Insight of the Functional and Biological Activities of Coconut (Cocos nucifera L.) Protein by Proteomics Analysis and Protein-Based Bioinformatics
Abstract
:1. Introduction
2. Results and Discussion
2.1. Coconut Meat Proteome
2.2. Functional and Bioinformatic Analysis
2.2.1. Molecular Functions
2.2.2. Cellular Components
2.2.3. Biological Processes
2.3. Network Analysis for Globulins and Antioxidant Proteins
2.4. Putative Bioactive Peptides for Globulins and Antioxidant Proteins
3. Materials and Methods
3.1. Materials
3.2. Transcriptome De Novo Assembly
3.3. Protein Extraction
3.4. SDS-Polyacrylamide Gel Electrophoresis
3.5. In-Solution Protein Digestion
3.6. Proteomic Identification
3.7. Bioinformatic Analysis
3.8. Bioactive Peptides Prediction
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yesiltas, B.; Gregersen, S.; Lægsgaard, L.; Brinch, M.L.; Olsen, T.H.; Marcatili, P.; Overgaard, M.T.; Hansen, E.B.; Jacobsen, C.; García-Moreno, P. Emulsifier peptides derived from seaweed, methanotrophic bacteria, and potato proteins identified by quantitative proteomics and bioinformatics. Food Chem. 2021, 362, 130217. [Google Scholar] [PubMed]
- Ghorab, H.; Lammi, C.; Arnoldi, A.; Kabouche, Z.; Aiello, G. Proteomic analysis of sweet algerian apricot kernels (Prunus armeniaca L.) by combinatorial peptide ligand libraries and LC-MS/MS. Food Chem. 2018, 239, 935–945. [Google Scholar] [PubMed]
- Ignacio, I.F.; Miguel, T.S. Research opportunities on the coconut (Cocos nucifera L.) using new technologies. S. Afr. J. Bot. 2021, 141, 414–420. [Google Scholar]
- Kumar, M.; Saini, S.S.; Agrawal, P.K.; Roy, P.; Sircar, D. Nutritional and metabolomics characterization of the coconut water at different nut developmental stages. J. Food Compos. Anal. 2021, 96, 103738. [Google Scholar] [CrossRef]
- Thaiphanit, S.; Anprung, P. Physicochemical and emulsion properties of edible protein concentrate from coconut (Cocos nucifera L.) processing by-products and the influence of heat treatment. Food Hydrocoll. 2016, 52, 756–765. [Google Scholar]
- Mepba, H.D.; Achinewhu, S.C. Effects of processing on protein nutritive quality of coconut Cocos nucifera products. Plant Foods Hum. Nutr. 2003, 58, 15–25. [Google Scholar]
- Garcia, R.N.; Arocena, R.V.; Laurena, A.; Tecson-Mendoza, E.M. 11S and 7S globulins of coconut (Cocos nucifera L.): Purification and characterization. J. Agric. Food Chem. 2005, 53, 1734–1739. [Google Scholar] [CrossRef]
- Balasundaresan, D.; Sugadev, R.; Ponnuswamy, M.N. Purification and crystallization of coconut globulin cocosin from Cocos nucifera. Biochim. Biophys. Acta 2002, 1601, 121–122. [Google Scholar]
- Nneli, R.O.; Woyike, O.A. Antiulcerogenic effects of coconut (Cocos nucifera) extract in rats. Phytother. Res. 2008, 22, 970–972. [Google Scholar]
- Li, S.; Fang, G.; Ping, W.; Lu, J.; Ma, M. Proteome analysis of the almond kernel (Prunus dulcis). J. Sci. Food Agric. 2016, 96, 3351–3357. [Google Scholar] [CrossRef]
- Cabello-Hurtado, F.; Keller, J.; Ley, J.; Sanchez-Lucas, R.; Jorrín-Novo, J.V.; Aïnouche, A. Proteomics for exploiting diversity of lupin seed storage proteins and their use as nutraceuticals for health and welfare. J. Proteom. 2016, 143, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, L.; Zhang, Z.; Dong, M.; Jin, W. iTRAQ-based quantitative proteomic analysis of transgenic and non-transgenic maize seeds. J. Food Compos. Anal. 2020, 92, 103564. [Google Scholar] [CrossRef]
- Juarez-Escobar, J.; Guerrero-Analco, J.A.; Zamora-Briseo, J.A.; Elizalde-Contreras, J.M.; Ruíz-May, E. Tissue-specific proteome characterization of avocado seed during postharvest shelf life. J. Proteom. 2021, 235, 104112. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ahsan, N. Soybean proteomics and its application to functional analysis. J. Proteom. 2009, 72, 325–336. [Google Scholar] [CrossRef]
- Galindo-Luján, R.; Pont, L.; Minic, Z.; Berezovski, M.V.; Benavente, F. Characterization and differentiation of quinoa seed proteomes by label-free mass spectrometry-based shotgun proteomics. Food Chem. 2021, 5, 130250. [Google Scholar] [CrossRef]
- D’Amato, A.; Fasoli, E.; Righetti, P.G. Harry Belafonte and the secret proteome of coconut milk. J. Proteom. 2012, 75, 914–920. [Google Scholar] [CrossRef]
- Huang, J.; Liu, X.Q.; Lan, Q.; Lai, X.; Yang, G.W. Proteomic profile of coconuts. Eur. Food Res. Technol. 2016, 242, 449–455. [Google Scholar] [CrossRef]
- DeMason, D.A.; Sekhar, K.N.C. Electrophoretic characterization and immunological localization of coconut (Cocos nucifera L.) endosperm storage proteins. Bot. Gaz. 1990, 151, 302–313. [Google Scholar] [CrossRef]
- Kwon, K.; Park, K.H.; Rhee, K.C. Fractionation and characterization of proteins from coconut (Cocos nucifera L.). J. Agric. Food Chem. 1996, 44, 1741–1745. [Google Scholar]
- Islas-Flores, I.; Oropeza, C.; Hernández-Sotomayor, S.M.T. Protein phosphorylation during coconut zygotic embryo development. Plant Physiol. 1998, 118, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Islas-Flores, I.; Chan, J.L. Occurrence of phosphorylated proteins and kinase activity in coconut tissues cultured in vitro in a medium that induces somatic embryogenesis. Plant Physiol. Biochem. 2000, 38, 825–836. [Google Scholar] [CrossRef]
- Tan, T.C.; Cheng, L.H.; Bhat, R.; Rusul, G.; Easa, A.M. Composition, physicochemical properties and thermal inactivation kinetics of polyphenol oxidase and peroxidase from coconut (Cocos nucifera) water obtained from immature, mature and overly mature coconut. Food Chem. 2014, 142, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Ejedegba, B.O.; Onyeneke, E.C.; Oviasogie, P.O. Characteristics of lipase isolated from coconut (Cocos nucifera Linn) seed under different nutrient treatments. Afr. J. Biotechnol. 2007, 6, 723–727. [Google Scholar]
- Ialicicco, M.; Viscosi, V.; Arena, S.; Scaloni, A.; Trupiano, D.; Rocco, M.; Chiatante, D.; Scippa, G.S. Lens culinaris Medik. seed proteome: Analysis to identify landrace markers. Plant Sci. 2012, 197, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kotecka-Majchrzak, K.; Sumara, A.; Fornal, E.; Montowska, M. Oilseed proteins -Properties and application as a food ingredient. Trends Food Sci. Technol. 2020, 106, 160–170. [Google Scholar] [CrossRef]
- Tang, C.H. Globular proteins as soft particles for stabilizing emulsions: Concepts and strategies. Food Hydrocoll. 2020, 103, 105664. [Google Scholar] [CrossRef]
- Sá, A.G.A.; Moreno, Y.M.F.; Carciofi, B.A.M. Plant proteins as high-quality nutritional source for human diet. Trends Food Sci. Technol. 2020, 97, 170–184. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Zhang, Y.; Xu, J.; Gao, G. Antioxidant activity of coconut (Cocos nucifera L.) protein fractions. Molecules 2018, 23, 707. [Google Scholar] [CrossRef] [Green Version]
- Osama, S.K.; Kerr, E.D.; Yousif, A.M.; Phung, T.K.; Kelly, A.M.; Fox, G.P.; Schulz, B.L. Proteomics reveals commitment to germination in barley seeds is marked by loss of stress response proteins and mobilisation of nutrient reservoirs. J. Proteom. 2021, 242, 104221. [Google Scholar] [CrossRef]
- Chang, Y.; Yin, C.; Peng, H.; Shi, Y. Differentially proteomic analysis of the hemocytes against Aeromonas hydrophila infection in oriental river prawn Macrobrachium nipponense by iTRAQ approach. Fish Shellfish Immunol. 2020, 104, 324–336. [Google Scholar] [CrossRef]
- Nascimento, J.R.S.; Neto, D.F.; Coutinho, Í.C.; Domont, G.B.; Nogueira, F.C.S.; Camposa, F.A.P. Proteome dynamics of the cotyledonary haustorium and endosperm in the course of germination of Euterpe oleracea seeds. Plant Sci. 2020, 298, 110569. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Darewicz, M.; Mogut, D.; Minkiewicz, P. Elucidation of the role of in silico methodologies in approaches to studying bioactive peptides derived from foods. J. Funct. Foods 2019, 61, 103486. [Google Scholar] [CrossRef]
- Pal, G.K.; Suresh, P.V. Physico-chemical characteristics and fibril-forming capacity of carp swim bladder collagens and exploration of their potential bioactive peptides by in silico approaches. Int. J. Biol. Macromol. 2017, 101, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant protein-derived antioxidant peptides: Isolation, identification, mechanism of action and application in food systems: A review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Chang, S.K.; Ismail, A.; Yanagita, T.; Esa, N.M.; Baharuldin, M. Antioxidant peptides purified and identified from the oil palm (Elaeis guineensis Jacq.) kernel protein hydrolysate. J. Funct. Foods 2015, 14, 63–75. [Google Scholar] [CrossRef]
- Pan, C.; Ma, J.; Tao, F.; Ji, C.; Zhao, Y.; Chen, S.; Yang, X. Novel insight into the antioxidant proteins derived from laver (porphyra haitanensis) by proteomics analysis and protein based bioinformatics. Food Biosci. 2021, 42, 101134. [Google Scholar] [CrossRef]
- Zakaria, W.N.A.W.; Loke, K.K.; Goh, H.H.; Noor, N.M. RNA-seq analysis for plant carnivory gene discovery in Nepenthes × ventrata. Genom. Data 2016, 7, 18–19. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Lukas, K.; Jesse, D.C.; Jason, W.; William, S.N.; Michael, J.M. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar]
- Carrera, M.; Ezquerra-Brauer, J.M.; Aubourg, S.P. Characterization of the jumbo squid (Dosidicus gigas) skin by-product by shotgun proteomics and protein-based bioinformatics. Mar. Drugs 2020, 18, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
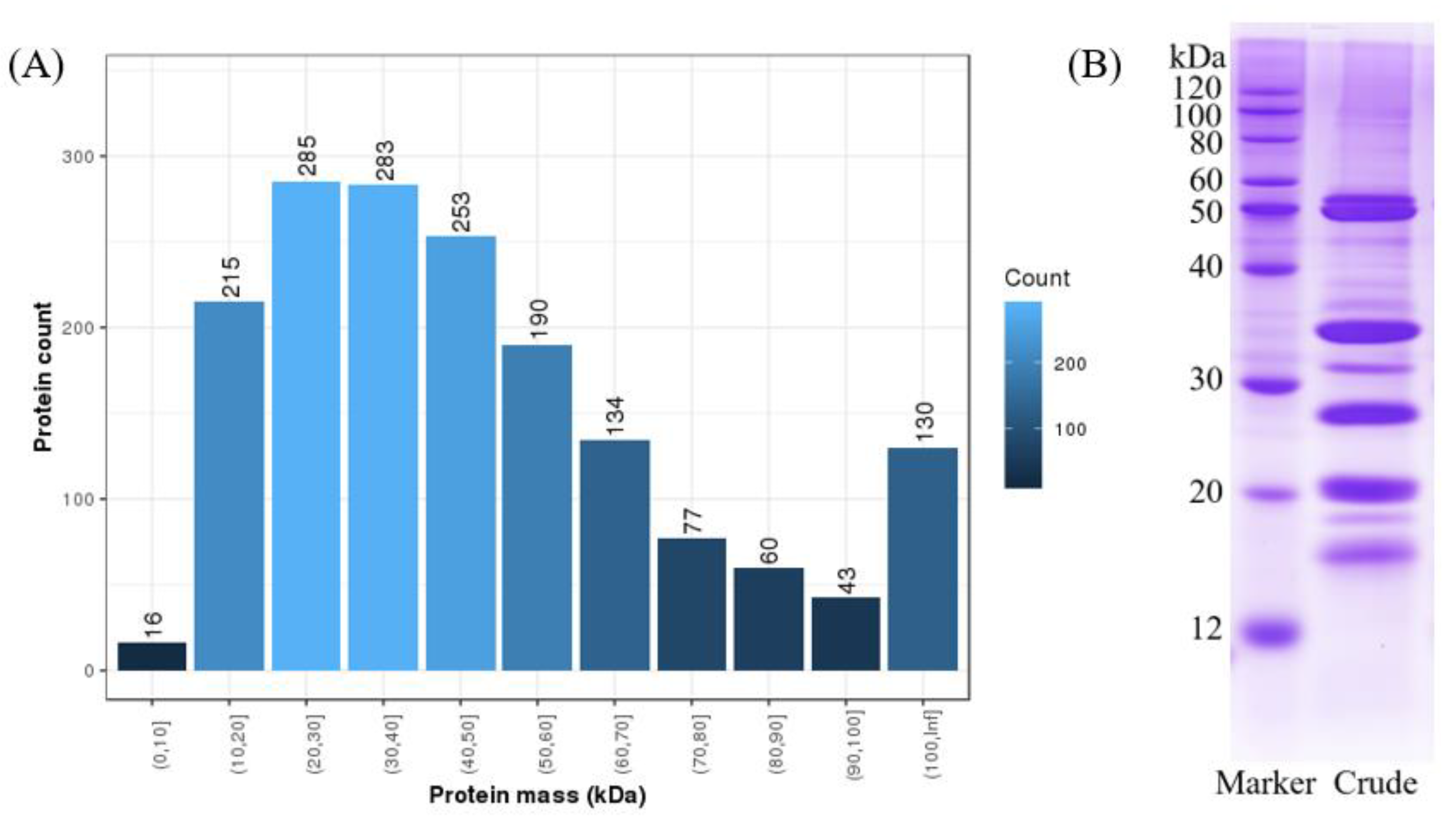
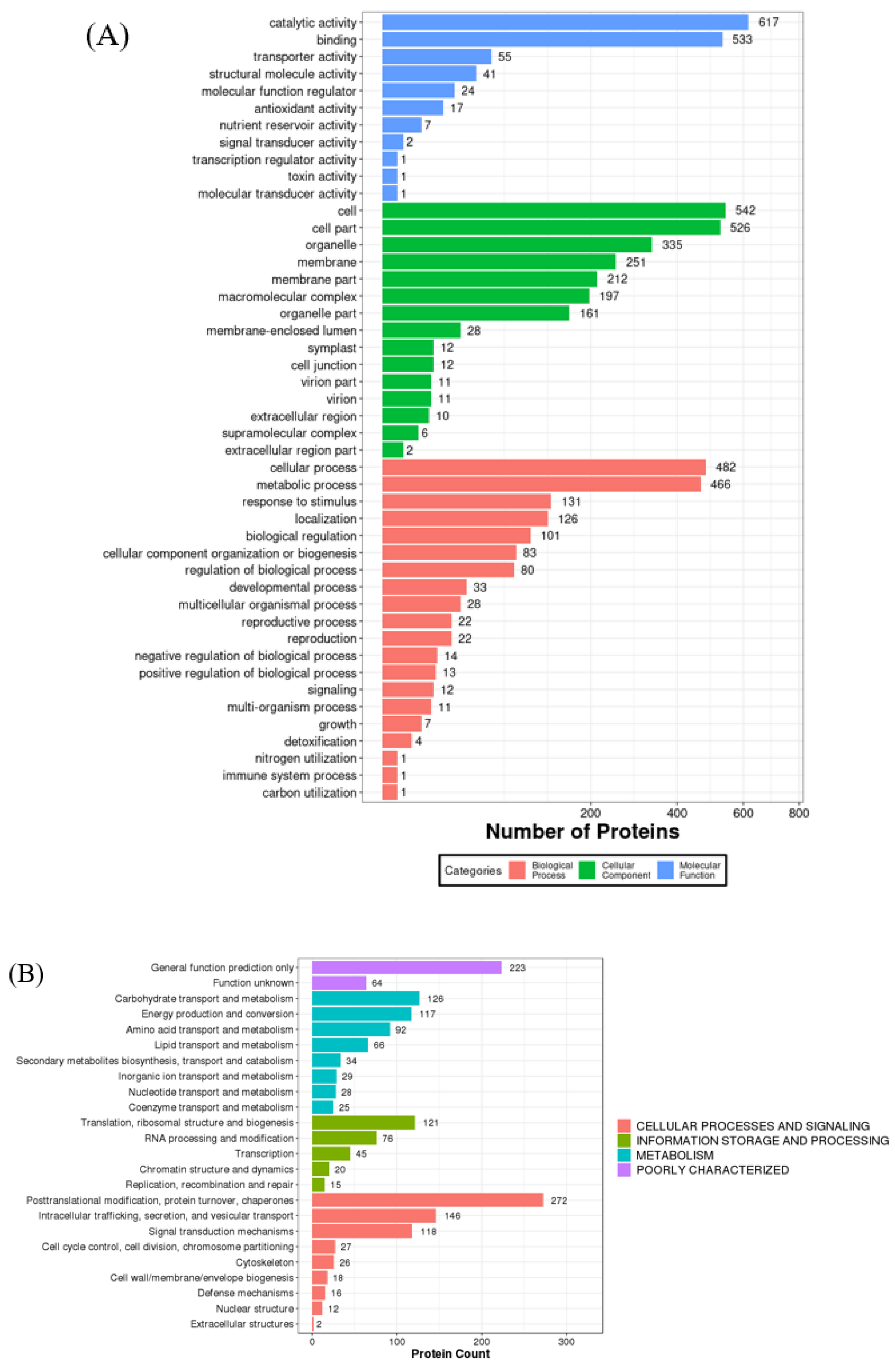

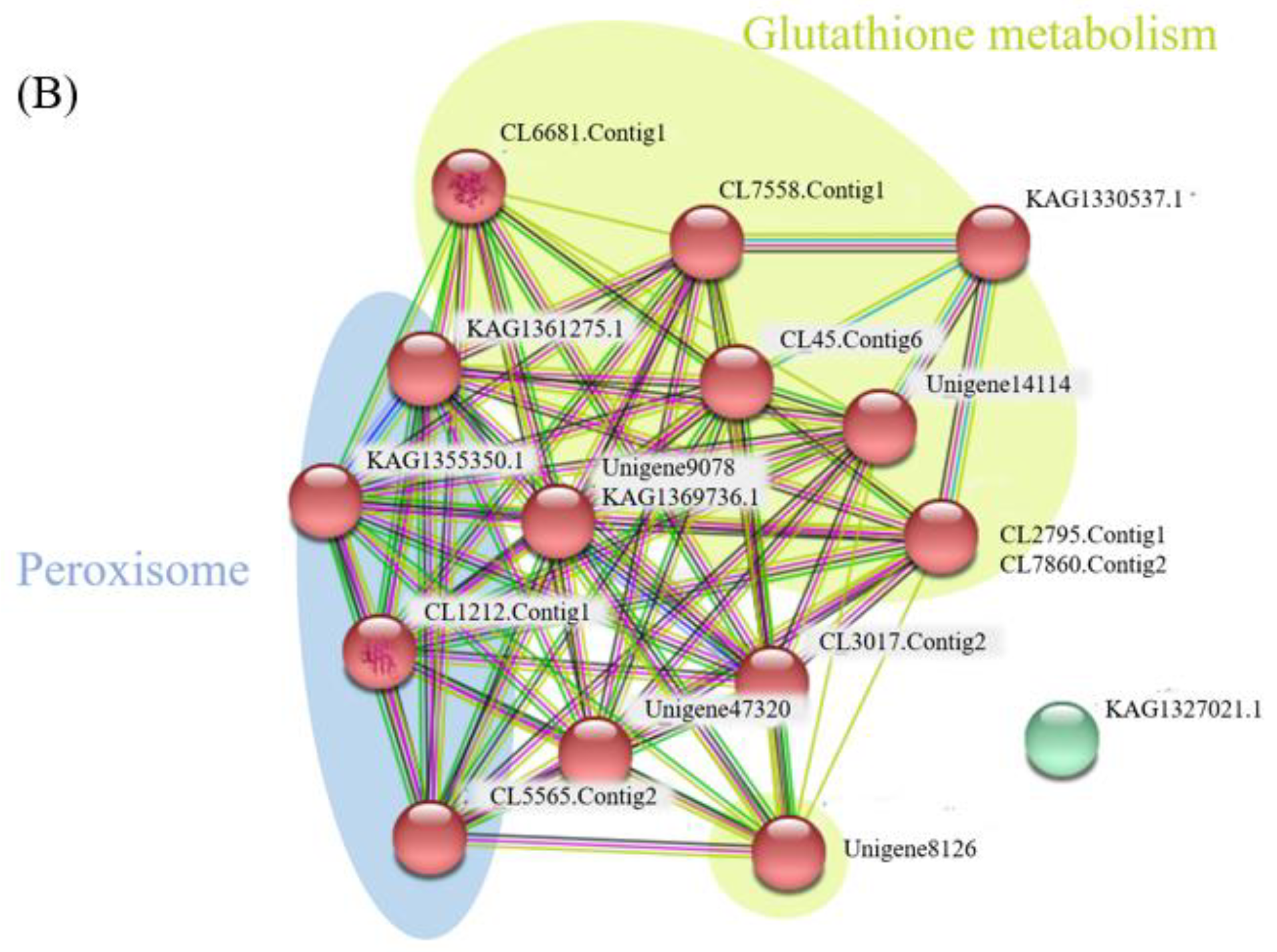
| N | Protein_ID | Protein Mass (kDa) | Description | Uni. Pep. | Cov. (%) |
|---|---|---|---|---|---|
| 1 | ALQ56981.1 | 54.81 | vicilin-like protein (Cocos nucifera) | 2 | 3.47 |
| 2 | CL8941.Contig1_coconut | 90.00 | vicilin-like seed storage protein At2g18540 (Elaeis guineensis) | 5 | 5.12 |
| 3 | KAG1361520.1 | 52.71 | cocosin 1 (Cocos nucifera) | 15 | 28.60 |
| 4 | ASQ40963.1 | 52.62 | cocosin (Cocos nucifera) | 12 | 50.43 |
| 5 | CL3433.Contig1_coconut | 52.48 | vicilin-like seed storage protein At2g28490 (Elaeis guineensis) | 8 | 12.16 |
| 6 | Unigene39216_coconut | 51.48 | cocosin 1 (Elaeis guineensis) | 32 | 55.97 |
| 7 | CL4719.Contig2_coconut | 64.75 | 7S globulin (Elaeis guineensis) | 11 | 41.83 |
| 8 | CL4719.Contig3_coconut | 65.98 | 7S globulin (Elaeis guineensis) | 19 | 34.32 |
| 9 | KAG1368674.1 | 50.38 | putative vicilin-like antimicrobial peptides 2-1 (Cocos nucifera) | 4 | 26.37 |
| N | Protein_ID | Protein Mass (kDa) | Description | Uni. Pep. | Cov. (%) |
|---|---|---|---|---|---|
| 1 | CL7558.Contig1_coconut | 26.66 | phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic (Cocos nucifera) | 2 | 6.58 |
| 2 | CL2795.Contig1_coconut | 28.12 | probable phospholipid hydroperoxide glutathione peroxidase (Elaeis guineensis) | 3 | 12.55 |
| 3 | Unigene9078_coconut | 22.16 | peroxiredoxin-2 (Capsaspora owczarzaki ATCC 30864) | 1 | 5.03 |
| 4 | CL7860.Contig2_coconut | 25.27 | probable glutathione peroxidase 5 (Elaeis guineensis) | 3 | 10.71 |
| 5 | Unigene47320_coconut | 18.82 | peroxiredoxin-2C (Elaeis guineensis) | 1 | 8.05 |
| 6 | CL1212.Contig1_coconut | 19.83 | superoxide dismutase [Cu-Zn], chloroplastic (Elaeis guineensis) | 2 | 14.67 |
| 7 | Unigene8126_coconut | 33.93 | probable L-ascorbate peroxidase 4, peroxisomal (Elaeis guineensis) | 3 | 7.77 |
| 8 | Unigene14114_coconut | 22.19 | probable phospholipid hydroperoxide glutathione peroxidase isoform X1 (Elaeis guineensis) | 2 | 8.67 |
| 9 | KAG1327021.1 | 36.53 | peroxidase 63 (Cocos nucifera) | 3 | 9.52 |
| 10 | KAG1355350.1 | 106.60 | putative Catalase isozyme 1 (Cocos nucifera) | 4 | 7.29 |
| 11 | KAG1361275.1 | 56.38 | catalase isozyme 2 (Cocos nucifera) | 3 | 8.81 |
| 12 | CL6681.Contig1_coconut | 29.17 | L-ascorbate peroxidase, cytosolic (Elaeis guineensis) | 1 | 3.37 |
| 13 | CL5565.Contig2_coconut | 32.61 | superoxide dismutase [Mn], mitochondrial (Elaeis guineensis) | 3 | 11.78 |
| 14 | KAG1330537.1 | 16.58 | microsomal glutathione S-transferase 3 (Cocos nucifera) | 2 | 11.49 |
| 15 | CL45.Contig6_coconut | 41.20 | glutathione reductase, cytosolic (Phoenix dactylifera) | 2 | 7.37 |
| 16 | KAG1369736.1 | 29.80 | 2-Cys peroxiredoxin BAS1, chloroplastic (Cocos nucifera) | 2 | 9.26 |
| 17 | CL3017.Contig2_coconut | 24.28 | 1-Cys peroxiredoxin (Elaeis guineensis) | 8 | 40.83 |
| N | Proteins | Peptides | Modifications | PeptideRanker Score | Toxin Prediction |
|---|---|---|---|---|---|
| 3 | KAG1361520.1 | QSFQQSESEQQGEKGQRRRSRDEHQRI | Gln->pyro-Glu (N-term Q) | 0.870457 | Non-Toxin |
| 7 | CL4719.Contig2_coconut | SEGWSRPF | 0.853573 | Non-Toxin | |
| 9 | KAG1368674.1 | QPVATPGMF | Gln->pyro-Glu (N-term Q) Oxidation (M) | 0.851509 | Non-Toxin |
| 8 | CL4719.Contig3_coconut | HPVATPGMF | Oxidation (M) | 0.840543 | Non-Toxin |
| 7 | CL4719.Contig2_coconut | HPVATPGMF | Oxidation (M) | 0.840543 | Non-Toxin |
| 6 | Unigene39216_coconut | MPGCPTTF | Oxidation (M) | 0.83791 | Non-Toxin |
| 3 | KAG1361520.1 | QFGRSPW | Gln->pyro-Glu (N-term Q) | 0.804836 | Non-Toxin |
| N | Proteins | Peptides | Modifications | PeptideRanker Score | Toxin Prediction |
|---|---|---|---|---|---|
| 5 | CL3433.Contig1_coconut | EMGAMGDRTAVMLLMLLLSSWCLTAVTGNR | Oxidation (M) | 0.998699 | Non-Toxin |
| 6 | Unigene39216_coconut | SSTASLLSFSLCLLLLCHSSLSQRECQLDR | 0.9969 | Non-Toxin | |
| 3 | KAG1361520.1 | AMATSAATLLPFSLCLLLLCRASLAQFGR | Oxidation (M) | 0.994361 | Non-Toxin |
| 3 | KAG1361520.1 | MKAMATSAATLLPFSLCLLLLCR | Met-loss+Acetyl (Protein N-term) Oxidation (M) | 0.99011 | Non-Toxin |
| 7 | CL4719.Contig2_coconut | AFVPFLLLLSILLVSATLTFSVTTEDPKR | 0.98242 | Non-Toxin | |
| 8 | CL4719.Contig3_coconut | AFVPFLLLLSILLVSATLTFSVTTEDPKR | 0.98242 | Non-Toxin | |
| 2 | CL8941.Contig1_coconut | MGKMASTLLATIYLWSLVAINGK | Oxidation (M) | 0.977959 | Non-Toxin |
| 1 | ALQ56981.1 | EMDSDDDDEAEQEEDDVWTWRCLLK | Oxidation (M) | 0.941081 | Non-Toxin |
| 7 | CL4719.Contig2_coconut | MTTKPRAFVPFLLLLSILLVSATLTFSVTTEDPK | Acetyl (Protein N-term) Oxidation (M) | 0.930602 | Non-Toxin |
| 8 | CL4719.Contig3_coconut | MTTKPRAFVPFLLLLSILLVSATLTFSVTTEDPK | Acetyl (Protein N-term) Oxidation (M) | 0.930602 | Non-Toxin |
| 7 | CL4719.Contig2_coconut | MTTKPRAFVPFLLLLSILLVSATLTFSVTTEDPK | Met-loss+Acetyl (Protein N-term) | 0.930602 | Non-Toxin |
| 8 | CL4719.Contig3_coconut | MTTKPRAFVPFLLLLSILLVSATLTFSVTTEDPK | Met-loss+Acetyl (Protein N-term) | 0.930602 | Non-Toxin |
| 9 | KAG1368674.1 | AYIPFLLLLSILFLSATLALSSNEQEDPELKQCK | 0.927218 | Non-Toxin | |
| 6 | Unigene39216_coconut | QLMATSRSSTASLLSFSLCLLLLCHSSLSQR | Oxidation (M) | 0.916637 | Non-Toxin |
| 6 | Unigene39216_coconut | QLMATSRSSTASLLSFSLCLLLLCHSSLSQR | Gln->pyro-Glu (N-term Q) | 0.916637 | Non-Toxin |
| 8 | CL4719.Contig3_coconut | SWPFGESRRPFNLFHK | 0.899487 | Non-Toxin | |
| 1 | ALQ56981.1 | TASAILALLLLSSWSLMVVMAYQGRGMEGR | Oxidation (M) | 0.879123 | Non-Toxin |
| 2 | CL8941.Contig1_coconut | MASTLLATIYLWSLVAINGKDFPSFGPLVTR | Oxidation (M) | 0.878424 | Non-Toxin |
| 7 | CL4719.Contig2_coconut | DGPLELFAF | 0.868349 | Non-Toxin | |
| 2 | CL8941.Contig1_coconut | SHPDPMR | Oxidation (M) | 0.828137 | Non-Toxin |
| 1 | ALQ56981.1 | NRPQFLVGKSSLLHSMR | Oxidation (M) | 0.823569 | Non-Toxin |
| 9 | KAG1368674.1 | QQPFYDEGMRR | Oxidation (M) | 0.823569 | Non-Toxin |
| 9 | KAG1368674.1 | QQPFYDEGMRR | Gln->pyro-Glu (N-term Q) | 0.815333 | Non-Toxin |
| 7 | CL4719.Contig2_coconut | EGDPYFFDK | 0.815321 | Non-Toxin | |
| 9 | KAG1368674.1 | EGDPYFFDK | 0.815321 | Non-Toxin |
| N | Proteins | Peptides | Modifications | PeptideRanker Score | Toxin Prediction |
|---|---|---|---|---|---|
| 15 | CL45.Contig6_coconut | SSEVVGGVGGTCVIRGCVPKKI | 0.988623 | Non-Toxin | |
| 2 | CL2795.Contig1_coconut | APPVCCRF | 0.969325 | Toxin | |
| 10 | KAG1355350.1 | SGGSPFPGL | 0.949596 | Non-Toxin | |
| 1 | CL7558.Contig1_coconut | PSGFPKSPF | 0.893866 | Non-Toxin | |
| 16 | KAG1369736.1 | FHPPMVSF | Oxidation (M) | 0.888879 | Toxin |
| 1 | CL7558.Contig1_coconut | SSKFPSGF | 0.872751 | Non-Toxin | |
| 2 | CL2795.Contig1_coconut | RGVSFPF | 0.866817 | Non-Toxin | |
| 13 | CL5565.Contig2_coconut | QGSGWVW | Gln->pyro-Glu (N-term Q) | 0.84448 | Non-Toxin |
| 17 | CL3017.Contig2_coconut | GISCDDVKCHMEW | Oxidation (M) | 0.820966 | Non-Toxin |
| 10 | KAG1355350.1 | VGNNFPVF | 0.818058 | Non-Toxin | |
| 13 | CL5565.Contig2_coconut | WKVMNW | Oxidation (M) | 0.812069 | Non-Toxin |
| 9 | KAG1327021.1 | MSDWRTRPF | Oxidation (M) | 0.801392 | Non-Toxin |
| N | Proteins | Peptides | Modifications | PeptideRanker Score | Toxin Prediction |
|---|---|---|---|---|---|
| 14 | KAG1330537.1 | LKIGGFNFLALFGLIICTASSGIHLLIR | 0.968709 | Non-Toxin | |
| 6 | CL1212.Contig1_coconut | FVTFCELLICFQWFHFAIGPTTVKVR | 0.960217 | Non-Toxin | |
| 1 | CL7558.Contig1_coconut | FPSGFPKSPFR | 0.954895 | Non-Toxin | |
| 9 | KAG1327021.1 | HLQHSSALLAAVLAVVVALSFPAPSAAKLTPDYYQR | 0.953075 | Non-Toxin | |
| 14 | KAG1330537.1 | IGGFNFLALFGLIICTASSGIHLLIREVL | 0.930888 | Non-Toxin | |
| 6 | CL1212.Contig1_coconut | GGHELSLTTGNAGGRLACGVVGLTPLE | 0.92959 | Non-Toxin | |
| 9 | KAG1327021.1 | FDNMYFK | Oxidation (M) | 0.920339 | Non-Toxin |
| 11 | KAG1361275.1 | EGNWDLLGNNFPVFFIR | 0.902904 | Non-Toxin | |
| 16 | KAG1369736.1 | IFHPPMVSFLR | Oxidation (M) | 0.890783 | Non-Toxin |
| 13 | CL5565.Contig2_coconut | MPVAAFIFHCR | Oxidation (M) | 0.8833 | Non-Toxin |
| 2 | CL2795.Contig1_coconut | GVSFPFPVSSSSAAPPVCCR | 0.882809 | Toxin | |
| 12 | CL6681.Contig1_coconut | NCAPLMLR | Oxidation (M) | 0.877201 | Non-Toxin |
| 4 | CL7860.Contig2_coconut | DLEILAFPCNQFLR | 0.876087 | Non-Toxin | |
| 9 | KAG1327021.1 | LFFHDCFVGGCDASILISSSAFNRAER | 0.86701 | Non-Toxin | |
| 8 | Unigene14114_coconut | MQPSLSWPVVFLGLALLFFFLRNPTPPDK | Met-loss+Acetyl (Protein N-term) | 0.855954 | Non-Toxin |
| 8 | Unigene14114_coconut | MQPSLSWPVVFLGLALLFFFLRNPTPPDK | Acetyl (Protein N-term) Oxidation (M) | 0.855954 | Non-Toxin |
| 1 | CL7558.Contig1_coconut | SSAGGFLGDLIKWNFEK | 0.851297 | Non-Toxin | |
| 7 | Unigene8126_coconut | NCAPIMLR | Oxidation (M) | 0.850796 | Non-Toxin |
| 13 | CL5565.Contig2_coconut | TLTLGLGFGPRGAVAASCPGLGSGLR | 0.849617 | Non-Toxin | |
| 11 | KAG1361275.1 | VHYVKFHWKPTCGVSCLLEEEATVVGGK | 0.840276 | Non-Toxin | |
| 9 | KAG1327021.1 | HLQHSSALLAAVLAVVVALSFPAPSAAK | 0.836338 | Non-Toxin | |
| 4 | CL7860.Contig2_coconut | NKDLEILAFPCNQFLR | 0.833597 | Non-Toxin | |
| 1 | CL7558.Contig1_coconut | SSLFQKNPSFAAKPLR | 0.830042 | Non-Toxin | |
| 2 | CL2795.Contig1_coconut | ACSLLPPMLYSSPLALSR | Acetyl (Protein N-term) Oxidation (M) | 0.826857 | Non-Toxin |
| 10 | KAG1355350.1 | EGNFDLVGNNFPVFFIR | 0.826634 | Non-Toxin | |
| 7 | Unigene8126_coconut | TSTVLAQSAFGVAVAAGVVILSYLYEVSRR | 0.82028 | Non-Toxin | |
| 6 | CL1212.Contig1_coconut | LDKFVTFCELLICFQWFHFAIGPTTVK | 0.818727 | Non-Toxin | |
| 16 | KAG1369736.1 | SFHGLRR | 0.814079 | Non-Toxin | |
| 10 | KAG1355350.1 | FHWRPTCGVKCLLEDEAVIVGGNNHSHATK | 0.809949 | Non-Toxin | |
| 9 | KAG1327021.1 | MARHLQHSSALLAAVLAVVVALSFPAPSAAK | Met-loss+Acetyl (Protein N-term) | 0.80904 | Non-Toxin |
| 9 | KAG1327021.1 | MARHLQHSSALLAAVLAVVVALSFPAPSAAK | Acetyl (Protein N-term) Oxidation (M) | 0.80904 | Non-Toxin |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Pan, C.; Chen, H.; Chen, W.; Chen, W.; Zhang, M.; Zhong, Q. Insight of the Functional and Biological Activities of Coconut (Cocos nucifera L.) Protein by Proteomics Analysis and Protein-Based Bioinformatics. Molecules 2022, 27, 2987. https://doi.org/10.3390/molecules27092987
Ma J, Pan C, Chen H, Chen W, Chen W, Zhang M, Zhong Q. Insight of the Functional and Biological Activities of Coconut (Cocos nucifera L.) Protein by Proteomics Analysis and Protein-Based Bioinformatics. Molecules. 2022; 27(9):2987. https://doi.org/10.3390/molecules27092987
Chicago/Turabian StyleMa, Jingrong, Chuang Pan, Haiming Chen, Weijun Chen, Wenxue Chen, Ming Zhang, and Qiuping Zhong. 2022. "Insight of the Functional and Biological Activities of Coconut (Cocos nucifera L.) Protein by Proteomics Analysis and Protein-Based Bioinformatics" Molecules 27, no. 9: 2987. https://doi.org/10.3390/molecules27092987
APA StyleMa, J., Pan, C., Chen, H., Chen, W., Chen, W., Zhang, M., & Zhong, Q. (2022). Insight of the Functional and Biological Activities of Coconut (Cocos nucifera L.) Protein by Proteomics Analysis and Protein-Based Bioinformatics. Molecules, 27(9), 2987. https://doi.org/10.3390/molecules27092987






