.; Hallewell, R.A.; Tainer, J.A. Atomic Structures of Wild-Type and Thermostable Mutant Recombinant Human Cu, Zn Superoxide Dismutase. Proc. Natl. Acad. Sci. USA 1992, 89, 6109–6113. [Google Scholar] [CrossRef]
- Ogihara, N.L.; Parge, H.E.; Hart, P.J.; Weiss, M.S.; Goto, J.J.; Crane, B.R.; Tsang, J.; Slater, K.; Roe, J.A.; Valentine, J.S.; et al. Unusual Trigonal-Planar Copper Configuration Revealed in the Atomic Structure of Yeast Copper-Zinc Superoxide Dismutase. Biochemistry 1996, 35, 2316–2321. [Google Scholar] [CrossRef]
- Hart, P.J.; Balbirnie, M.M.; Ogihara, N.L.; Nersissian, A.M.; Weiss, M.S.; Valentine, J.S.; Eisenberg, D. A Structure-Based Mechanism for Copper-Zinc Superoxide Dismutase. Biochemistry 1999, 38, 2167–2178. [Google Scholar] [CrossRef] [PubMed]
- Green, S.L.; Tolwani, R.J.; Varma, S.; Quignon, P.; Galibert, F.; Cork, L.C. Structure, Chromosomal Location, and Analysis of the Canine Cu/Zn Superoxide Dismutase (SOD1) Gene. J. Hered. 2002, 93, 119–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kimura, S.; Kamatari, Y.O.; Kuwahara, Y.; Hara, H.; Yamato, O.; Maeda, S.; Kamishina, H.; Honda, R. Canine SOD1 Harboring E40K or T18S Mutations Promotes Protein Aggregation without Reducing the Global Structural Stability. PeerJ 2020, 8, e9512. [Google Scholar] [CrossRef] [PubMed]
- Kamatari, Y.O.; Konno, T.; Kataoka, M.; Akasaka, K. The Methanol-Induced Globular and Expanded Denatured States of Cytochrome c: A Study by CD Fluorescence, NMR and Small-Angle X-Ray Scattering. J. Mol. Biol. 1996, 259, 512–523. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein Folding. Solid Evidence for Molten Globules. Curr. Biol. 1994, 4, 636–640. [Google Scholar] [CrossRef]
- Kuwajima, K. The Molten Globule State as a Clue for Understanding the Folding and Cooperativity of Globular-Protein Structure. Proteins 1989, 6, 87–103. [Google Scholar] [CrossRef]
- Kimura, S.; Kamishina, H.; Hirata, Y.; Furuta, K.; Furukawa, Y.; Yamato, O.; Maeda, S.; Kamatari, Y.O. Novel oxindole compounds inhibit the aggregation of amyloidogenic proteins associated with neurodegenerative diseases. Biochim. Biophys. Acta Gen. Subj. 2022, 1866, 130114. [Google Scholar] [CrossRef]
- Furukawa, Y.; O’Halloran, T.V. Amyotrophic Lateral Sclerosis Mutations Have the Greatest Destabilizing Effect on the Apo- and Reduced Form of SOD1, Leading to Unfolding and Oxidative Aggregation. J. Biol. Chem. 2005, 280, 17266–17274. [Google Scholar] [CrossRef]
- Hart, P.J.; Liu, H.; Pellegrini, M.; Nersissian, A.M.; Gralla, E.B.; Valentine, J.S.; Eisenberg, D. Subunit Asymmetry in the Three-Dimensional Structure of a Human CuZnSOD Mutant Found in Familial Amyotrophic Lateral Sclerosis. Protein Sci. 1998, 7, 545–555. [Google Scholar] [CrossRef]
- Khare, S.D.; Dokholyan, N.V. Common Dynamical Signatures of Familial Amyotrophic Lateral Sclerosis-Associated Structurally Diverse Cu, Zn Superoxide Dismutase Mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 3147–3152. [Google Scholar] [CrossRef]
- Deng, H.X.; Hentati, A.; Tainer, J.A.; Iqbal, Z.; Cayabyab, A.; Hung, W.Y.; Getzoff, E.D.; Hu, P.; Herzfeldt, B.; Roos, R.P. Amyotrophic Lateral Sclerosis and Structural Defects in Cu, Zn Superoxide Dismutase. Science 1993, 261, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.S.; Richardson, D.C. Natural Beta-Sheet Proteins Use Negative Design to Avoid Edge-to-Edge Aggregation. Proc. Natl. Acad. Sci. USA 2002, 99, 2754–2759. [Google Scholar] [CrossRef] [PubMed]
- Shipp, E.L.; Cantini, F.; Bertini, I.; Valentine, J.S.; Banci, L. Dynamic Properties of the G93A Mutant of Copper−Zinc Superoxide Dismutase as Detected by NMR Spectroscopy: Implications for the Pathology of Familial Amyotrophic Lateral Sclerosis. Biochemistry 2003, 42, 1890–1899. [Google Scholar] [CrossRef] [PubMed]
- Borchelt, D.R.; Lee, M.K.; Slunt, H.S.; Guarnieri, M.; Xu, Z.S.; Wong, P.C.; Brown, R.H., Jr.; Price, D.L.; Sisodia, S.S.; Cleveland, D.W. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. USA 1994, 91, 8292–8296. [Google Scholar] [CrossRef]
- Chiti, F.; Calamai, M.; Taddei, N.; Stefani, M.; Ramponi, G.; Dobson, C.M. Studies of the Aggregation of Mutant Proteins in Vitro Provide Insights into the Genetics of Amyloid Diseases. Proc. Natl. Acad. Sci. USA 2002, 99, 16419–16426. [Google Scholar] [CrossRef] [PubMed]
- Calamai, M.; Taddei, N.; Stefani, M.; Ramponi, G.; Chiti, F. Relative Influence of Hydrophobicity and Net Charge in the Aggregation of Two Homologous Proteins. Biochemistry 2003, 42, 15078–15083. [Google Scholar] [CrossRef] [PubMed]
- Draper, A.C.E.; Wilson, Z.; Maile, C.; Faccenda, D.; Campanella, M.; Piercy, R.J. Species-Specific Consequences of an E40K Missense Mutation in Superoxide Dismutase 1 (SOD1). FASEB J 2020, 34, 458–473. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Boca, M.; Calderone, V.; Cantini, F.; Girotto, S.; Vieru, M. Structural and Dynamic Aspects Related to Oligomerization of Apo SOD1 and Its Mutants. Proc. Natl. Acad. Sci. USA 2009, 106, 6980–6985. [Google Scholar] [CrossRef]



| Protein Name | Tm of the Tertiary Structure Monitored by DSF |
| cSOD1(WT) | 50.94 ± 0.95 |
| cSOD1(E40K) | 46.28 ± 0.51 |
| cSOD1(E40K,K91E) | 51.78 ± 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wakayama, K.; Kimura, S.; Kobatake, Y.; Kamishina, H.; Nishii, N.; Takashima, S.; Honda, R.; Kamatari, Y.O. Molecular Mechanisms of Aggregation of Canine SOD1 E40K Amyloidogenic Mutant Protein. Molecules 2023, 28, 156. https://doi.org/10.3390/molecules28010156
Wakayama K, Kimura S, Kobatake Y, Kamishina H, Nishii N, Takashima S, Honda R, Kamatari YO. Molecular Mechanisms of Aggregation of Canine SOD1 E40K Amyloidogenic Mutant Protein. Molecules. 2023; 28(1):156. https://doi.org/10.3390/molecules28010156
Chicago/Turabian StyleWakayama, Kento, Shintaro Kimura, Yui Kobatake, Hiroaki Kamishina, Naohito Nishii, Satoshi Takashima, Ryo Honda, and Yuji O. Kamatari. 2023. "Molecular Mechanisms of Aggregation of Canine SOD1 E40K Amyloidogenic Mutant Protein" Molecules 28, no. 1: 156. https://doi.org/10.3390/molecules28010156
APA StyleWakayama, K., Kimura, S., Kobatake, Y., Kamishina, H., Nishii, N., Takashima, S., Honda, R., & Kamatari, Y. O. (2023). Molecular Mechanisms of Aggregation of Canine SOD1 E40K Amyloidogenic Mutant Protein. Molecules, 28(1), 156. https://doi.org/10.3390/molecules28010156







