Gastric Cancer Pre-Stage Detection and Early Diagnosis of Gastritis Using Serum Protein Signatures
Abstract
1. Introduction
2. Results
2.1. Patients and Samples
2.2. Biopsy Analysis
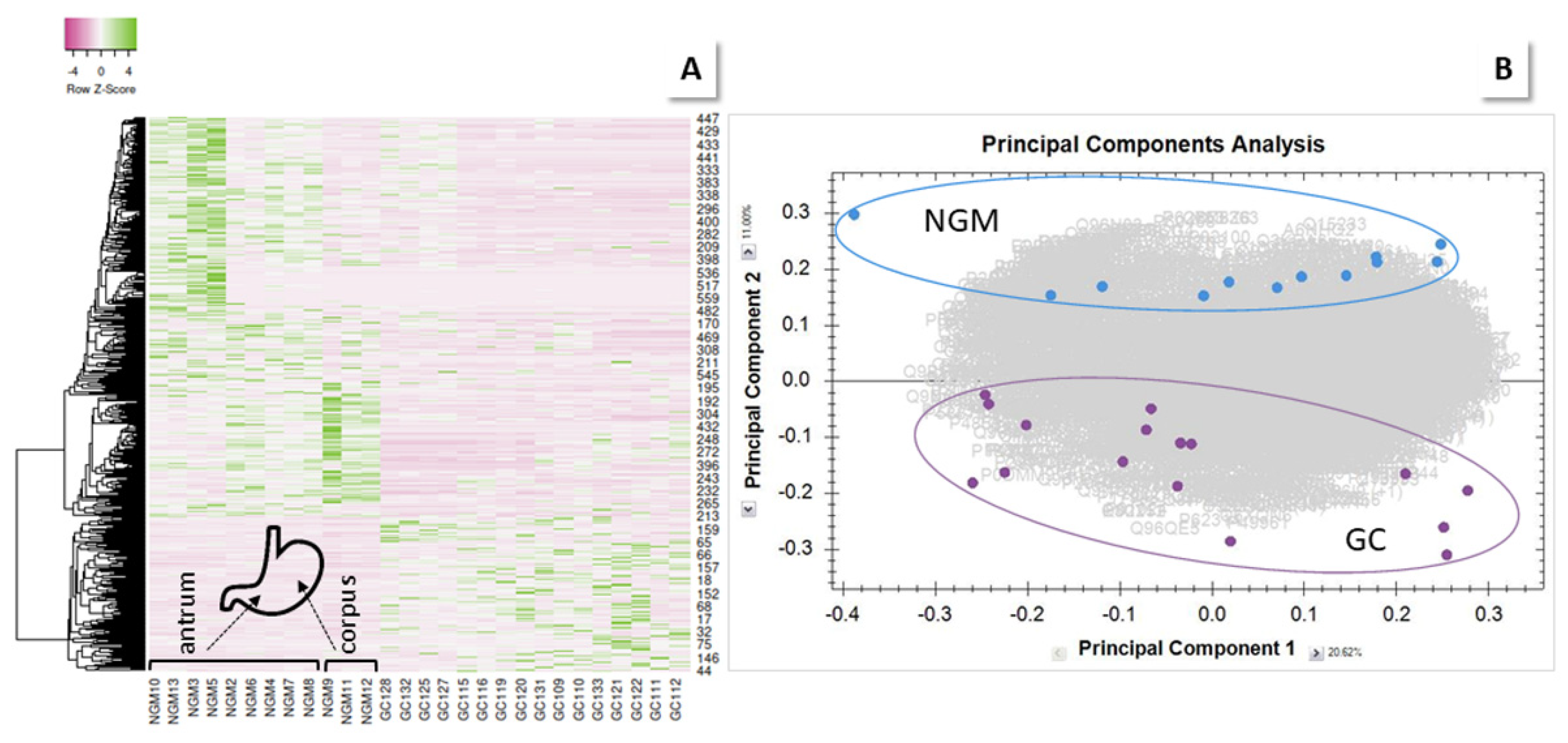
2.2.1. GC vs. NGM
HPpD
GC N vs. D Sites
Gender
1st and Advanced Stage GC
Antrum Samples
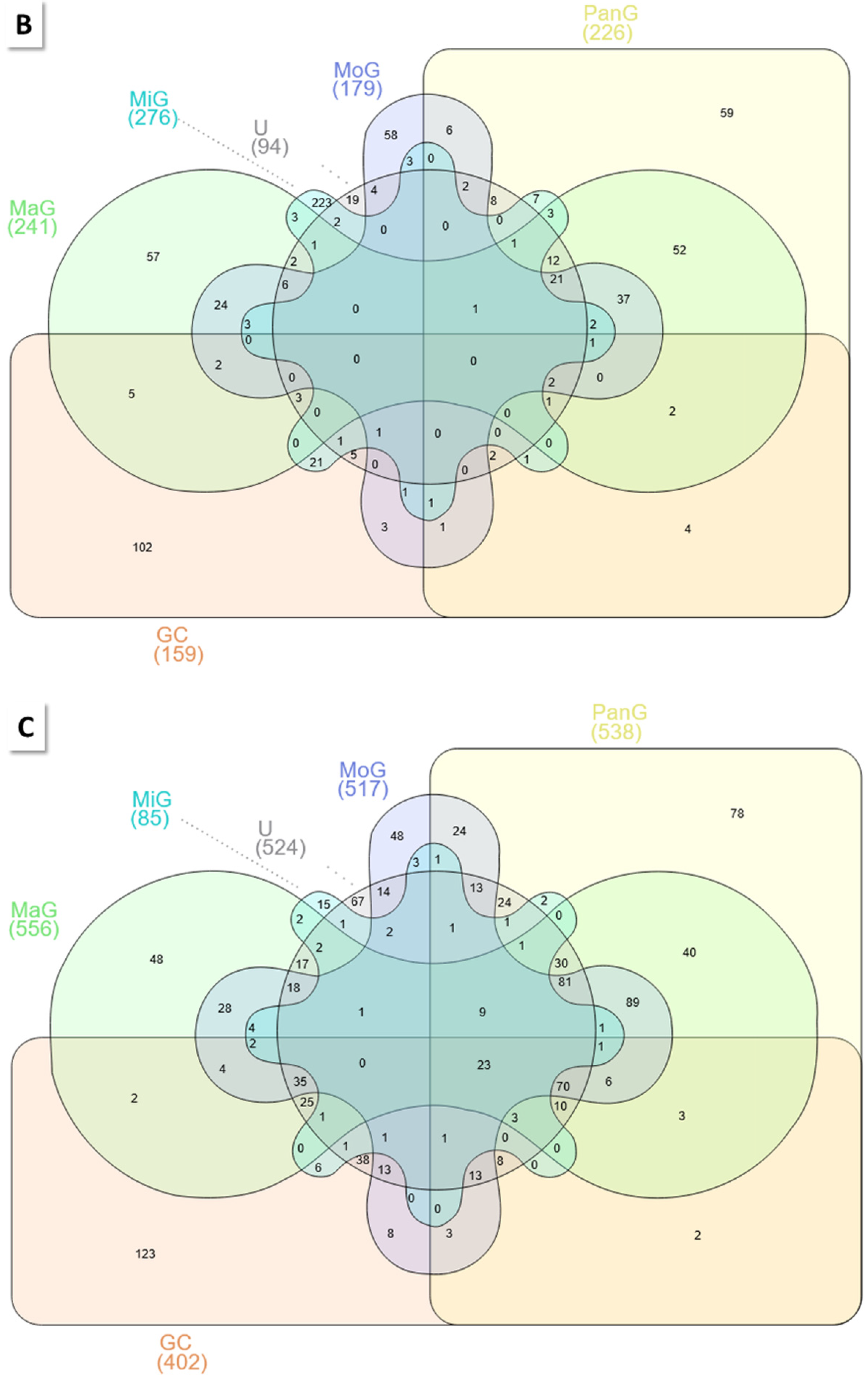
2.2.2. Other Group Comparisons
2.3. Comparative Serum Analysis
2.3.1. MiG
2.3.2. Fibrinogen to Albumin Ratio (FAR)
2.3.3. NGM vs. V
2.3.4. GC vs. V and NGM
2.3.5. GC
2.3.6. Other Comparisons
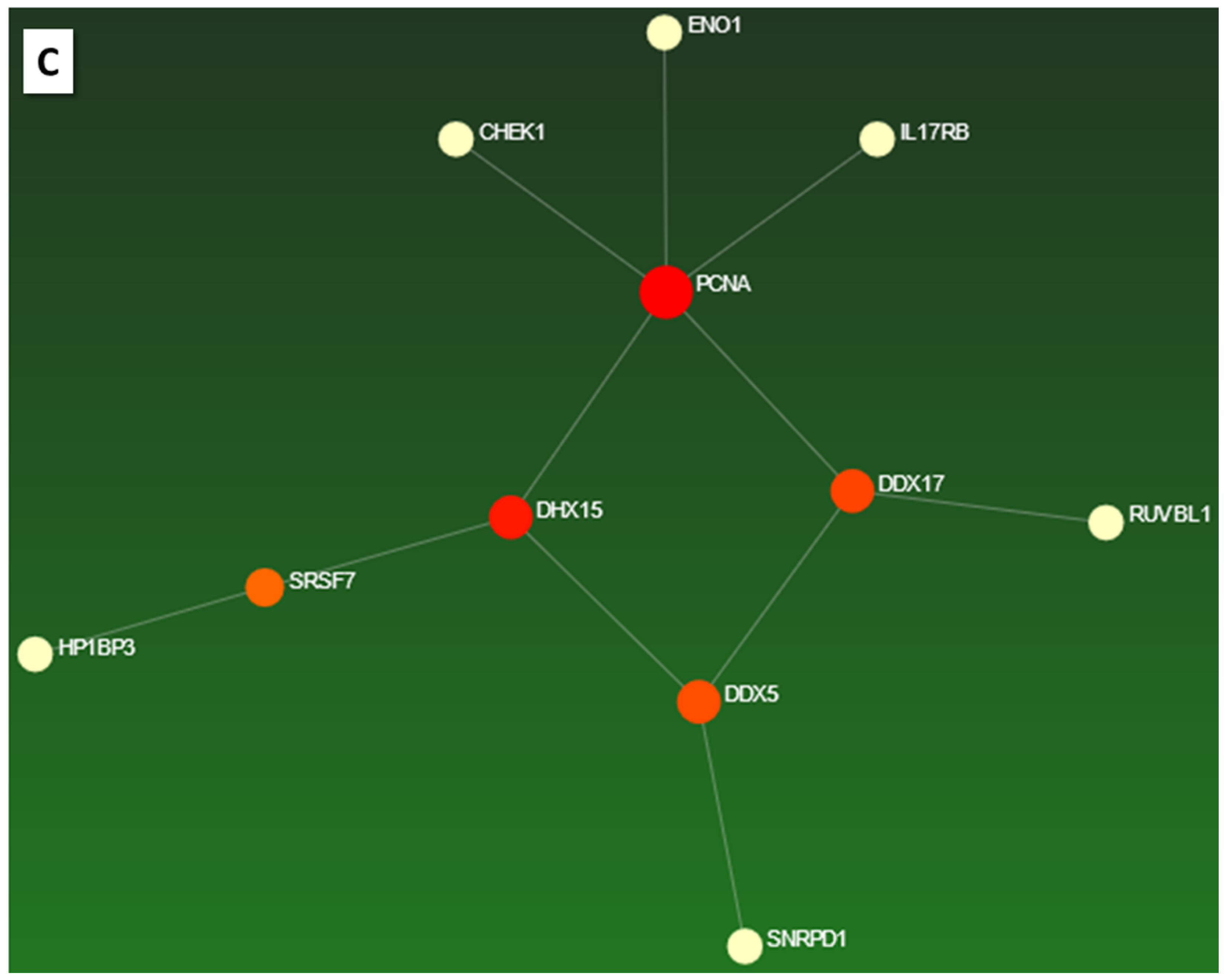
2.3.7. Selection of Marker Proteins
3. Discussion
4. Experimental Section
4.1. Permissions
4.2. Protein Isolation and Analysis
4.3. Data Analysis/Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- American Cancer Society. Available online: www.cancer.org/cancer/stomach-cancer/about/key-statistics.html (accessed on 1 April 2022).
- Wong, M.C.S.; Huang, J.; Chan, P.S.F.; Choi, P.; Lao, X.Q.; Chan, S.M.; Teoh, A.; Liang, P. Global incidence and mortality of gastric cancer 1980–2018. JAMA Netw. Open 2021, 4, e2118457. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, S.; Yalcin, S. Recent advances in systemic treatments for HER-2 positive advanced gastric cancer. OncoTargets Ther. 2021, 14, 4149. [Google Scholar] [CrossRef]
- Peixoto, A.; Silva, M.; Pereira, P.; Macedo, G. Biopsies in gastrointestinal endoscopy: When and how. GE Port. J. Gastroenterol. 2016, 23, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818–2832. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Program. Available online: www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (accessed on 20 September 2021).
- Ni, X.; Tan, Z.; Ding, C.; Zhang, C.; Song, L.; Yang, S.; Liu, M.; Jia, R.; Zhao, C.; Song, L.; et al. A region-resolved mucosa proteome of the human stomach. Nat. Commun. 2019, 10, 39. [Google Scholar] [CrossRef]
- Abyadeh, M.; Meyfour, A.; Gupta, V.; Zabet, M.M.; Fitzhenry, M.J.; Shahbazian, S.; Hosseini Salekdeh, G.; Mirzaei, M. Recent advances of functional proteomics in gastrointestinal cancers—A path towards the identification of candidate diagnostic, prognostic, and therapeutic molecular biomarkers. Int. J. Mol. Sci. 2020, 21, 8532. [Google Scholar] [CrossRef]
- Dhakras, P.; Uboha, N.; Horner, V.; Reinig, E.; Matkowskyj, K.A. Gastrointestinal cancers: Current biomarkers in esophageal and gastric adenocarcinoma. Transl. Gastroenterol. Hepatol. 2020, 5, 55. [Google Scholar] [CrossRef]
- Muzaheed. Helicobacter pylori oncogenicity: Mechanism, prevention, and risk factors. Sci. World J. 2020, 2020, 3018326. [Google Scholar] [CrossRef]
- Aziz, S.; König, S.; Umer, M.; Iqbal, S.; Akhter, T.S.; Ahmad, T.; Zahra, R.; Ibrar, M.; Rasheed, F. Risk factor profiles for gastric cancer prediction with respect to Helicobacter pylori: A study of a tertiary care hospital in Pakistan. BMC Gastroenterol. 2022; submitted. [Google Scholar]
- Forrest, J.H.; Finlayson, N.D.C.; Shearman, D.J.C. Endoscopy in gastrointestinal bleeding. Lancet 1974, 304, 394–397. [Google Scholar] [CrossRef]
- Rasheed, F.; Ahmad, T.; Bilal, R. Frequency of Helicobacter pylori infection using 13C-UBT in asymptomatic individuals of Barakaho, Islamabad, Pakistan. J. Coll. Physicians Surg. Pak. 2011, 21, 379–381. [Google Scholar] [PubMed]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and grading of gastritis: The updated Sydney system. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef] [PubMed]
- Rugge, M.; Correa, P.; Di Mario, F.; El-Omar, E.; Fiocca, R.; Geboes, K.; Vieth, M. OLGA staging for gastritis: A tutorial. Dig. Liver Dis. 2008, 40, 650–658. [Google Scholar] [CrossRef]
- Rasheed, F.; Ahmad, T.; Bilal, R. Prevalence and risk factors of Helicobacter pylori infection among Pakistani population. Pak. J. Med. Sci. 2012, 28, 661–665. [Google Scholar]
- Aziz, S.; König, S.; Noor, H.; Nadeem, M.; Zahra, R.; Rasheed, F. Isolated dextrogastria with eventration of right hemidiaphragm and hiatal hernia in an adult male—A case report. BMS Gastroenterol. 2022, 22, 56. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Aziz, S.; Ahmad, I.; Saadia, A.; Zahra, R.; May, F.E.B.; Pritchard, D.M.; Rasheed, F. Gastric metastasis before diagnosis of primary invasive lobular breast carcinoma: A rare case presentation from Pakistan. Women Health 2021, 61, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Torresano, L.; Nuevo-Tapioles, C.; Santacatterina, F.; Cuezva, J.M. Metabolic reprogramming and disease progression in cancer patients. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165721. [Google Scholar] [CrossRef]
- Almaguel, F.A.; Sanchez, T.W.; Ortiz-Hernandez, G.L.; Casiano, C.A. Alpha-Enolase: Emerging tumor-associated antigen, cancer biomarker, and oncotherapeutic target. Front. Genet. 2021, 11, 614726. [Google Scholar] [CrossRef]
- Heerma van Voss, M.R.; van Diest, P.J.; Raman, V. Targeting RNA helicases in cancer: The translation trap. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 510–520. [Google Scholar] [CrossRef]
- Du, Z.; Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 2018, 17, 58. [Google Scholar] [CrossRef]
- Bastid, J.; Dejou, C.; Docquier, A.; Bonnefoy, N. The emerging role of the IL-17B/IL-17RB pathway in cancer. Front. Immun. 2020, 11, 718. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Downing, S.; Tzanakakis, E.S. Four decades after the discovery of regenerating islet-derived (Reg) proteins: Current understanding and challenges. Front. Cell Dev. Biol. 2019, 7, 235. [Google Scholar] [CrossRef] [PubMed]
- Hirashima, Y.; Yamada, Y.; Matsubara, J.; Takahari, D.; Okita, N.; Takashima, A.; Kato, K.; Hamaguchi, T.; Shirao, K.; Shimada, Y.; et al. Impact of vascular endothelial growth factor receptor 1, 2, and 3 expression on the outcome of patients with gastric cancer. Cancer Sci. 2009, 100, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Eynden, J.V.D.; SahebAli, S.; Horwood, N.; Carmans, S.; Brône, B.; Hellings, N.; Steels, P.; Harvey, R.J.; Rigo, J.-M. Glycine and glycine receptor signalling in non-neuronal cells. Front. Mol. Neurosci. 2009, 2, 9. [Google Scholar] [CrossRef]
- Xiao, Z.Y.; Ru, Y.; Sun, J.T.; Gao, S.G.; Wang, Y.F.; Wang, L.D.; Feng, X.S. Expression of CDX2 and villin in gastric cardiac intestinal metaplasia and the relation with gastric cardiac carcinogenesis. Asian Pac. J. Cancer Prev. 2012, 13, 247–250. [Google Scholar] [CrossRef][Green Version]
- Park, J.M.; Kim, J.W.; Hahm, K.B. HSPA4, the “Evil Chaperone” of the HSP family, delays gastric ulcer healing. Dig. Dis. Sci. 2015, 60, 824–826. [Google Scholar] [CrossRef]
- Bodoor, K.; Jalboush, S.A.; Matalka, I.; Abu-Sheikha, A.; Waq, R.A.; Ebwaini, H.; Abu-Awad, A.; Fayyad, L.; Al-Arjat, J.; Haddad, Y. Heat shock protein association with clinico-pathological characteristics of gastric cancer in Jordan: HSP70 is predictive of poor prognosis. Asian Pac. J. Cancer Prev. 2016, 17, 3929–3937. [Google Scholar]
- Wang, Z. ErbB receptors and cancer. Methods Mol. Biol. 2017, 1652, 3–35. [Google Scholar]
- Ahrens, T.D.; Bang-Christensen, S.R.; Jørgensen, A.M.; Løppke, C.; Spliid, C.B.; Sand, N.T.; Clausen, T.M.; Salanti, A.; Agerbæk, M.Ø. The role of proteoglycans in cancer metastasis and circulating tumor cell analysis. Front. Cell Dev. Biol. 2020, 8, 749. [Google Scholar] [CrossRef]
- Lytovchenko, O.; Kunji, E.R.S. Expression and putative role of mitochondrial transport proteins in cancer. Biochim. Biophys. Acta (BBA)-Bioenerg. 2017, 1858, 641–654. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, D.; Lin, Y.; Chen, H.; Zou, M.; Zhong, S.; Yi, X.; Han, S. Proteasome β-4 subunit contributes to the development of melanoma and is regulated by miR-148b. Tumor Biol. 2017, 39, 1010428317705767. [Google Scholar] [CrossRef] [PubMed]
- Sequeira, I.; Neves, J.F.; Carrero, D.; Peng, Q.; Palasz, N.; Liakath-Ali, K.; Lord, G.M.; Morgan, P.R.; Lombardi, G.; Watt, F.M. Immunomodulatory role of Keratin 76 in oral and gastric cancer. Nat. Commun. 2018, 9, 3437. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.S.; de Rooy, C.; Levinger, I.; Rana, K.; Clarke, M.V.; How, J.M.; Garnham, A.; McLean, C.; Zajac, J.D.; Davey, R.A.; et al. Actin α cardiac muscle 1 gene expression is upregulated in the skeletal muscle of men undergoing androgen deprivation therapy for prostate cancer. J. Steroid Biochem. Mol. Biol. 2017, 174, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.; Zhang, C.; Li, L.; Wang, H.; Li, Q.; Ni, L.; Lin, Y.; Huang, R.; Yang, Z.; Zhang, Y.; et al. Integrative analysis of complement system to prognosis and immune infiltrating in colon cancer and gastric cancer. Front. Oncol. 2021, 10, 553297. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Guo, J.; Wang, Z.; North, B.J.; Tao, K.; Dai, X.; Wei, W. Functional analysis of Cullin 3 E3 ligases in tumorigenesis. Biochimica Biophysica Acta Rev. Cancer 2018, 1869, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Izawa, I.; Li, P.; Inagaki, M. Novel regulation of checkpoint kinase 1: Is checkpoint kinase 1 a good candidate for anti-cancer therapy? Cancer Sci. 2012, 103, 1195–1200. [Google Scholar] [CrossRef]
- Feng, H.; Jin, Z.; Liu, K.; Peng, Y.; Jiang, S.; Wang, C.; Hu, J.; Shen, X.; Qiu, W.; Cheng, X.; et al. Identification and validation of critical alternative splicing events and splicing factors in gastric cancer progression. J. Cell. Mol. Med. 2020, 24, 12667–12680. [Google Scholar] [CrossRef]
- Du, C.; Li, D.-Q.; Li, N.; Chen, L.; Li, S.-S.; Yang, Y.; Hou, M.-X.; Xie, M.-J.; Zheng, Z.-D. DDX5 promotes gastric cancer cell proliferation in vitro and in vivo through mTOR signaling pathway. Sci. Rep. 2017, 7, 42876. [Google Scholar] [CrossRef]
- Xue, Y.; Jia, X.; Li, C.; Zhang, K.; Li, L.; Wu, J.; Yuan, J.; Li, Q. DDX17 promotes hepatocellular carcinoma progression via inhibiting Klf4 transcriptional activity. Cell Death Discov. 2019, 10, 814. [Google Scholar] [CrossRef]
- Fahey, M.; Daly, P.; Bardhordarian, F.; Smyth, C.; Kay, E.; Murray, F.E. Gastric atypia—A cause for concern? Endoscopy 2006, 38, 37. [Google Scholar] [CrossRef]
- Pimentel-Nunes, P.; Libânio, D.; Marcos-Pinto, R.; Areia, M.; Leja, M.; Esposito, G.; Dinis-Ribeiro, M. Management of epithelial precancerous conditions and lesions in the stomach (maps II): European Society of gastrointestinal endoscopy (ESGE), European Helicobacter and microbiota Study Group (EHMSG), European Society of pathology (ESP), and Sociedade Portuguesa de Endoscopia Digestiva (SPED) guideline update 2019. Endoscopy 2019, 51, 365–388. [Google Scholar] [PubMed]
- Kennedy, L.; Sandhu, J.K.; Harper, M.E.; Cuperlovic-Culf, M. Role of glutathione in cancer: From mechanisms to therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef] [PubMed]
- Goroshinskaya, I.A.; Surikova, E.I.; Frantsiyants, E.M.; Nemashkalova, L.A.; Kachesova, P.S.; Chudilova, A.V.; Medvedeva, D.E.; Maslov, A.A.; Malinin, S.A.; Kit, O.I.; et al. Glutathione system in the blood of gastric cancer patients with various tumor histotypes and prevalence of the disease. J. Clin. Oncol. 2020, 38 (Suppl. 15), e16534. [Google Scholar] [CrossRef]
- Kwon, C.H.; Moon, H.J.; Park, H.J.; Choi, J.H.; Park, D.Y. S100A8 and S100A9 promotes invasion and migration through p38 mitogen-activated protein kinase-dependent NF-κB activation in gastric cancer cells. Mol. Cells 2013, 35, 226–234. [Google Scholar] [CrossRef]
- Cui, M.; Huang, J.; Zhang, S.; Liu, Q.; Liao, Q.; Qiu, X. Immunoglobulin expression in cancer cells and its critical roles in tumorigenesis. Front. Immunol. 2021, 12, 613530. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.Y.; Li, L.X.; Zhou, R.C.; Sikong, Y.; Gu, X.; Jin, B.Y.; Li, B.; Li, Y.Q.; Zuo, X.L. S100A10 accelerates aerobic glycolysis and malignant growth by activating mTOR-signaling pathway in gastric cancer. Front. Cell Dev. Biol. 2020, 26. [Google Scholar] [CrossRef]
- Sapio, L.; Di Maiolo, F.; Illiano, M.; Esposito, A.; Chiosi, E.; Spina, A.; Naviglio, S. Targeting protein kinase A in cancer therapy: An update. EXCLI J. 2014, 13, 843–855. [Google Scholar]
- Han, Z.; Zhang, W.; Ning, W.; Wang, C.; Deng, W.; Li, Z.; Shang, Z.; Shen, X.; Liu, X.; Baba, O.; et al. Model-based analysis uncovers mutations altering autophagy selectivity in human cancer. Nat. Commun 2021, 12, 3258. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Xiao, J.; Zhang, Z.; Li, H.; Wang, Y.; Dong, Q.; Piao, H.; Wang, Q.; Bi, F.; et al. Prognostic value of fibrinogen-to-albumin ratio in patients with gastric cancer receiving first-line chemotherapy. Oncol. Lett. 2020, 20, 10. [Google Scholar] [CrossRef]
- Madisch, A.; Aust, D.; Morgner, A.; Grossmann, D.; Schmelz, R.; Kropp, J.; Ehninger, G.; Baretton, G.; Miehlke, S. Resolution of gastrointestinal protein loss after Helicobacter pylori eradication in a patient with hypertrophic lymphocytic gastritis. Helicobacter 2004, 9, 629–631. [Google Scholar] [CrossRef]
- Meuwissen, S.G.; Ridwan, B.U.; Hasper, H.J.; Innemee, G. Hypertrophic protein-losing gastropathy. A retrospective analysis of 40 cases in The Netherlands. The Dutch Ménétrier Study Group. Scand. J. Gastroenterol. Suppl. 1992, 194, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Furuta, T.; Shirai, N.; Xiao, F.; Takashima, M.; Hanai, H. Effect of Helicobacter pylori infection and its eradication on nutrition. Aliment Pharmacol. Ther. 2002, 16, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Repetto, O.; De Re, V. Coagulation and fibrinolysis in gastric cancer. Ann. N. Y. Acad. Sci. 2017, 1404, 27–48. [Google Scholar] [CrossRef] [PubMed]
- Om Kolsoum, E.; Olfat, H.; Iman, O.; Hesham, S.; Nemin, E.; Mone, H. Helicobacter associated chronic gastritis and thrombotic risk. New Egypt. J. Med. 2006, 35 (Suppl. 6), 34–41. [Google Scholar]
- Todorovic, V.; Sokic-Milutinovic, A.; Drndarevic, N.; Micev, M.; Mitrovic, O.; Nikolic, I.; Wex, T.; Milosavljevic, T.; Malfertheiner, P. Expression of cytokeratins in Helicobacter pylori-associated chronic gastritis of adult patients infected with cagA+ strains: An immunohistochemical study. World J. Gastroenterol. 2006, 12, 1865–1873. [Google Scholar] [CrossRef]
- Berstad, A.E.; Brandtzaeg, P.; Stave, R.; Halstensen, T.S. Epithelium related deposition of activated complement in Helicobacter pylori associated gastritis. Gut 1997, 40, 196–203. [Google Scholar] [CrossRef]
- Craig, V.J.; Arnold, I.; Gerke, C.; Huynh, M.Q.; Wündisch, T.; Neubauer, A.; Renner, C.; Falkow, S.; Müller, A. Gastric MALT lymphoma B cells express polyreactive, somatically mutated immunoglobulins. Blood 2010, 115, 581–591. [Google Scholar] [CrossRef]
- Subbannayya, Y.; Mir, S.A.; Renuse, S.; Manda, S.S.; Pinto, S.M.; Puttamallesh, V.N.; Solanki, H.S.; Manju, H.C.; Syed, N.; Sharma, R.; et al. Identification of differentially expressed serum proteins in gastric adenocarcinoma. J. Proteomics. 2015, 127 Pt A, 80–88. [Google Scholar] [CrossRef]
- Ohbatake, Y.; Fushida, S.; Tsukada, T.; Kinoshita, J.; Oyama, K.; Hayashi, H.; Miyashita, T.; Tajima, H.; Takamura, H.; Ninomiya, I.; et al. Elevated α1-acid glycoprotein in gastric cancer patients inhibits the anticancer effects of paclitaxel, effects restored by co-administration of erythromycin. Clin. Exp. Med. 2016, 16, 585–592. [Google Scholar] [CrossRef]
- Corda, G.; Sala, A. Non-canonical WNT/PCP signalling in cancer: Fzd6 takes centre stage. Oncogenesis 2017, 6, e364. [Google Scholar] [CrossRef]
- Chao, J.; Li, P.; Chao, L. Kallistatin suppresses cancer development by multi-factorial actions. Crit. Rev. Oncol. Hematol. 2017, 113, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Repetto, O.; Maiero, S.; Magris, R.; Miolo, G.; Cozzi, M.R.; Steffan, A.; Canzonieri, V.; Cannizzaro, R.; De Re, V. Quantitative proteomic approach targeted to fibrinogen β chain in tissue gastric carcinoma. Int. J. Mol. Sci. 2018, 19, 759. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lin, Y. Evaluation of ficolin-3 as a potential prognostic serum biomarker in Chinese patients with esophageal cancer. Genet. Test. Mol. Biomark. 2019, 23, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.; Jin, H.; Liu, X.; Siu, J.M.T.; Wong, Y.P.; Ng, E.K.O.; Yu, J.; Leung, W.-K.; Sung, J.J.Y.; Chan, F.K.L. Fibulin 1 is downregulated through promoter hypermethylation in gastric cancer. Br. J. Cancer 2008, 99, 2083–2087. [Google Scholar] [CrossRef]
- Huang, K.; Chen, S.; Xie, R.; Jiang, P.; Yu, C.; Fang, J.; Liu, X.; Yu, F. Identification of three predictors of gastric cancer progression and prognosis. FEBS Open Biol. 2020, 10, 1891–1899. [Google Scholar] [CrossRef]
- Ma, F.; Li, X.; Fang, H.; Jin, Y.; Sun, Q.; Li, X. Prognostic value of ANXA8 in gastric carcinoma. J. Cancer 2020, 11, 3551–3558. [Google Scholar] [CrossRef]
- Hsu, P.I.; Chen, C.H.; Hsieh, C.S.; Chang, W.C.; Lai, K.H.; Lo, G.H.; Hsu, P.N.; Tsay, F.W.; Chen, Y.S.; Hsiao, M.; et al. α1-antitrypsin precursor in gastric juice is a novel biomarker for gastric cancer and ulcer. Clin. Cancer Res. 2007, 1, 876–883. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef]
- Repetto, O.; De Re, V.; Giuffrida, P.; Lenti, M.V.; Magris, R.; Venerito, M.; Steffan, A.; Di Sabatino, A.; Cannizzaro, R. Proteomics signature of autoimmune atrophic gastritis: Towards a link with gastric cancer. Gastric Cancer 2021, 24, 666–679. [Google Scholar] [CrossRef]
- Jiang, B.; Li, S.; Jiang, Z.; Shao, P. Gastric cancer associated genes identified by an integrative analysis of gene expression data. Biomed. Res. Int. 2017, 2017, 7259097. [Google Scholar] [CrossRef]
- Geiger, T.; Cox, J.; Mann, M. Proteomic changes resulting from gene copy number variations in cancer cells. PLoS Genet. 2010, 6, e1001090. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Wu, H.; Qu, K.; Sun, Q.; Li, F.; Shi, C.; Li, Y.; Xiong, X.; Qin, Q.; Yu, T.; et al. Identification of serum proteins AHSG, FGA and APOA-I as diagnostic biomarkers for gastric cancer. Clin. Proteom. 2018, 30, 18. [Google Scholar] [CrossRef] [PubMed]
- König, S. Spectral quality overrides software score—A brief tutorial on the analysis of peptide fragmentation data for mass spectrometry laymen. J. Mass Spectrom. 2021, 56, e4616. [Google Scholar] [PubMed]
- Cem, G.; Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 1999, 340, 448–454. [Google Scholar]
- Distler, U.; Kuharev, J.; Navarro, P.; Tenzer, S. Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Prot. 2016, 11, 795–812. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucl. Acids Res. 2020, 49, D394–D403. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucl. Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Available online: www.newchoicehealth.com/endoscopy/cost (accessed on 10 November 2021).
- Aziz, S.; Zahra, R.; Berg, A.M.; Ackermann, D.; Rasheed, F.; König, S. Albumin aggregation and the re-solubilization of dried serum proteins. Mercator J. Biomol. Anal. 2021, 5, 1–3. [Google Scholar]
- Jiang, Y.; Zheng, X.; Jiao, D.; Chen, P.; Xu, Y.; Wei, H.; Qian, Y. Peptidase inhibitor 15 as a novel blood diagnostic marker for cholangiocarcinoma. EBioMedicine 2019, 40, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Li, X.; Yuan, Y.; Jia, C.; Chen, L.; Gao, Q.; Hou, Y. ITGB1 enhances the proliferation, survival, and motility in gastric cancer cells. Microsc. Microanal. 2021, 27, 1192–1201. [Google Scholar] [CrossRef]
- Murray, G.I.; Taylor, M.C.; Burke, M.D.; Melvin, W.T. Enhanced expression of cytochrome P450 in stomach cancer. Br. J. Cancer 1998, 77, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.A.J.; Wang, M.; Farley, S.; Lee, L.Y.; Lee, L.C.; Sawicki, M.P. Menin promotes the Wnt signaling pathway in pancreatic endocrine cells. Mol. Cancer Res. 2008, 6, 1894–1907. [Google Scholar] [CrossRef]
- Sugihara, T.; Kaul, S.C.; Kato, J.; Reddel, R.R.; Nomura, H.; Wadhwa, R. Pex19p dampens the p19ARF-p53-p21WAF1 tumor suppressor pathway. J. Biol. Chem. 2001, 276, 18649–18652. [Google Scholar] [CrossRef]
- Moh, M.C.; Lee, L.H.; Yang, X.; Shen, S. HEPN1, a novel gene that is frequently down-regulated in hepatocellular carcinoma, suppresses cell growth and induces apoptosis in HepG2 cells. J. Hepatol. 2003, 39, 580–586. [Google Scholar] [CrossRef]
- Tuupanen, S.; Hänninen, U.A.; Kondelin, J.; Nandelstadh, P.v.o.n.; Cajuso, T.; Gylfe, A.E.; Katainen, R.; Tanskanen, T.; Ristolainen, H.; Böhm, J.; et al. Identification of 33 candidate oncogenes by screening for base-specific mutations. Br. J. Cancer 2014, 111, 1657–1662. [Google Scholar] [CrossRef]
- Huang, H.; Fu, J.; Zhang, L.; Xu, J.; Li, D.; Onwuka, J.U.; Zhang, D.; Zhao, L.; Sun, S.; Zhu, L.; et al. Integrative analysis of identifying methylation-driven genes signature predicts prognosis in colorectal carcinoma. Front. Oncol. 2021, 11, 629860. [Google Scholar] [CrossRef]
- Boudhraa, Z.; Carmona, E.; Provencher, D.; Mes-Masson, A.M. Ran GTPase: A Key Player in Tumor Progression and Metastasis. Front. Cell Dev. Biol. 2020, 8, 345. [Google Scholar] [CrossRef]
- Gao, Y.; Yuan, C.Y.; Yuan, W. Will targeting PI3K/Akt/mTOR signaling work in hematopoietic malignancies? Stem Cell Investig. 2016, 3, 31. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, R.; Zhong, G.; Chen, X.; Cheng, Y.; Li, W.; Yang, Y. ST6GAL2 downregulation inhibits cell adhesion and invasion and is associated with improved patient survival in breast cancer. OncoTargets 2020, 13, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity. Cell Death Discov. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiang, H.; Lu, Y.; Wu, T.; Ji, G. The role and therapeutic implication of CPTs in fatty acid oxidation and cancers progression. Am. J. Cancer Res. 2021, 11, 2477–2494. [Google Scholar] [PubMed]
- Available online: www.proteinatlas.org/ENSG00000164172-MOCS2/pathology/renal+cancer. (accessed on 26 October 2021).
- Bastide, A.; David, A. The ribosome, (slow) beating heart of cancer (stem) cell. Oncogenesis 2018, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, F.; Lisanti, M.P. Mitochondrial biomarkers predict tumor progression and poor overall survival in gastric cancers: Companion diagnostics for personalized medicine. Oncotarget 2017, 8, 67117–67128. [Google Scholar] [CrossRef]
- Vlachos, A. Acquired ribosomopathies in leukemia and solid tumors. Hematol. Am. Soc. Hematol. Educ. Program 2017, 2017, 716–719. [Google Scholar] [CrossRef]
- Zhao, J.; Peng, H.; Gao, J.; Nong, A.; Hua, H.; Yang, S.; Chen, L.; Wu, X.; Hao, Z.; Wang, J. Current insights into the expression and functions of tumor-derived immunoglobulins. Cell Death Discov. 2021, 7, 148. [Google Scholar] [CrossRef]
- Martorelli, D.; Guidoboni, M.; De Re, V.; Muraro, E.; Turrini, R.; Merlo, A.; Pasini, E.; Caggiari, L.; Romagnoli, L.; Spina, M.; et al. IGKV3 proteins as candidate “off-the-shelf” vaccines for kappa-light chain-restricted B-cell non-Hodgkin lymphomas. Clin. Cancer Res. 2012, 18, 4080–4091. [Google Scholar] [CrossRef]
- Zhu, D.; Lossos, C.; Chapman-Fredricks, J.R.; Matthews, J.M.; Ikpatt, O.F.; Ruiz, P.; Lossos, I.S. Biased use of the IGHV4 family and evidence for antigen selection in Chlamydophila psittaci-negative ocular adnexal extranodal marginal zone lymphomas. PLoS ONE 2011, 6, e29114. [Google Scholar] [CrossRef]
- Tong, W.L.; Tu, Y.N.; Samy, M.D.; Sexton, W.J.; Blanck, G. Identification of immunoglobulin V(D)J recombinations in solid tumor specimen exome files: Evidence for high level B-cell infiltrates in breast cancer. Hum. Vaccin Immunother. 2017, 13, 501–506. [Google Scholar] [CrossRef]
- Available online: www.proteinatlas.org/ENSG00000204438-GPANK1/pathology/stomach+cancer. (accessed on 25 October 2021).
- Aissani, B.; Boehme, A.K.; Wiener, H.W.; Shrestha, S.; Jacobson, L.P.; Kaslow, R.A. SNP screening of central MHC-identified HLA-DMB as a candidate susceptibility gene for HIV-related Kaposi’s sarcoma. Genes Immun. 2014, 15, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.H.; Mao, T.; Tao, Y.; Dong, T.; Tang, X.; Ge, G.; Xu, Z. Pan-cancer analysis identifies ITIH1 as a novel prognostic indicator for hepatocellular carcinoma. Aging 2021, 13, 11096–11119. [Google Scholar] [CrossRef] [PubMed]
- Hamm, A.; Veeck, J.; Bektas, N.; Wild, P.J.; Hartmann, A.; Heindrichs, U.; Kristiansen, G.; Werbowetski-Ogilvie, T.; Maestro, R.D.; Knuechel, R.; et al. Frequent expression loss of inter-α-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: A systematic expression analysis. BMC Cancer 2008, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Choy, T.K.; Wang, C.Y.; Phan, N.N.; Khoa Ta, H.D.; Anuraga, G.; Liu, Y.H.; Wu, Y.F.; Lee, K.H.; Chuang, J.Y.; Kao, T.J. Identification of dipeptidyl peptidase (DPP) family genes in clinical breast cancer patients via an integrated bioinformatics approach. Diagnostics 2021, 11, 1204. [Google Scholar] [CrossRef]
- Chao, J.; Bledsoe, G.; Chao, L. Kallistatin: A novel biomarker for hypertension, organ injury and cancer. Austin Biomark. Diagn. 2015, 2, 1019. [Google Scholar]
- Lee, H.S.; Choi, J.; Son, T.; Lee, E.J.; Kim, J.G.; Ryu, S.H.; Lee, D.; Jang, M.K.; Yu, E.; Chung, Y.H.; et al. A kinase anchoring protein 12 is downregulated in human hepatocellular carcinoma and its deficiency in mice aggravates thioacetamide induced liver injury. Oncol. Lett. 2018, 16, 5907–5915. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, H.; Xu, G.; Wen, H.; Gu, B.; Liu, J.; Mao, S.; Na, R.; Jing, Y.; Ding, Q.; et al. Proteins S100A8 and S100A9 are potential biomarkers for renal cell carcinoma in the early stages: Results from a proteomic study integrated with bioinformatics analysis. Mol. Med. Rep. 2015, 11, 4093–4100. [Google Scholar] [CrossRef]
- Misawa, A.; Takayama, K.I.; Fujimura, T.; Homma, Y.; Suzuki, Y.; Inoue, S. Androgen-induced lncRNA POTEF-AS1 regulates apoptosis-related pathway to facilitate cell survival in prostate cancer cells. Cancer Sci. 2017, 108, 373–379. [Google Scholar] [CrossRef]
- Shen, Z.; Feng, X.; Fang, Y.; Li, Y.; Li, Z.; Zhan, Y.; Lin, M.; Li, G.; Ding, Y.; Deng, H. POTEE drives colorectal cancer development via regulating SPHK1/p65 signaling. Cell Death Discov. 2019, 10, 863. [Google Scholar] [CrossRef]
- Haworth, A.S.; Brackenbury, W.J. Emerging roles for multifunctional ion channel auxiliary subunits in cancer. Cell Calcium 2019, 80, 125–140. [Google Scholar] [CrossRef]
- Iorio, F.; Garcia-Alonso, L.; Brammeld, J.; Martincorena, I.; Wille, D.R.; McDermott, U.; Saez-Rodriguez, J. Pathway-based dissection of the genomic heterogeneity of cancer hallmarks’ acquisition with SLAPenrich. Sci. Rep. 2018, 8, 6713. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.A.; Ghiselli, G. Hinderin, a five-domains protein including coiled-coil motifs that binds to SMC3. BMC Cell Biol. 2005, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Pan, Z.; Zhang, J. CircSMC3 regulates gastric cancer tumorigenesis by targeting miR-4720-3p/TJP1 axis. Cancer Med. 2020, 9, 4299–4309. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Sun, C.F.; Pai, H.J.; Wang, W.K.; Hsieh, C.C.; Kuo, L.M.; Wang, C.S. Preoperative serum C-reactive protein and gastric cancer; clinical-pathological correlation and prognostic significance. Chang Gung Med. J. 2010, 33, 301–312. [Google Scholar]
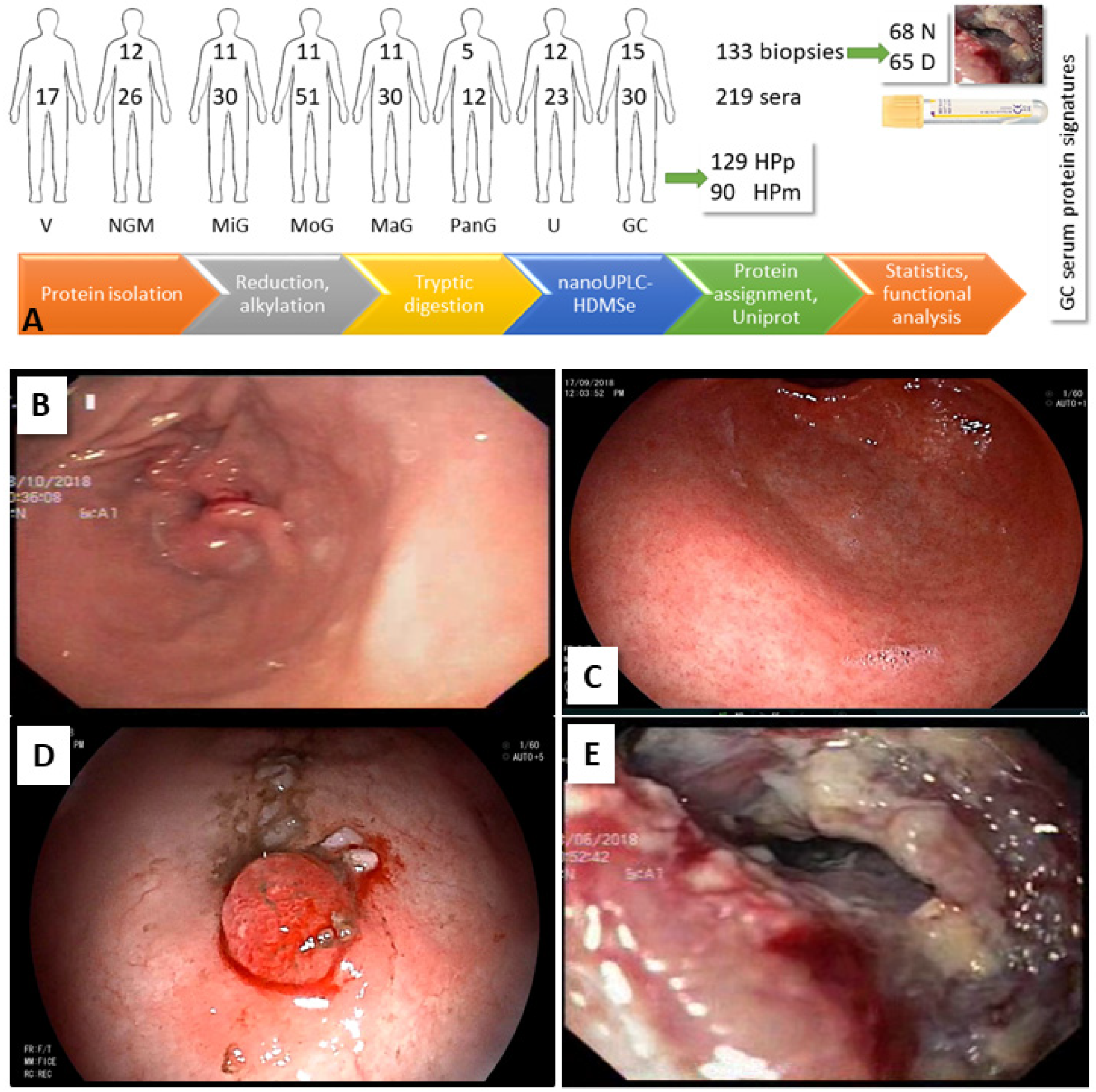

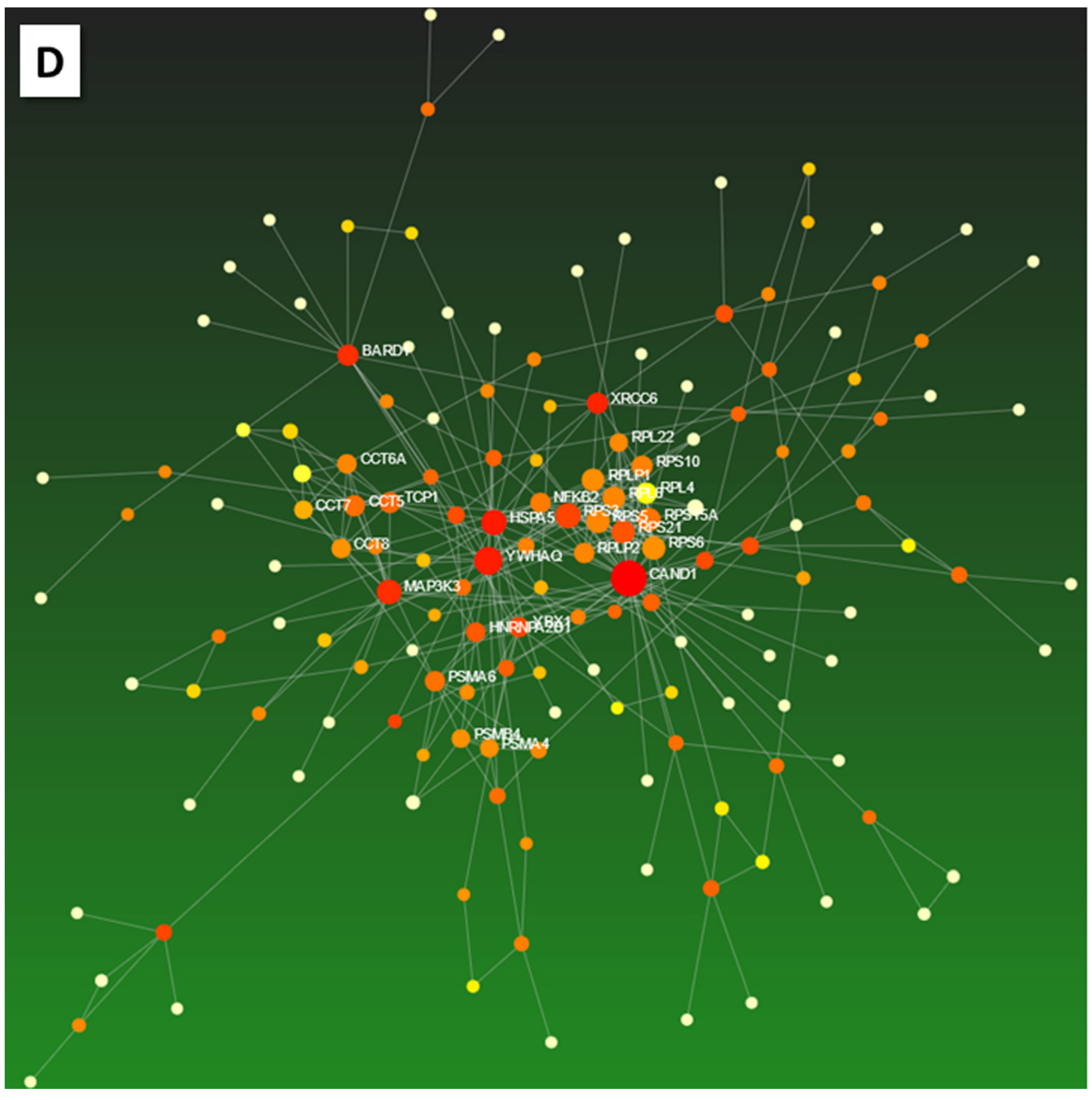
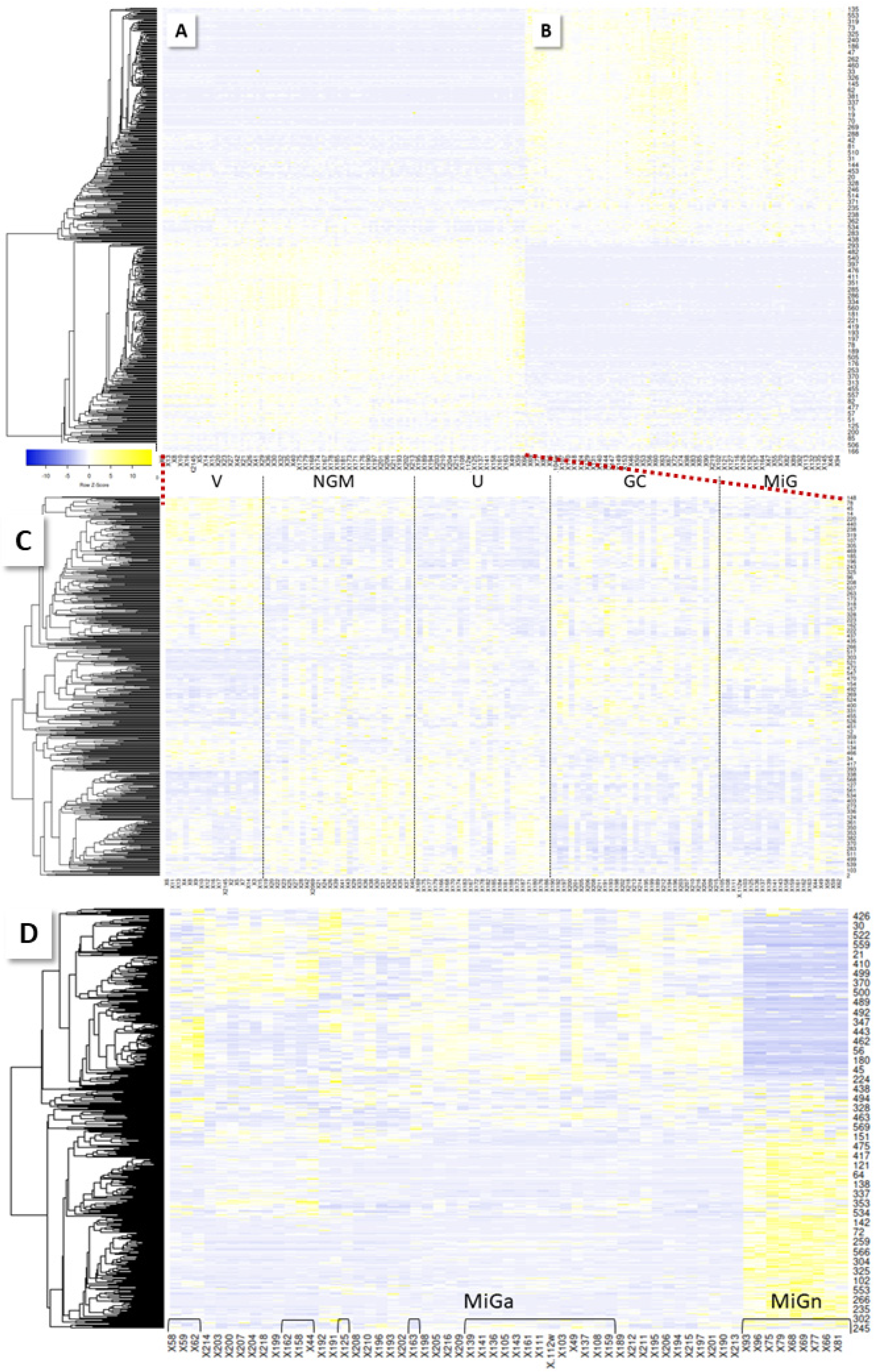
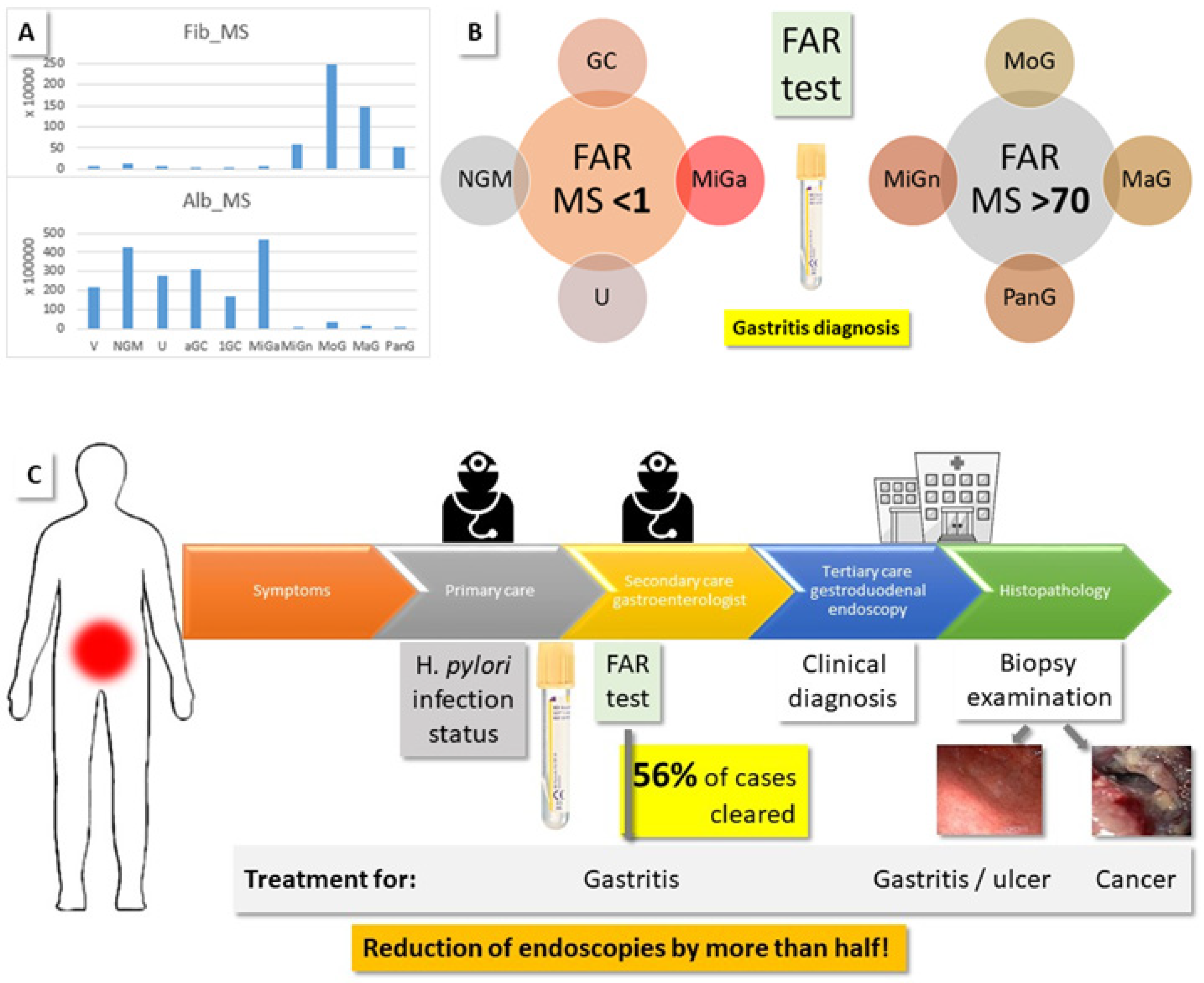
| Biopsies | NGM | MiG | MoG | MaG | PanG | U | GC | Total | % | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | 12 | 11 | 11 | 11 | 5 | 12 | 13 | 75 | |||||
| Female | 4 | 8 | 6 | 3 | 2 | 4 | 5 | 32 | 43 | ||||
| Male | 8 | 3 | 5 | 8 | 3 | 8 | 8 | 43 | 57 | ||||
| Age < 46 | 9 | 7 | 7 | 10 | - | 3 | 6 | 43 | 56 | ||||
| Age > 45 | 3 | 4 | 4 | 1 | 5 | 9 | 7 | 34 | 44 | ||||
| Samples | 12 | 22 | 22 | 22 | 5 | 24 | 26 | 133 | |||||
| Female | 4 | 16 | 12 | 6 | 2 | 8 | 11 | 59 | 44 | ||||
| Male | 8 | 6 | 10 | 16 | 3 | 16 | 15 | 74 | 56 | ||||
| Normal mucosa—N site | 12 | 11 | 11 | 11 | - | 12 | 11 | 68 | 51 | ||||
| Diseased mucosa—D site | - | 11 | 11 | 11 | 5 | 12 | 15 | 65 | 49 | ||||
| H. pylori positive—HPp | 9 | 14 | 10 | 16 | 3 | 20 | 21 | 93 | 70 | ||||
| H. pylori negative—HPn | 3 | 8 | 12 | 6 | 2 | 4 | 5 | 40 | 30 | ||||
| Serum | V | NGM | MiG | MoG | MaG | PanG | GU | DU | PU | 1GC | aGC | Total | % |
| Patients | 17 | 26 | 30 | 51 | 30 | 12 | 16 | 5 | 2 | 10 | 20 | 219 | |
| Female | 9 | 14 | 13 | 30 | 11 | 3 | 6 | 2 | - | 4 | 6 | 98 | 45 |
| Male | 8 | 12 | 17 | 21 | 19 | 9 | 10 | 3 | 2 | 6 | 14 | 121 | 55 |
| Age < 46 | 17 | 21 | 23 | 40 | 14 | 5 | 6 | 2 | 2 | 7 | 4 | 141 | 64 |
| Age > 45 | 5 | 7 | 11 | 16 | 7 | 10 | 3 | - | 3 | 16 | 78 | 36 | |
| HPp | 10 | 15 | 16 | 25 | 18 | 8 | 9 | 4 | 1 | 7 | 16 | 129 | 59 |
| HPn | 7 | 11 | 14 | 26 | 12 | 4 | 7 | 1 | 1 | 3 | 4 | 90 | 41 |
| Accession | Name | Description | Peptide Count | Unique Peptides | Confidence Score | Max Fold Change |
|---|---|---|---|---|---|---|
| O95872 | GPANK1 | G patch domain and ankyrin repeat-containing protein 1 | 7 | 2 | 40 | 2.5 |
| P0DP01 | IGHV1-8 | Immunoglobulin heavy variable 1–8 | 6 | 2 | 46 | 2.3 |
| P18564 | ITGB6 | Integrin β-6 | 5 | 5 | 34 | 2.1 |
| P04433 | IGKV3-11 | Immunoglobulin κ variable 3–11 | 5 | 2 | 46 | 4.7 |
| P22352 | GPX3 | Glutathione peroxidase 3 | 5 | 2 | 30 | 2.3 |
| Q5UE93 | PIK3R6 | Phosphoinositide 3-kinase regulatory subunit 6 | 5 | 1 | 25 | 3.3 |
| P0DP08 | IGHV4-38-2 | Immunoglobulin heavy variable 4-38-2 | 5 | 1 | 45 | 2.4 |
| P23786 | CPT2 | Carnitine O-palmitoyltransferase 2_ mitochondrial | 4 | 1 | 23 | 3.1 |
| Q64ET8 | FRG2 | Protein FRG2 | 4 | 1 | 28 | 4.6 |
| Q9H0U6 | MRPL18 | 39S ribosomal protein L18_ mitochondrial | 4 | 2 | 20 | 4.1 |
| P41247 | PNPLA4 | Patatin-like phospholipase domain-containing protein 4 | 4 | 1 | 18 | 2.4 |
| Q15032 | R3HDM1 | R3H domain-containing protein 1 | 4 | 2 | 30 | 18.2 |
| Q9NR63 | CYP26B1 | Cytochrome P450 26B1 | 3 | 1 | 19 | 3.1 |
| O00255 | MEN1 | Menin | 3 | 1 | 18 | 2.1 |
| P08865 | RPSA | 40S ribosomal protein SA | 3 | 1 | 15 | 7.9 |
| P40855 | PEX19 | Peroxisomal biogenesis factor 19 | 3 | 2 | 16 | 2.7 |
| O96007 | MOCS2 | Molybdopterin synthase catalytic subunit | 3 | 1 | 24 | 2.5 |
| Q92841 | DDX17 | Probable ATP-dependent RNA helicase DDX17 | 3 | 1 | 17 | 2.6 |
| Q6WQI6 | HEPN1 | Putative cancer susceptibility gene HEPN1 protein | 3 | 2 | 24 | 2.3 |
| O43692 | PI15 | Peptidase inhibitor 15 | 2 | 1 | 24 | 2.9 |
| Q96JF0 | ST6GAL2 | Β-galactoside α-2_6-sialyltransferase 2 | 2 | 2 | 17 | 2.4 |
| Q5VZM2 | RRAGB | Ras-related GTP-binding protein B | 9 | 1 | 55 | 8.4 |
| P06702 | S100A9 | Protein S100-A9 | 7 | 6 | 57 | 2.0 |
| P0DP09 | IGKV1-13 | Immunoglobulin kappa variable 1-13 | 7 | 4 | 74 | 3.5 |
| P30711 | GSTT1 | Glutathione S-transferase theta-1 | 9 | 5 | 54 | 2.1 |
| Albumin | Fibrinogen | FAR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Cmp/g/L | MS int./counts | Factor to V | Cmp/g/L | MS int./counts | Factor to V | Cmp/g/L | MS | |
| Ref. | V | 45.0 | 21659682 | 3.0 | 53631 | 6.7 | 0.25 | ||
| 1GC | 35.3 | 16989199 | 0.8 | 1.1 | 19117 | 0.4 | 3.0 | 0.11 | |
| High albumin | NGM | 88.9 | 42806672 | 2.0 | 6.8 | 120974 | 2.3 | 7.6 | 0.28 |
| U | 57.2 | 27549815 | 1.3 | 3.4 | 60796 | 1.1 | 5.9 | 0.22 | |
| aGC | 64.6 | 31103607 | 1.4 | 2.9 | 50985 | 1.0 | 4.4 | 0.16 | |
| MiGa | 97.1 | 46721703 | 2.2 | 3.0 | 54459 | 1.0 | 3.1 | 0.12 | |
| Low albumin/high fibrinogen | MiGn | 1.6 | 756160 | 0.03 | 31.9 | 570425 | 10.6 | 2031.1 | 75.44 |
| MoG | 7.0 | 3364998 | 0.16 | 139.4 | 2491549 | 46.5 | 1993.6 | 74.04 | |
| MaG | 3.1 | 1494820 | 0.07 | 81.9 | 1463330 | 27.3 | 2635.7 | 97.89 | |
| PanG | 1.3 | 633851 | 0.03 | 29.5 | 528261 | 9.8 | 2243.9 | 83.34 | |
| Accession | Name | Description | Peptide Count | Unique Peptides | Confidence Score | Max Fold Change |
|---|---|---|---|---|---|---|
| P19827 | ITIH1 | Inter-α-trypsin inhibitor heavy chain H1 | 80 | 60 | 596 | 3.1 |
| O60353 | FZD6 | Frizzled-6 | 27 | 19 | 198 | 2.3 |
| Q8N608 | DPP10 | Inactive dipeptidyl peptidase 10 | 14 | 10 | 103 | 3.4 |
| P29622 | SERPINA4 | Kallistatin | 22 | 17 | 147 | 2.6 |
| Q02952 | AKAP12 | A-kinase anchor protein 12 | 15 | 6 | 98 | 2.8 |
| P06702 | S100A9 | Protein S100-A9 | 7 | 6 | 57 | 2.6 |
| A5A3E0 | POTEF | POTE ankyrin domain family member F | 43 | 4 | 295 | 2.3 |
| Q02641 | CACNB1 | Voltage-dependent L-type calcium channel subunit β-1 | 12 | 5 | 82 | 2.0 |
| P02741 | CRP | C-reactive protein | 11 | 7 | 83 | 3.4 |
| Q86T90 | KIAA1328 | Protein hinderin | 9 | 3 | 62 | 2.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aziz, S.; Rasheed, F.; Zahra, R.; König, S. Gastric Cancer Pre-Stage Detection and Early Diagnosis of Gastritis Using Serum Protein Signatures. Molecules 2022, 27, 2857. https://doi.org/10.3390/molecules27092857
Aziz S, Rasheed F, Zahra R, König S. Gastric Cancer Pre-Stage Detection and Early Diagnosis of Gastritis Using Serum Protein Signatures. Molecules. 2022; 27(9):2857. https://doi.org/10.3390/molecules27092857
Chicago/Turabian StyleAziz, Shahid, Faisal Rasheed, Rabaab Zahra, and Simone König. 2022. "Gastric Cancer Pre-Stage Detection and Early Diagnosis of Gastritis Using Serum Protein Signatures" Molecules 27, no. 9: 2857. https://doi.org/10.3390/molecules27092857
APA StyleAziz, S., Rasheed, F., Zahra, R., & König, S. (2022). Gastric Cancer Pre-Stage Detection and Early Diagnosis of Gastritis Using Serum Protein Signatures. Molecules, 27(9), 2857. https://doi.org/10.3390/molecules27092857







