The Role of the Nucleotides in the Insertion of the bis-Molybdopterin Guanine Dinucleotide Cofactor into apo-Molybdoenzymes
Abstract
1. Introduction
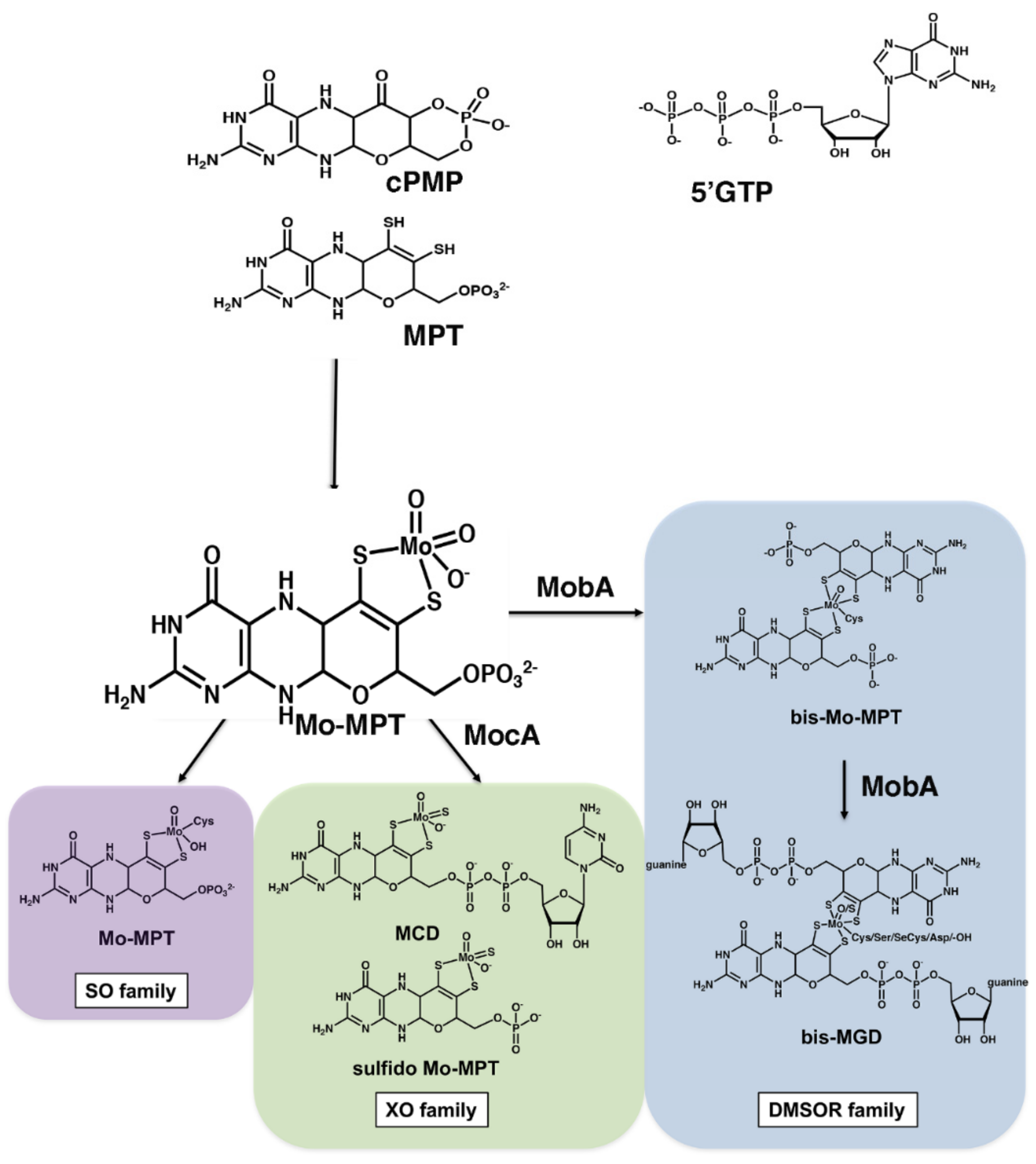
- The synthesis of cyclic pyranopterin monophosphate from 5′GTP (cPMP).
- Conversion of cPMP into MPT by introduction of two sulfur atoms.
- Insertion of molybdate to form Moco (Mo-MPT).
- In most bacteria, Mo-MPT is further modified by covalent attachment of GMP or other nucleotides (like CMP, IMP or AMP) to the terminal phosphate group of MPT via a pyrophosphate link, to form the molybdopterin dinucleotide cofactors [14]. Further, the bis-form containing two MGD or MPT moieties is formed (Figure 1). The roles of these nucleotides in enzyme activity are not clear to date.
2. Results
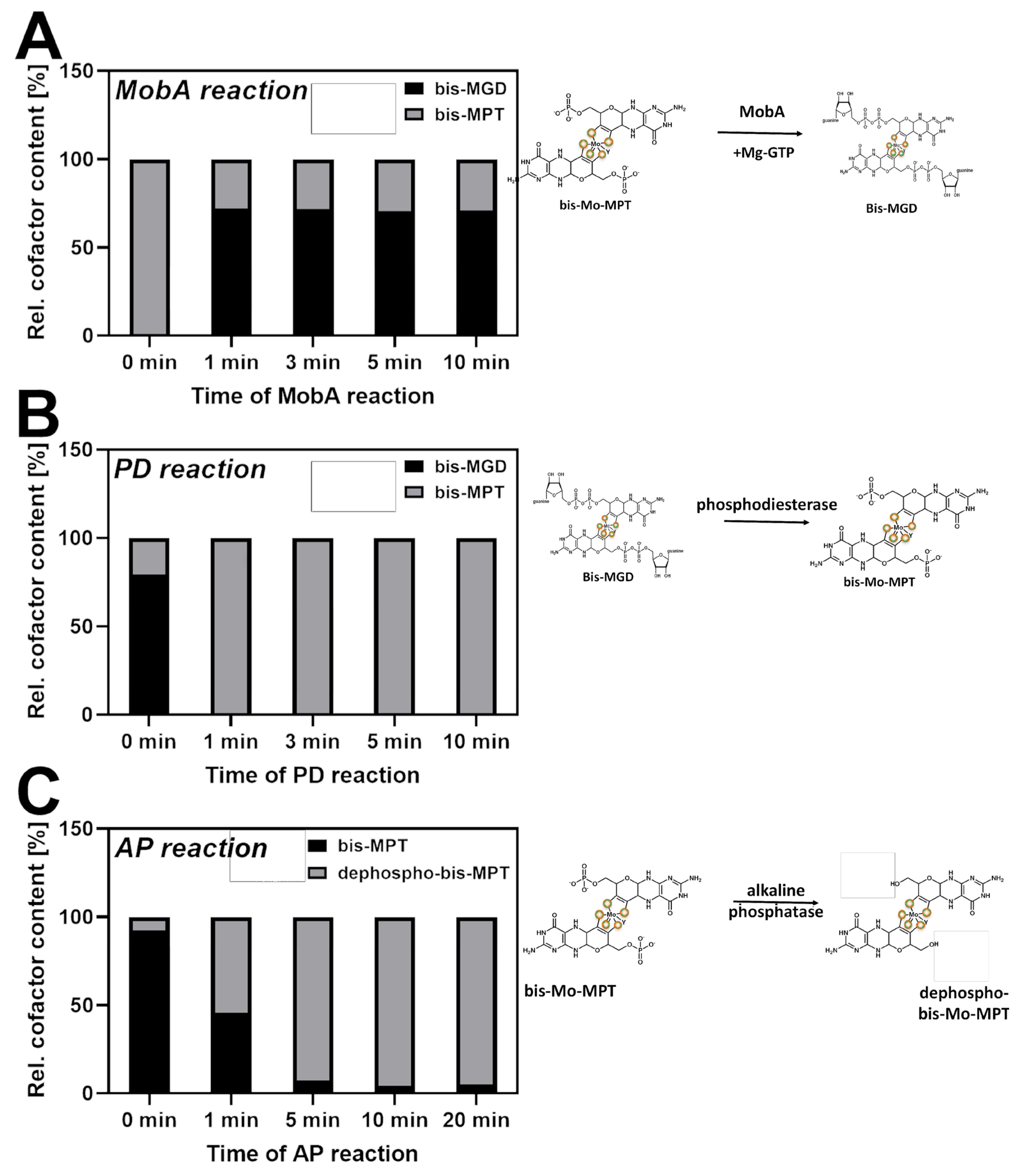
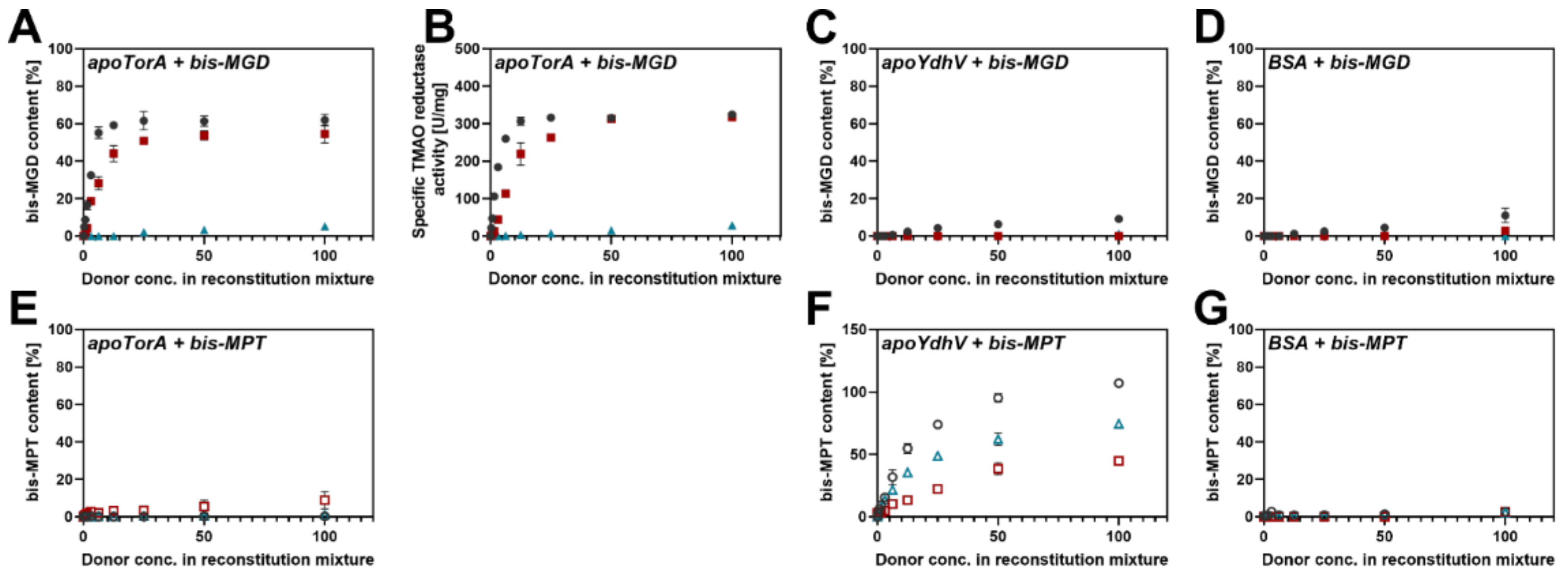
2.1. The Specificity of the Cofactor Insertion into the Respective Target Enzyme: Production of bis-MGD and bis-Mo-MPT
2.2. bis-Mo-MPT Insertion into apo-TorA, apo-YdhV and BSA
2.3. Attempts to Facilitate bis-MPT Insertion into apo-TorA
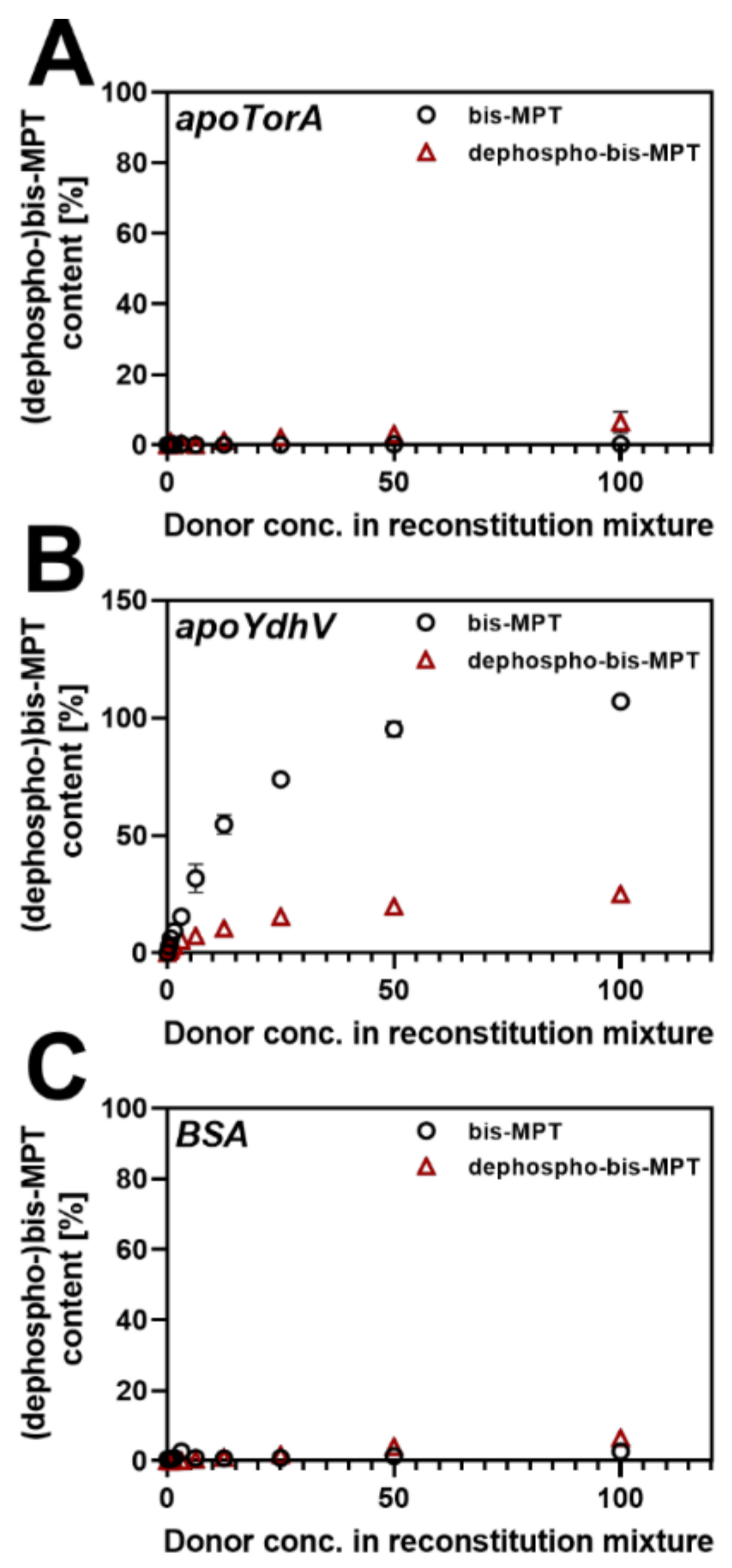
2.4. Production of Dephospho-bis-Mo-MPT and Insertion into apo-TorA and apo-YdhV
3. Materials and Methods
3.1. Expression and Purification Conditions
3.2. Protein Purification
3.3. Cofactor Analysis
3.4. Activity Assay
3.5. Reconstitution Assay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Mendel, R.R. Metabolism of molybdenum. Met. Ions Life Sci. 2013, 12, 503–528. [Google Scholar] [CrossRef] [PubMed]
- Grunden, A.M.; Shanmugam, K.T. Molybdate transport and regulation in bacteria. Arch. Microbiol. 1997, 168, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.V.; Johnson, J.L. The pterin molybdenum cofactors. J. Biol. Chem. 1992, 267, 10199–10202. [Google Scholar] [CrossRef]
- Schwarz, G.; Mendel, R.R.; Ribbe, M.W. Molybdenum cofactors, enzymes and pathways. Nature 2009, 460, 839–847. [Google Scholar] [CrossRef]
- Leimkühler, S.; Iobbi-Nivol, C. Bacterial molybdoenzymes: Old enzymes for new purposes. FEMS Microbiol. Rev. 2016, 40, 1–18. [Google Scholar] [CrossRef]
- Hille, R.; Hall, J.; Basu, P. The Mononuclear Molybdenum Enzymes. Chem. Rev. 2014, 114, 3963–4038. [Google Scholar] [CrossRef]
- Hille, R. The mononuclear molybdenum enzymes. Chem. Rev. 1996, 96, 2757–2816. [Google Scholar] [CrossRef]
- Mendel, R.R.; Kruse, T. Cell biology of molybdenum in plants and humans. Biochim. Biophys Acta 2012, 1823, 1568–1579. [Google Scholar] [CrossRef]
- Rajagopalan, K.V. Biosynthesis of the molybdenum cofactor. In Escherichia coli and Salmonella. Cellular and Molecular Biology; Neidhardt, F.C., Ed.; ASM Press: Washington, DC, USA, 1996; Volume I, pp. 674–679. [Google Scholar]
- Zhang, Y.; Gladyshev, V.N. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 2008, 379, 881–899. [Google Scholar] [CrossRef]
- Zhang, Y.; Gladyshev, V.N. General trends in trace element utilization revealed by comparative genomic analyses of Co, Cu, Mo, Ni, and Se. J. Biol. Chem. 2010, 285, 3393–3405. [Google Scholar] [CrossRef]
- Zhang, Y.; Rump, S.; Gladyshev, V.N. Comparative Genomics and Evolution of Molybdenum Utilization. Coord. Chem. Rev. 2011, 255, 1206–1217. [Google Scholar] [CrossRef]
- Hagen, F. Cellular uptake of molybdenum and tungsten. Coord. Chem. Rev. 2011, 255, 1117–1128. [Google Scholar] [CrossRef]
- Börner, G.; Karrasch, M.; Thauer, R.K. Molybdopterin adenine dinucleotide and molybdopterin hypoxanthine dinucleotide in formylmethanofuran dehydrogenase from Methanobacterium thermoautotrophicum (Marburg). FEBS Lett. 1991, 290, 31–34. [Google Scholar] [CrossRef]
- Palmer, T.; Santini, C.-L.; Iobbi-Nivol, C.; Eaves, D.J.; Boxer, D.H.; Giordano, G. Involvement of the narJ and mob gene products in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol. Microbiol. 1996, 20, 875–884. [Google Scholar] [CrossRef]
- Lake, M.W.; Temple, C.A.; Rajagopalan, K.V.; Schindelin, H. The crystal structure of the Escherichia coli MobA protein provides insight into molybdopterin guanine dinucleotide biosynthesis. J. Biol. Chem. 2000, 275, 40211–40217. [Google Scholar] [CrossRef]
- Reschke, S.; Sigfridsson, K.G.; Kaufmann, P.; Leidel, N.; Horn, S.; Gast, K.; Schulzke, C.; Haumann, M.; Leimkühler, S. Identification of a Bis-molybdopterin Intermediate in Molybdenum Cofactor Biosynthesis in Escherichia coli. J. Biol. Chem. 2013, 288, 29736–29745. [Google Scholar] [CrossRef]
- Stevenson, C.E.; Sargent, F.; Buchanan, G.; Palmer, T.; Lawson, D.M. Crystal structure of the molybdenum cofactor biosynthesis protein MobA from Escherichia coli at near-atomic resolution. Struct. Fold. Des. 2000, 8, 1115–1125. [Google Scholar] [CrossRef]
- Genest, O.; Mejean, V.; Iobbi-Nivol, C. Multiple roles of TorD-like chaperones in the biogenesis of molybdoenzymes. FEMS Microbiol. Lett. 2009, 297, 1–9. [Google Scholar] [CrossRef]
- Genest, O.; Neumann, M.; Seduk, F.; Stocklein, W.; Mejean, V.; Leimkuhler, S.; Iobbi-Nivol, C. Dedicated metallochaperone connects apoenzyme and molybdenum cofactor biosynthesis components. J. Biol. Chem. 2008, 283, 21433–21440. [Google Scholar] [CrossRef]
- Genest, O.; Ilbert, M.; Mejean, V.; Iobbi-Nivol, C. TorD, an essential chaperone for TorA molybdoenzyme maturation at high temperature. J. Biol. Chem. 2005, 280, 15644–15648. [Google Scholar] [CrossRef]
- Genest, O.; Seduk, F.; Theraulaz, L.; Mejean, V.; Iobbi-Nivol, C. Chaperone protection of immature molybdoenzyme during molybdenum cofactor limitation. FEMS Microbiol. Lett. 2006, 265, 51–55. [Google Scholar] [CrossRef][Green Version]
- Nason, A.; Lee, K.-Y.; Pan, S.-S.; Erickson, R.H. Evidence for a molybdenum cofactor common to all molybdenum enzymes based on the in vitro assembly of assimilatory NADPH-nitrate reductase using the Neurospora mutant nit-1. J. Less Common Met. 1974, 36, 449–459. [Google Scholar] [CrossRef]
- Krüger, B.; Meyer, O. Structural elements of bactopterin from Pseudomonas carboxydoflava carbon monoxide dehydrogenase. Biochim. Biophys. Acta 1987, 912, 357–364. [Google Scholar] [CrossRef]
- Krüger, B.; Meyer, O.; Nagel, M.; Andreesen, J.R.; Meincke, M.; Bock, E.; Blümle, S.; Zumft, W.G. Evidence for the presence of bactopterin in the eubacterial molybdoenzymes nicotinic acid dehydrogenase, nitrite oxidoreductase, and respiratory nitrate reductase. FEMS Microbiol. Lett. 1987, 48, 225–227. [Google Scholar] [CrossRef]
- Satoh, T.; Kurihara, F.N. Purification and properties of dimethylsulfoxide reductase containing a molybdenum cofactor from a photonitrifier, Rhodopseudomonas sphaeroides f.s. denitrificans. J. Biochem. 1987, 102, 191–197. [Google Scholar] [CrossRef]
- Johnson, J.L.; Bastian, N.R.; Rajagopalan, K.V. Molybdopterin guanine dinucleotide: A modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc. Natl. Acad. Sci. USA 1990, 87, 3190–3194. [Google Scholar] [CrossRef]
- Rajagopalan, K.V. Novel aspects of the biochemistry of the molybdenum cofactor. In Advances in Enzymology and Related Areas of Molecular Biology; Meister, A., Ed.; John Wiley and Sons: New York, NY, USA, 1991; Volume 64, pp. 215–290. [Google Scholar]
- Leimkühler, S.; Wuebbens, M.M.; Rajagopalan, K.V. The History of the Discovery of the Molybdenum Cofactor and Novel Aspects of its Biosynthesis in Bacteria. Coord. Chem. Rev. 2011, 255, 1129–1144. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Rajagopalan, K.V. Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc. Natl. Acad. Sci. USA 1982, 79, 6856–6860. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.K.; Mukund, S.; Kletzin, A.; Adams, M.W.W.; Rees, D.C. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science 1995, 267, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Boyington, J.C.; Gladyshev, V.N.; Khangulov, S.V.; Stadtman, T.C.; Sun, P.D. Crystal structure of formate dehydrogenase H: Catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 1997, 275, 1305–1308. [Google Scholar] [CrossRef]
- Schneider, F.; Löwe, J.; Huber, R.; Schindelin, H.; Kisker, C.; Knäblein, J. Crystal structure of dimethyl sulfoxide reducatase from Rhodobacter capsulatus at 1.88 Å resolution. J. Mol. Biol. 1996, 263, 53–69. [Google Scholar] [CrossRef]
- Giordano, G.; Santini, C.L.; Saracino, L.; Iobbi, C. Involvement of a protein with molybdenum cofactor in the in vitro activation of nitrate reductase from a chlA mutant of Escherichia coli K12. Biochim. Biophys. Acta 1987, 914, 220–232. [Google Scholar] [CrossRef]
- Kaufmann, P.; Iobbi-Nivol, C.; Leimkuhler, S. Reconstitution of Molybdoenzymes with Bis-Molybdopterin Guanine Dinucleotide Cofactors. Methods Mol. Biol. 2019, 1876, 141–152. [Google Scholar] [CrossRef]
- Kaufmann, P.; Duffus, B.R.; Mitrova, B.; Iobbi-Nivol, C.; Teutloff, C.; Nimtz, M.; Jansch, L.; Wollenberger, U.; Leimkuhler, S. Modulating the Molybdenum Coordination Sphere of Escherichia coli Trimethylamine N-Oxide Reductase. Biochemistry 2018, 57, 1130–1143. [Google Scholar] [CrossRef]
- Reschke, S.; Duffus, B.R.; Schrapers, P.; Mebs, S.; Teutloff, C.; Dau, H.; Haumann, M.; Leimkuhler, S. Identification of YdhV as the First Molybdoenzyme Binding a Bis-Mo-MPT Cofactor in Escherichia coli. Biochemistry 2019, 58, 2228–2242. [Google Scholar] [CrossRef]
- Culka, M.; Huwiler, S.G.; Boll, M.; Ullmann, G.M. Breaking Benzene Aromaticity-Computational Insights into the Mechanism of the Tungsten-Containing Benzoyl-CoA Reductase. J. Am. Chem. Soc. 2017, 139, 14488–14500. [Google Scholar] [CrossRef]
- Wang, H.; Chen, X.; Li, C.; Liu, Y.; Yang, F.; Wang, C. Sequence-Based Prediction of Cysteine Reactivity Using Machine Learning. Biochemistry 2018, 57, 451–460. [Google Scholar] [CrossRef]
- Rothery, R.A.; Stein, B.; Solomonson, M.; Kirk, M.L.; Weiner, J.H. Pyranopterin conformation defines the function of molybdenum and tungsten enzymes. Proc. Natl. Acad. Sci. USA 2012, 109, 14773–14778. [Google Scholar] [CrossRef]
- Stolz, J.F.; Basu, P. Evolution of nitrate reductase: Molecular and structural variations on a common function. Chembiochem 2002, 3, 198–206. [Google Scholar] [CrossRef]
- Iobbi-Nivol, C.; Leimkuhler, S. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta 2013, 1827, 1086–1101. [Google Scholar] [CrossRef]
- Temple, C.A.; Rajagopalan, K.V. Mechanism of assembly of the Bis(Molybdopterin guanine Dinucleotide)Molybdenum cofactor in Rhodobacter sphaeroides dimethyl sulfoxide reductase. J. Biol. Chem. 2000, 275, 40202–40210. [Google Scholar] [CrossRef]
- Leimkühler, S.; Rajagopalan, K.V. In vitro incorporation of nascent molybdenum cofactor into human sulfite oxidase. J. Biol. Chem. 2001, 276, 1837–1844. [Google Scholar] [CrossRef]
- Neumann, M.; Stöcklein, W.; Leimkühler, S. Transfer of the Molybdenum Cofactor Synthesized by Rhodobacter capsulatus MoeA to XdhC and MobA. J. Biol. Chem. 2007, 282, 28493–28500. [Google Scholar] [CrossRef]
- Leimkühler, S.; Klipp, W. The molybdenum cofactor biosynthesis protein MobA from Rhodobacter capsulatus is required for the activity of molybdenum enzymes containing MGD, but not for xanthine dehydrogenase harboring the MPT cofactor. FEMS Microbiol. Lett. 1999, 174, 239–246. [Google Scholar] [CrossRef]
- Hänzelmann, P.; Meyer, O. Effect of molybdate and tungstate on the biosynthesis of CO dehydrogenase and the molybdopterin cytosine-dinucleotide-type of molybdenum cofactor in Hydrogenophaga pseudoflava. Eur. J. Biochem. 1998, 255, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Santini, C.L.; Iobbi-Nivol, C.; Romane, C.; Boxer, D.H.; Giordano, G. Molybdoenzyme biosynthesis in E. coli: In vitro activation of purified nitrate reductase from a chlB mutant. J. Bacteriol. 1992, 174, 7934–7940. [Google Scholar] [CrossRef][Green Version]
- Rothery, R.A.; Magalon, A.; Giordano, G.; Guigliarelli, B.; Blasco, F.; Weiner, J.H. The molybdenum cofactor of Escherichia coli nitrate reductase A (NarGHI). Effect of a mobAB mutation and interactions with [Fe-S] clusters. J. Biol. Chem. 1998, 273, 7462–7469. [Google Scholar] [CrossRef] [PubMed]
- Rothery, R.A.; Simala Grant, J.L.; Johnson, J.L.; Rajagopalan, K.V.; Weiner, J.H. Association of molybdopterin guanine dinucleotide with Escherichia coli dimethyl sulfoxide reductase: Effect of tungstate and a mob mutation. J. Bacteriol. 1995, 177, 2057–2063. [Google Scholar] [CrossRef]
- Czjzek, M.; Dos Santos, J.P.; Pommier, J.; Giordano, G.; Mejean, V.; Haser, R. Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 A resolution. J. Mol. Biol. 1998, 284, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Redelberger, D.; Genest, O.; Arabet, D.; Mejean, V.; Ilbert, M.; Iobbi-Nivol, C. Quality control of a molybdoenzyme by the Lon protease. FEBS Lett. 2013, 587, 3935–3942. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiedemann, K.; Iobbi-Nivol, C.; Leimkühler, S. The Role of the Nucleotides in the Insertion of the bis-Molybdopterin Guanine Dinucleotide Cofactor into apo-Molybdoenzymes. Molecules 2022, 27, 2993. https://doi.org/10.3390/molecules27092993
Tiedemann K, Iobbi-Nivol C, Leimkühler S. The Role of the Nucleotides in the Insertion of the bis-Molybdopterin Guanine Dinucleotide Cofactor into apo-Molybdoenzymes. Molecules. 2022; 27(9):2993. https://doi.org/10.3390/molecules27092993
Chicago/Turabian StyleTiedemann, Kim, Chantal Iobbi-Nivol, and Silke Leimkühler. 2022. "The Role of the Nucleotides in the Insertion of the bis-Molybdopterin Guanine Dinucleotide Cofactor into apo-Molybdoenzymes" Molecules 27, no. 9: 2993. https://doi.org/10.3390/molecules27092993
APA StyleTiedemann, K., Iobbi-Nivol, C., & Leimkühler, S. (2022). The Role of the Nucleotides in the Insertion of the bis-Molybdopterin Guanine Dinucleotide Cofactor into apo-Molybdoenzymes. Molecules, 27(9), 2993. https://doi.org/10.3390/molecules27092993





