Synthetic Receptors Based on Abiotic Cyclo(pseudo)peptides
Abstract
1. Introduction
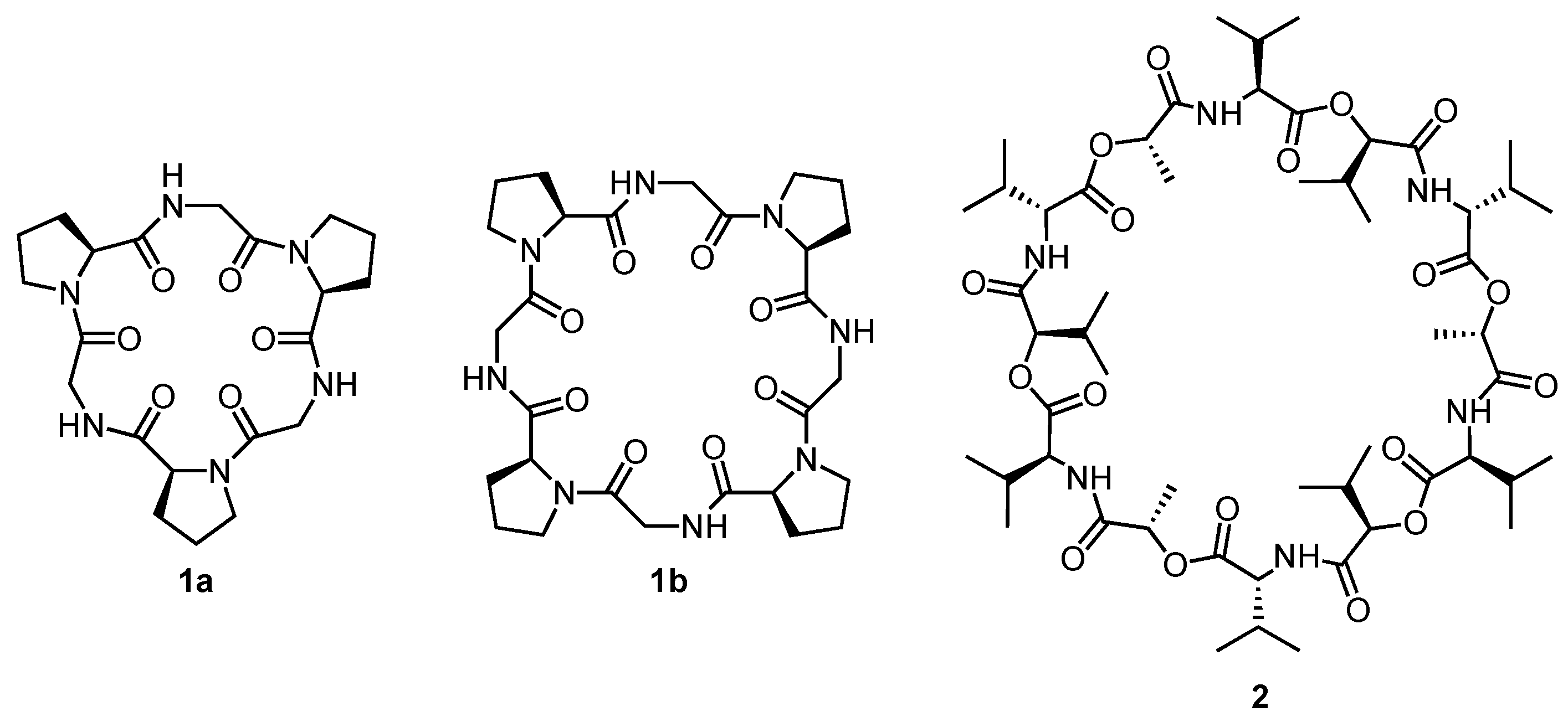
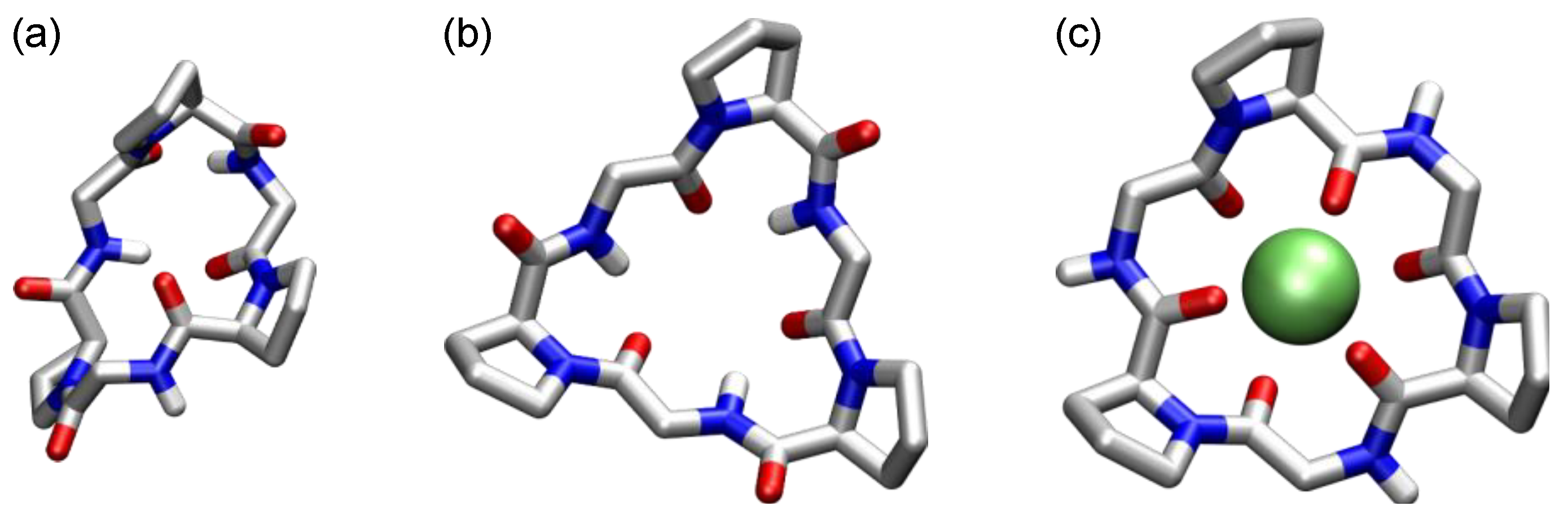
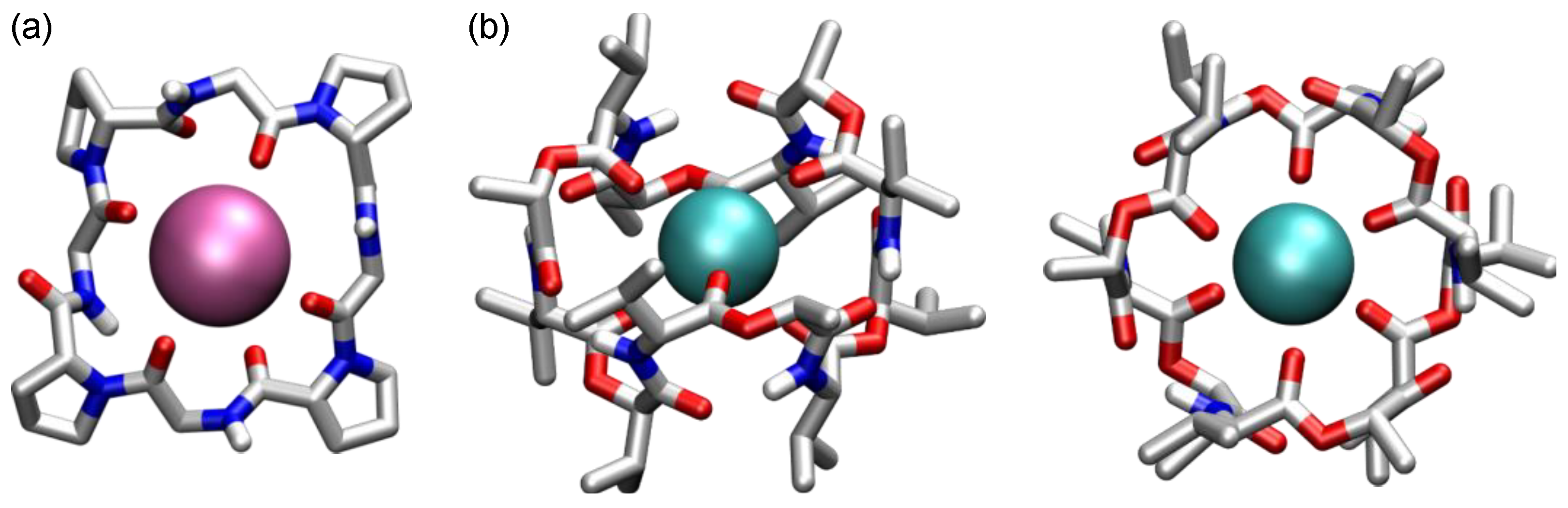
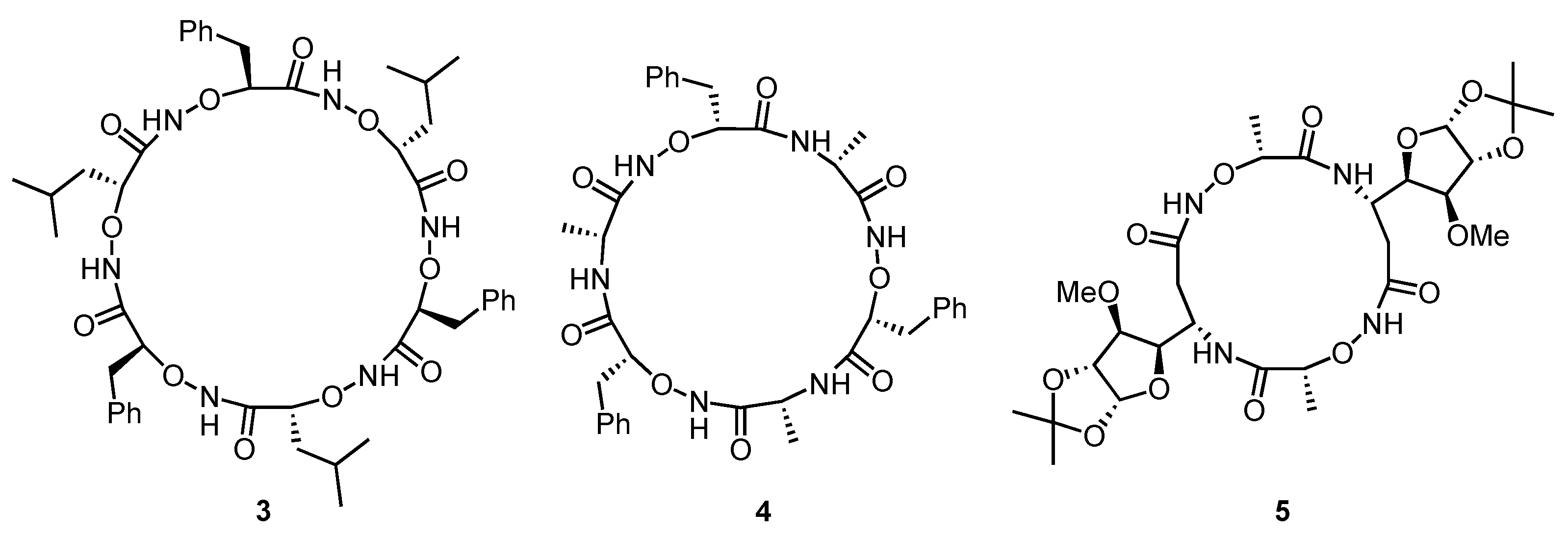
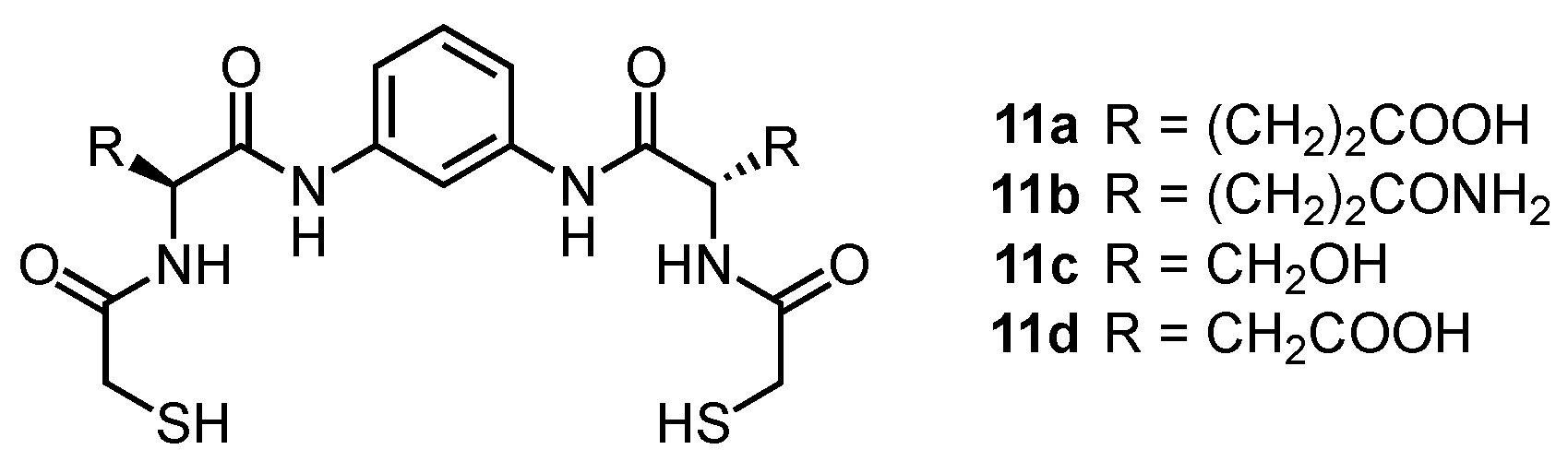
2. Cyclic Peptides and Pseudopeptides Containing Only α-Amino Acid-Derived Subunits
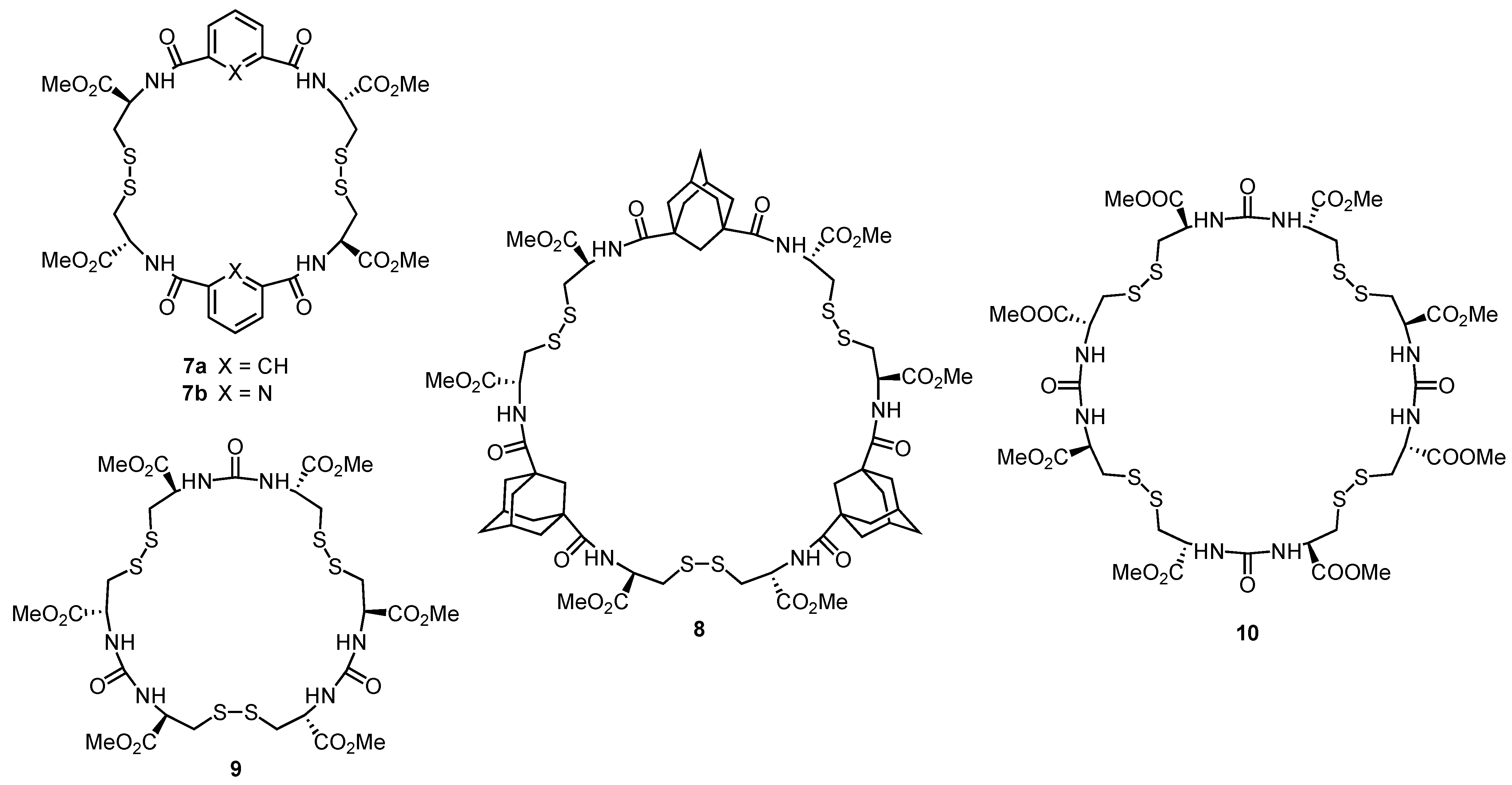
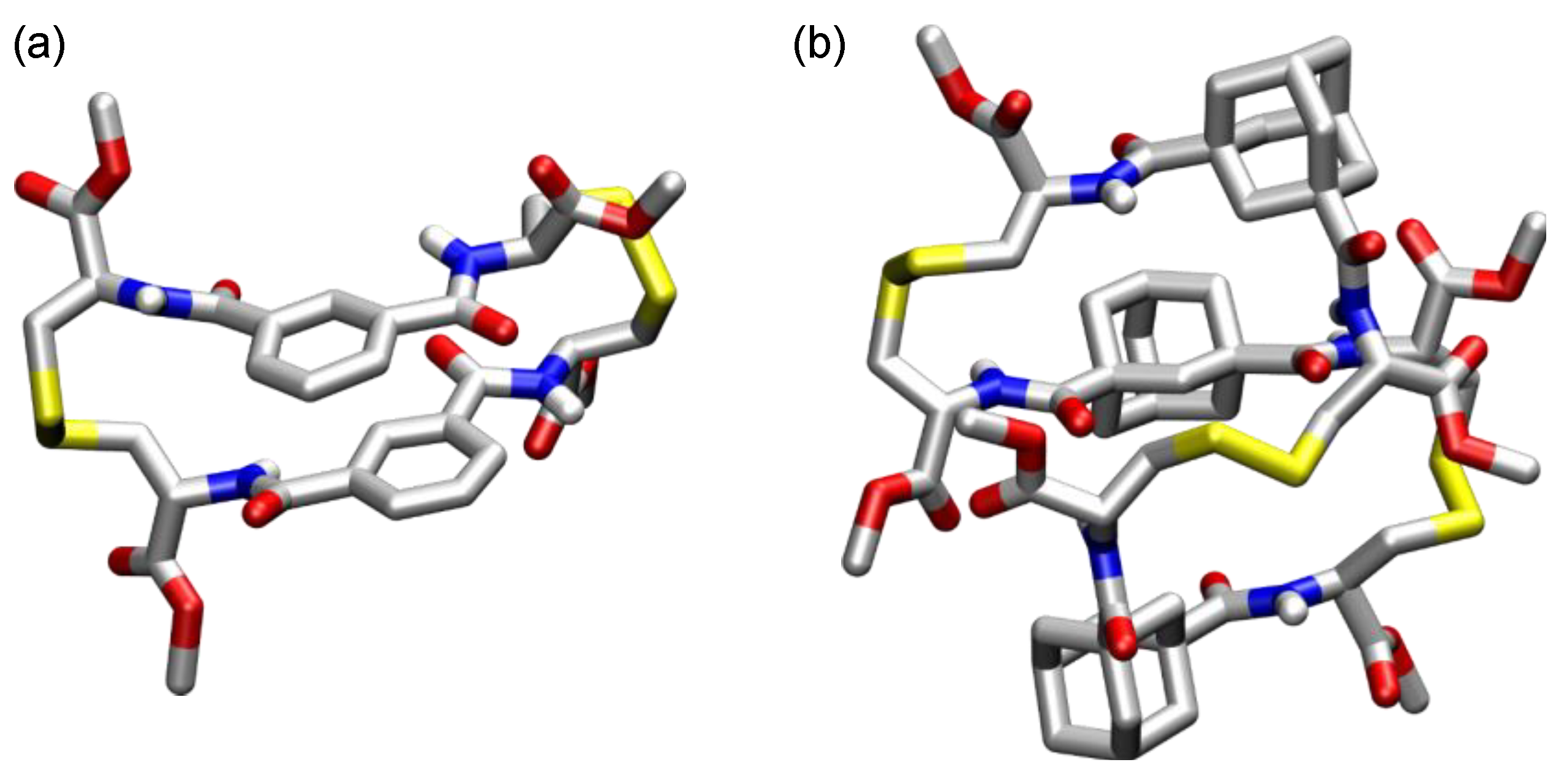
3. Cyclic Pseudopeptides Containing Cystine Subunits
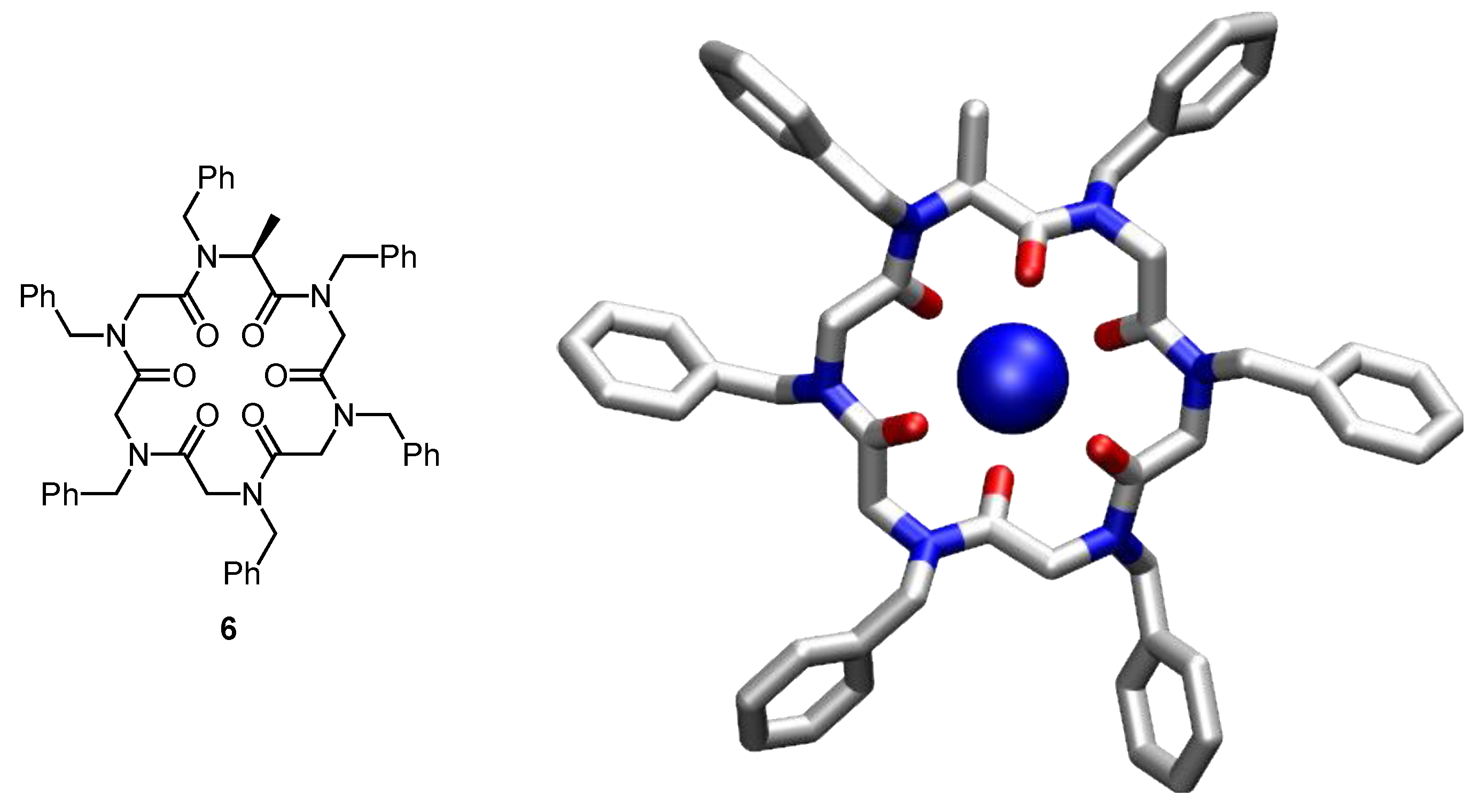
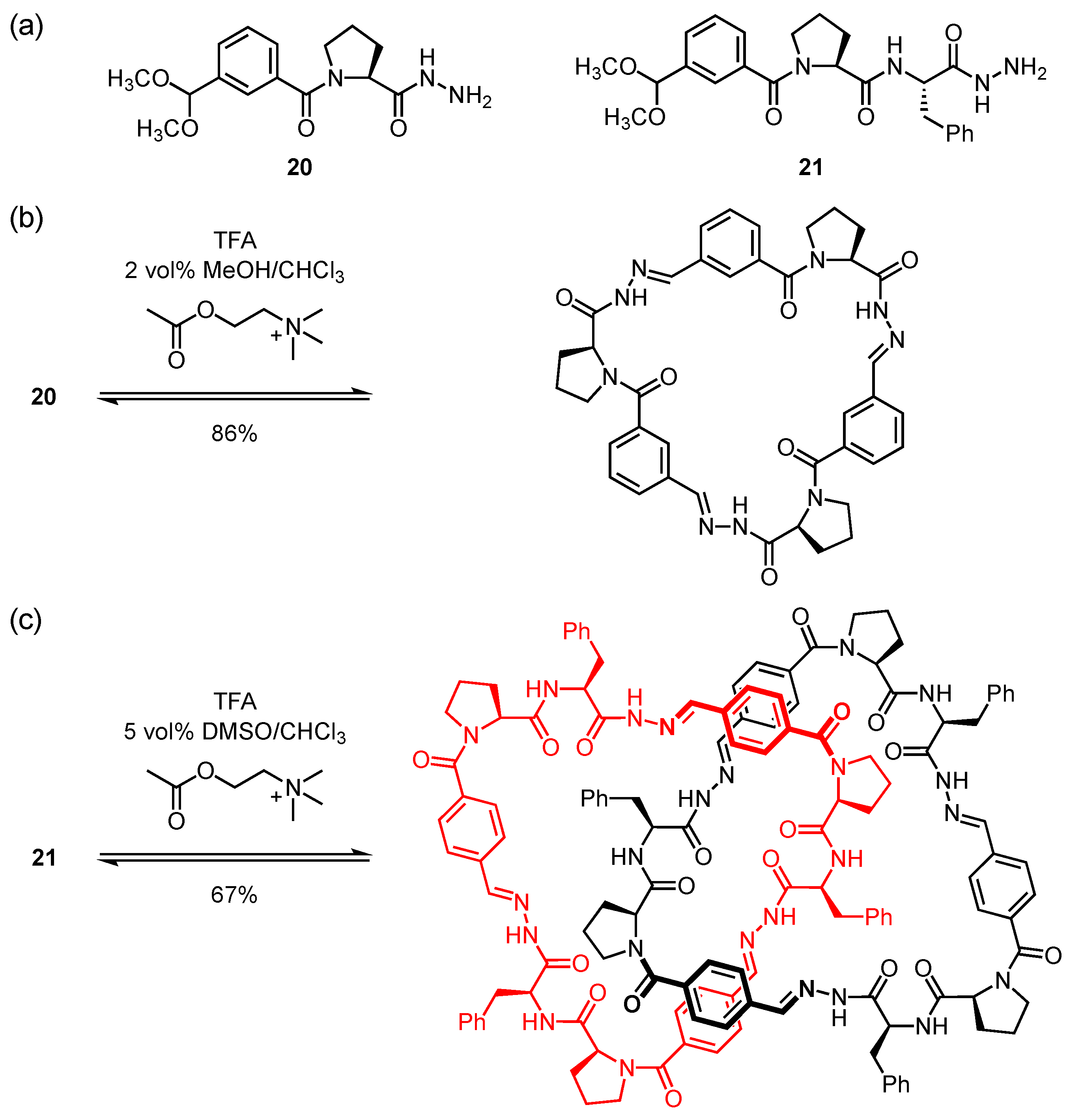
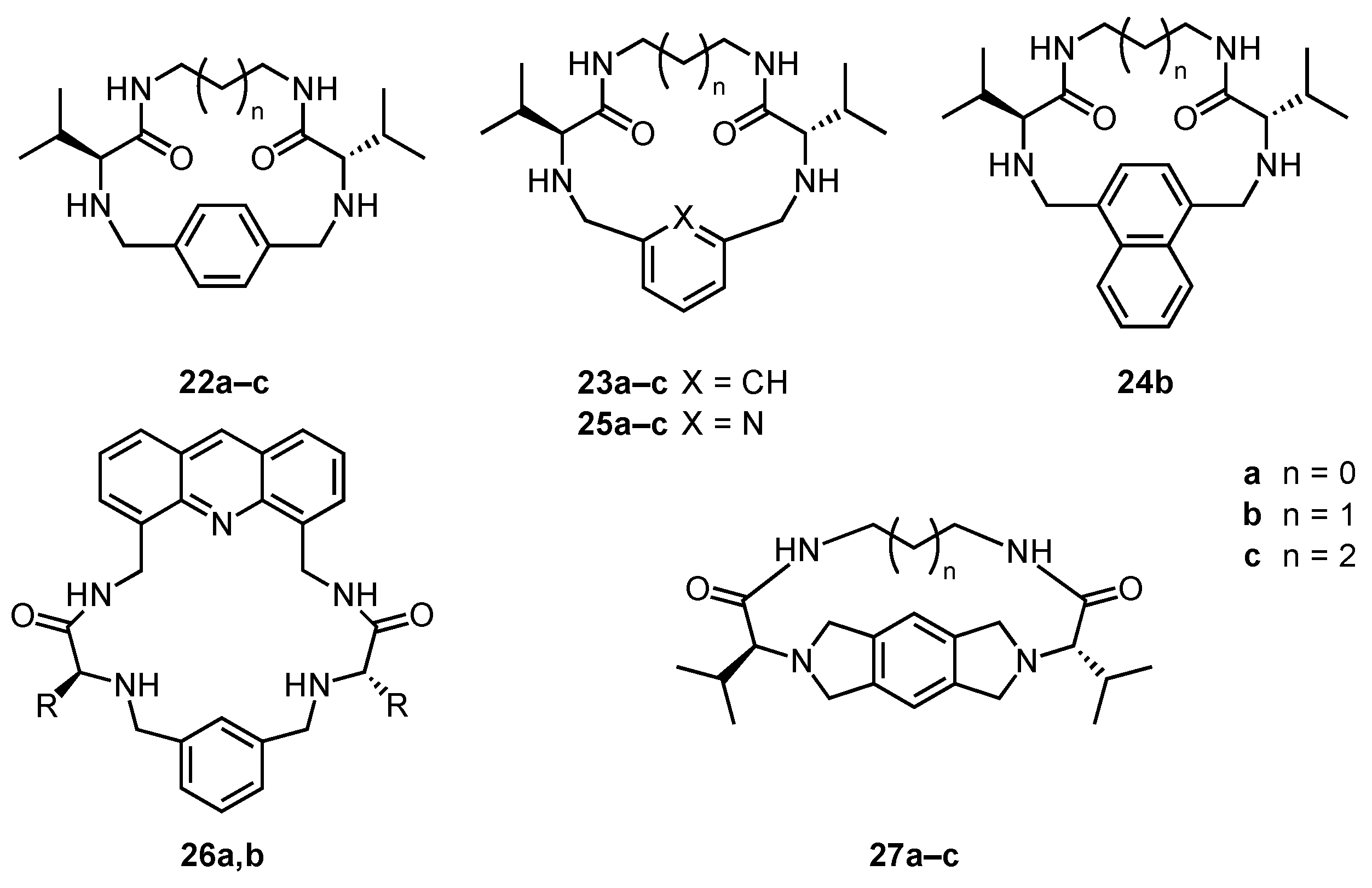
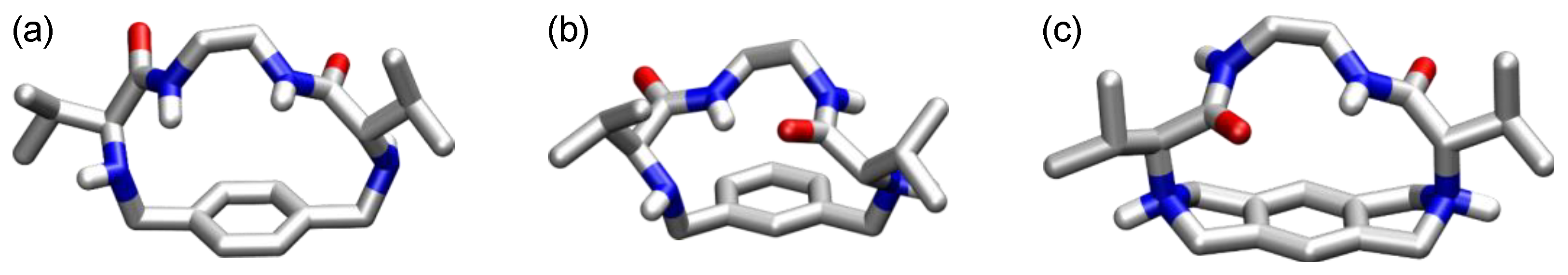
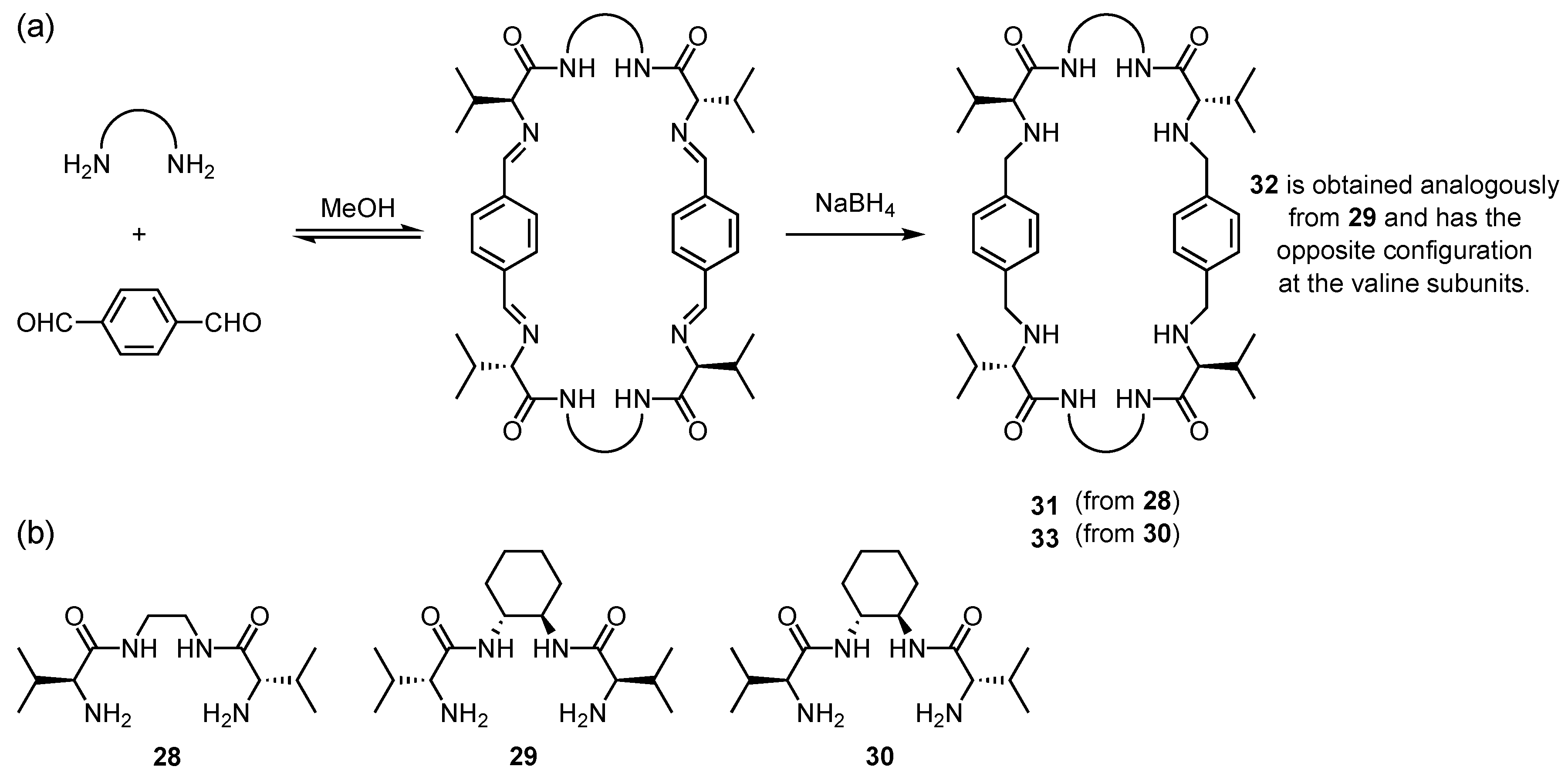
4. Cyclic Pseudopeptides Containing Nonnatural Aliphatic Subunits
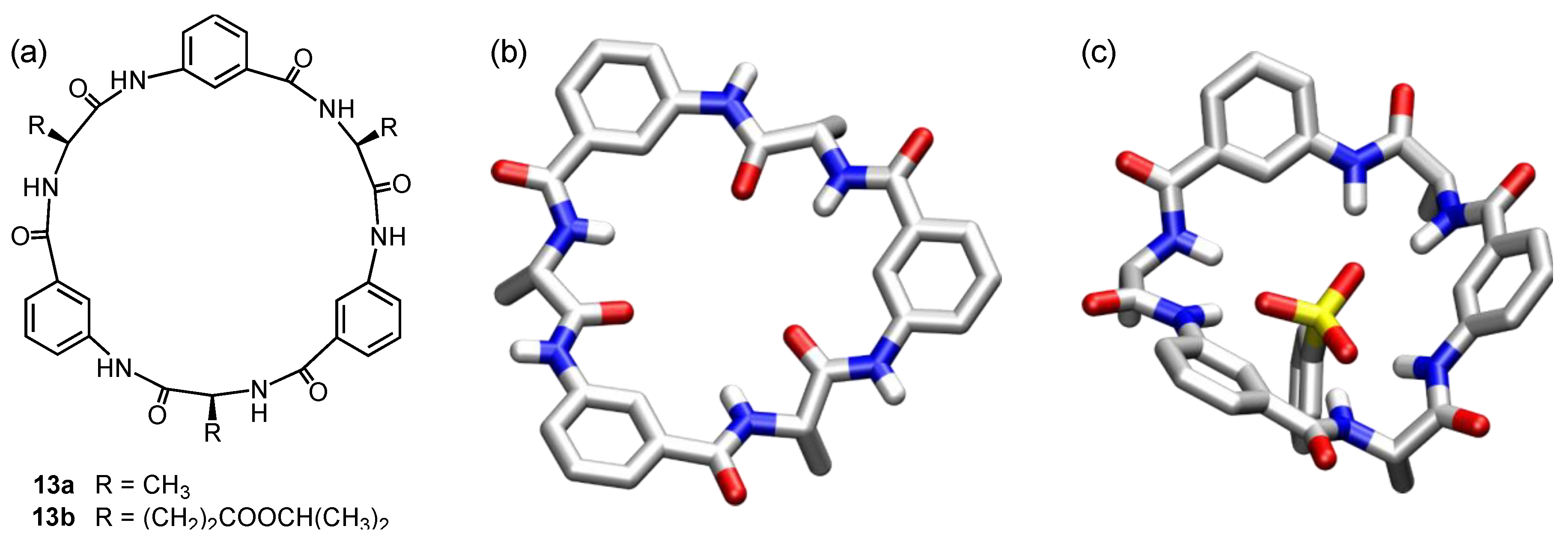
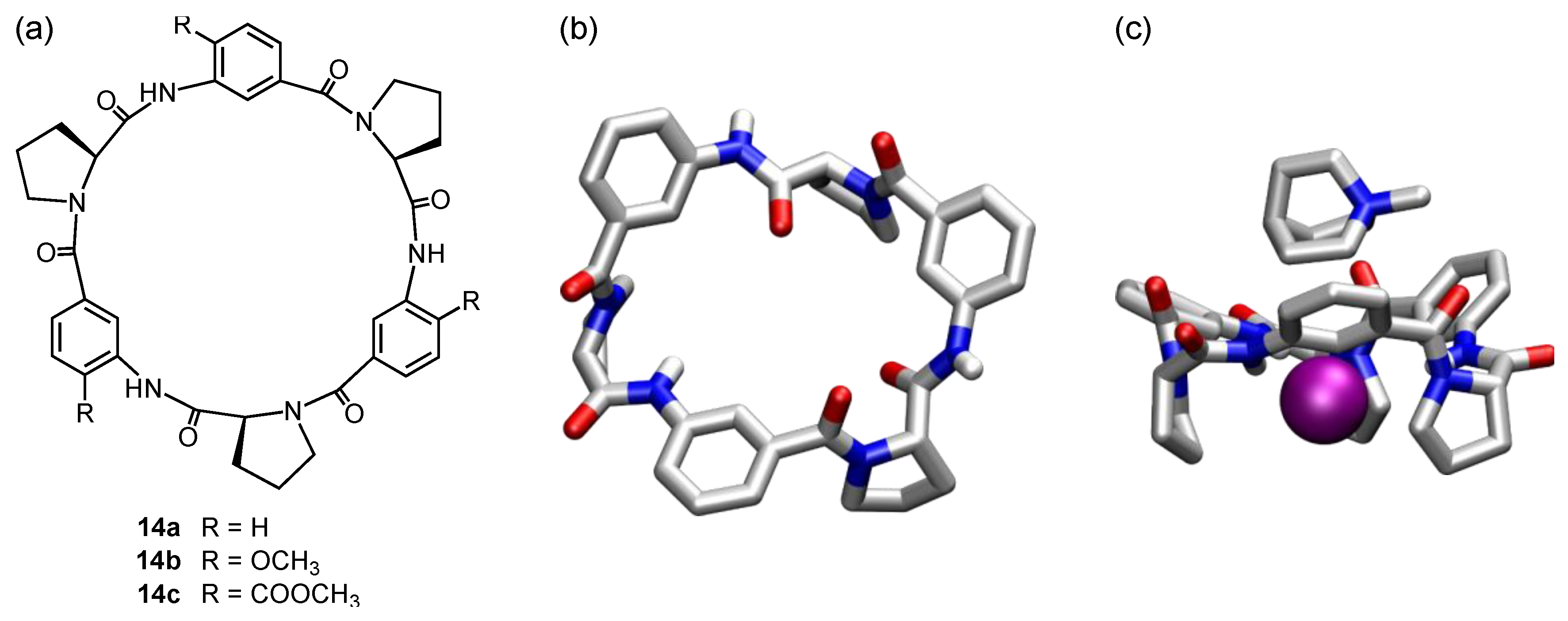
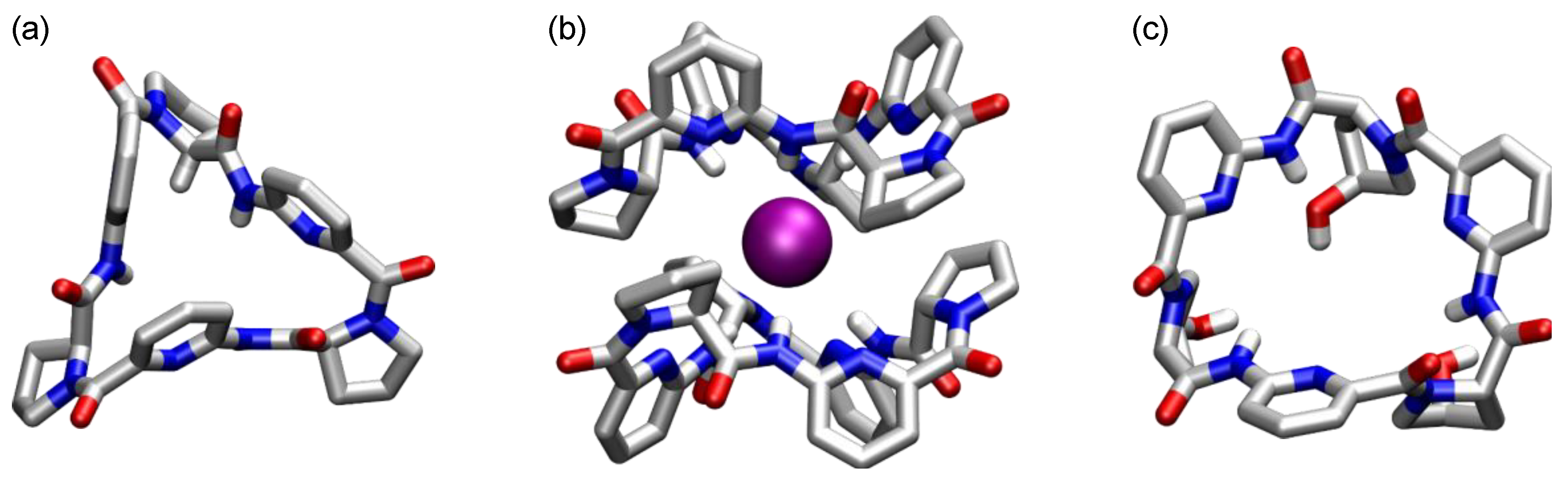
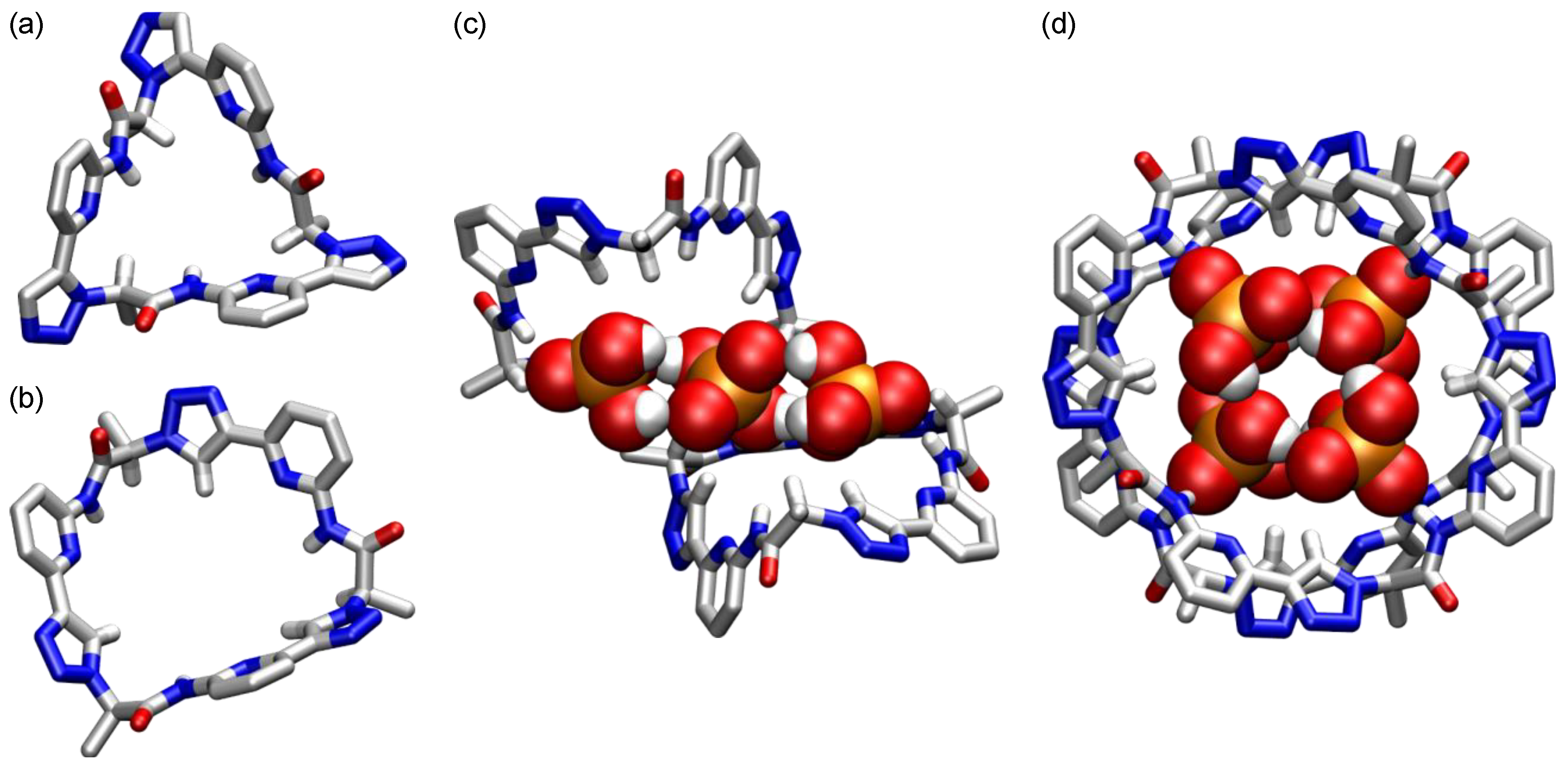
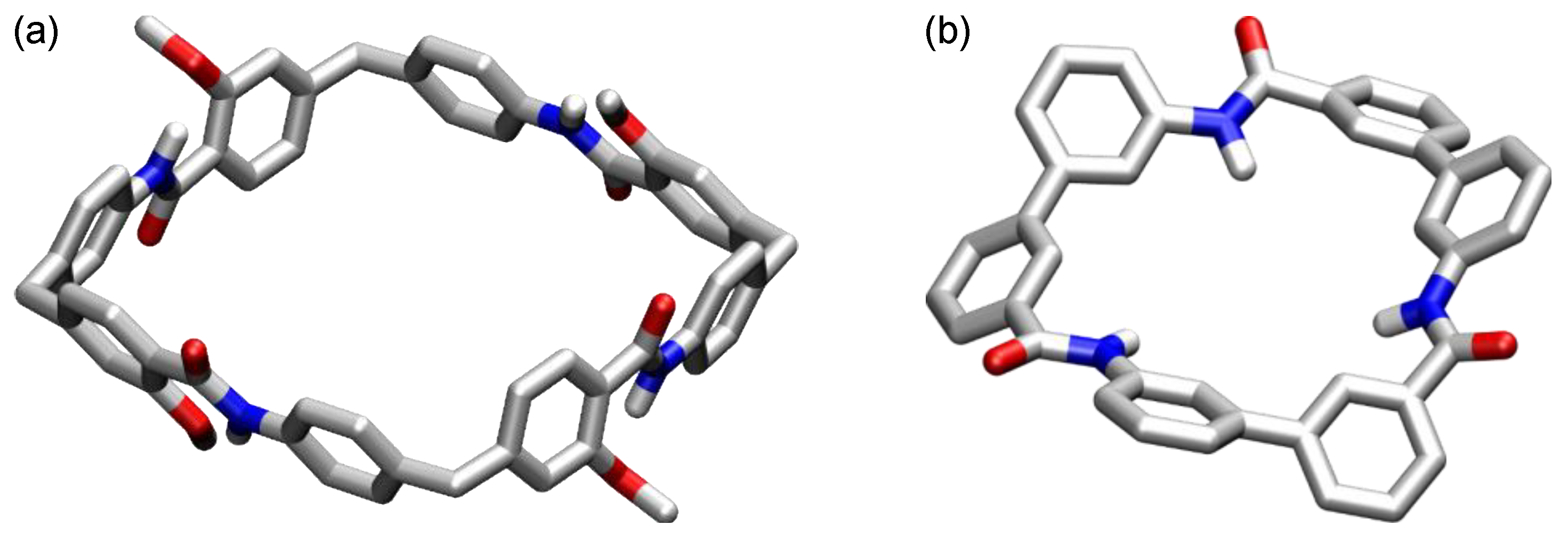
5. Cyclic Peptides and Pseudopeptides Containing Six-Membered Aromatic Subunits
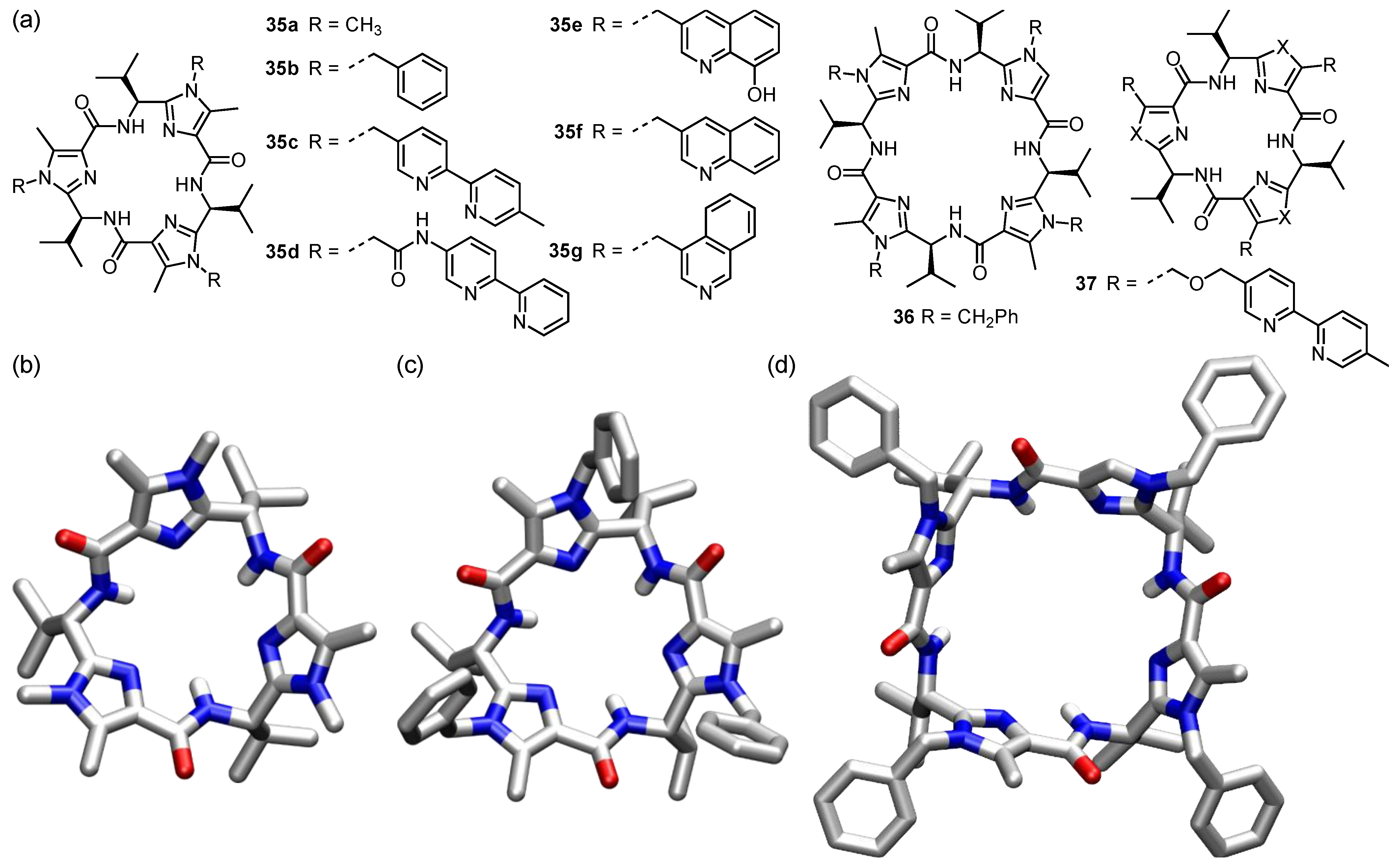
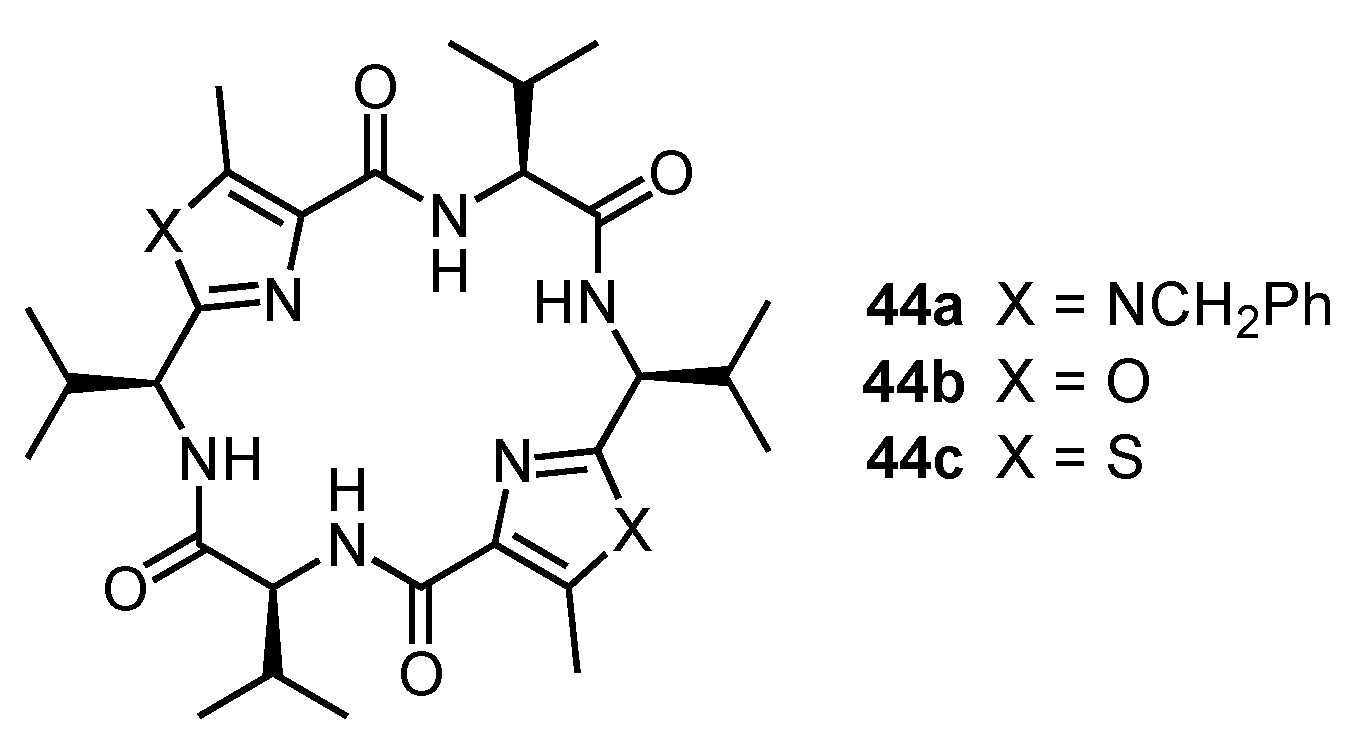
6. Cyclic Peptides Containing Five-Membered Heterocyclic Subunits
7. Cyclic Peptides Not Containing α-Amino Acid-Derived Subunits
8. Conclusions and Outlook
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deber, C.M.; Madison, V.; Blout, E.R. Why cyclic peptides—Complementary approaches to conformations. Acc. Chem. Res. 1976, 9, 106–113. [Google Scholar] [CrossRef]
- Madison, V.; Deber, C.M.; Blout, E.R. Cyclic peptides. 17. Metal and amino acid complexes of cyclo(Pro-Gly)4 and analogs studied by nuclear magnetic resonance and circular dichroism. J. Am. Chem. Soc. 1977, 99, 4788–4798. [Google Scholar] [CrossRef] [PubMed]
- Lehn, J.-M. Supramolecular Chemistry; Wiley-VCH: Weinheim, Germany, 1995. [Google Scholar]
- Gibson, S.E.; Lecci, C. Amino acid derived macrocycles—An area driven by synthesis or application? Angew. Chem. Int. Ed. 2006, 45, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Kubik, S. Artificial Receptors for Chemical Sensors; Mirsky, V.M., Yatsimirsky, A.K., Eds.; Wiley-VCH: Weinheim, Germany, 2010; pp. 135–167. [Google Scholar] [CrossRef]
- Kubik, S. Supramolecular Chemistry: From Molecules to Nanomaterials; Gale, P.A., Steed, J.W., Eds.; J. Wiley & Sons: Chichester, UK, 2012; Volume 3, pp. 1179–1203. [Google Scholar] [CrossRef]
- Luis, S.V.; Alfonso, I. Bioinspired chemistry based on minimalistic pseudopeptides. Acc. Chem. Res. 2014, 47, 112–124. [Google Scholar] [CrossRef]
- Elmes, R.B.P.; Jolliffe, K.A. Anion recognition by cyclic peptides. Chem. Commun. 2015, 51, 4951–4968. [Google Scholar] [CrossRef]
- Alfonso, I. From simplicity to complex systems with bioinspired pseudopeptides. Chem. Commun. 2016, 52, 239–250. [Google Scholar] [CrossRef]
- Tapia, L.; Alfonso, I.; Solà, J. Molecular cages for biological applications. Org. Biomol. Chem. 2021, 19, 9527–9540. [Google Scholar] [CrossRef]
- Bong, D.T.; Clark, T.D.; Granja, J.R.; Ghadiri, M.R. Self-assembling organic nanotubes. Angew. Chem. Int. Ed. 2001, 40, 988–1011. [Google Scholar] [CrossRef]
- Brea, R.J.; Reiriz, C.; Granja, J.R. Towards functional bionanomaterials based on self-assembling cyclic peptide nanotubes. Chem. Soc. Rev. 2010, 39, 1448–1456. [Google Scholar] [CrossRef]
- Song, Q.; Cheng, Z.; Kariuki, M.; Hall, S.C.L.; Hill, S.K.; Rho, J.Y.; Perrier, S. Molecular self-assembly and supramolecular chemistry of cyclic peptides. Chem. Rev. 2021, 121, 13936–13995. [Google Scholar] [CrossRef]
- Madison, V.; Atreyi, M.; Deber, C.M.; Blout, E.R. Cyclic peptides. IX. Conformations of a synthetic ion-binding cyclic peptide, cyclo(Pro-Gly)3, from circular dichroism and 1H and 13C nuclear magnetic resonance. J. Am. Chem. Soc. 1974, 96, 6725–6734. [Google Scholar] [CrossRef]
- Chiu, Y.-Y.; Brown, L.D.; Lipscomb, W.N. Crystal and molecular structure of complex between cyclo(L-prolylglycyl)4 and RbSCN. J. Am. Chem. Soc. 1977, 99, 4799–4803. [Google Scholar] [CrossRef] [PubMed]
- Bartman, B.; Deber, C.M.; Blout, E.R. Cyclic peptides. 16. Carbon-13 NMR relaxation studies of complexes between cyclo(L-Pro-Gly)3 and amino acids. Conformational aspects of stepwise binding. J. Am. Chem. Soc. 1977, 99, 1028–1033. [Google Scholar] [CrossRef] [PubMed]
- Deber, C.M.; Blout, E.R. Amino acid cyclic peptide complexes. J. Am. Chem. Soc. 1974, 96, 7566–7568. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, C.J. Cyclic polyethers and their complexes with metal salts. J. Am. Chem. Soc. 1967, 89, 7017–7036. [Google Scholar] [CrossRef]
- Pressman, B.C.; Harris, E.J.; Jagger, W.S.; Johnson, J.H. Antibiotic-mediated transport of alkali ions across lipid barriers. Proc. Natl. Acad. Sci. USA 1967, 58, 1949–1956. [Google Scholar] [CrossRef]
- Pletnev, V.Z.; Tsygannik, I.N.; Fonarev, Y.D.; Mikhailova, I.Y.; Kulikov, Y.V.; Ivanov, V.T.; Langs, D.A.; Duax, W.L. Crystal and molecular structure of K+ complex of meso-valinomycin, cyclo[-(D-Val-L-Hyi-L-Val-D-Hyi)(3)-]·KAuCl4. Bioorg. Khim. 1995, 21, 828–833. [Google Scholar] [PubMed]
- Rose, M.C.; Henkens, R.W. Stability of sodium and potassium complexes of valinomycin. Biochim. Biophys. Acta 1974, 372, 426–435. [Google Scholar] [CrossRef]
- Blout, E.R. Cyclic peptides—Past, present, and future. Biopolymers 1981, 20, 1901–1912. [Google Scholar] [CrossRef]
- Ovchinnikov, Y.A.; Ivanov, V.T. Conformational states and biological activity of cyclic peptides. Tetrahedron 1975, 31, 2177–2209. [Google Scholar] [CrossRef]
- Gisin, B.F.; Ting-Beall, H.P.; Davis, D.G.; Grell, E.; Tosteson, D.C. Selective ion binding and membrane activity of synthetic cyclopeptides. Biochim. Biophys. Acta 1978, 509, 201–217. [Google Scholar] [CrossRef]
- Saviano, G.; Rossi, F.; Benedetti, E.; Pedone, C.; Mierke, D.F.; Maione, A.; Zanotti, G.; Tancredi, T.; Saviano, M. Structural consequences of metal complexation of cyclo[Pro-Phe-Phe-Ala-Xaa]2 decapeptides. Chem. Eur. J. 2001, 7, 1176–1183. [Google Scholar] [CrossRef]
- Farkas, E.; Sóvágó, I. Amino Acids, Peptides and Proteins: Amino Acids, Peptides And Proteins; RSC: Cambridge, UK, 2006; Volume 35, pp. 353–434. [Google Scholar] [CrossRef]
- Yang, D.; Qu, J.; Zhang, Y.-H.; Ren, Y.; Wang, D.-P.; Wu, Y.-D. Cyclic hexapeptide of D,L-α-aminoxy acids as a selective receptor for chloride ion. J. Am. Chem. Soc. 2002, 124, 12410–12411. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.-D.; Yang, D. α-Aminoxy acids: New possibilities from foldamers to anion receptors and channels. Acc. Chem. Res. 2008, 41, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Avan, I.; Hall, C.D.; Katritzky, A.R. Peptidomimetics via modifications of amino acids and peptide bonds. Chem. Soc. Rev. 2014, 43, 3575–3594. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Li, X.; Sha, Y.; Wu, Y.-D. A cyclic hexapeptide comprising alternating R-aminoxy acids and R-amino acids is a selective chloride ion receptor. Chem. Eur. J. 2005, 11, 3005–3009. [Google Scholar] [CrossRef]
- Sharma, G.V.M.; Manohar, V.; Dutta, S.K.; Sridhar, B.; Ramesh, V.; Srinivas, R.; Kunwar, A.C. Self-assembling cyclic tetrapeptide from alternating C-linked carbo-beta-amino acid [(S)-beta-Caa] and alpha-aminoxy acid [(R)-Ama]: A selective chloride ion receptor. J. Org. Chem. 2010, 75, 1087–1094. [Google Scholar] [CrossRef]
- Sun, J.; Zuckermann, R.N. Peptoid polymers: A highly designable bioinspired material. ACS Nano 2013, 7, 4715–4732. [Google Scholar] [CrossRef]
- Shin, S.B.Y.; Yoo, B.; Todaro, L.J.; Kirshenbaum, K. Cyclic peptoids. J. Am. Chem. Soc. 2007, 129, 3218–3225. [Google Scholar] [CrossRef]
- Yoo, B.; Shin, S.B.Y.; Huang, M.L.; Kirshenbaum, K. Peptoid macrocycles: Making the rounds with peptidomimetic oligomers. Chem. Eur. J. 2010, 16, 5528–5537. [Google Scholar] [CrossRef]
- Webster, A.M.; Cobb, S.L. Recent advances in the synthesis of peptoid macrocycles. Chem. Eur. J. 2018, 24, 7560–7573. [Google Scholar] [CrossRef] [PubMed]
- De Riccardis, F. The challenge of conformational isomerism in cyclic peptoids. Eur. J. Org. Chem. 2020, 2981–2994. [Google Scholar] [CrossRef]
- D’Amato, A.; Pierri, G.; Costabile, C.; Della Sala, G.; Tedesco, C.; Izzo, I.; De Riccardis, F. Cyclic peptoids as topological templates: Synthesis via central to conformational chirality induction. Org. Lett. 2018, 20, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Maulucci, N.; Izzo, I.; Bifulco, G.; Aliberti, A.; De Cola, C.; Comegna, D.; Gaeta, C.; Napolitano, A.; Pizza, C.; Tedesco, C.; et al. Synthesis, structures, and properties of nine-, twelve-, and eighteen-membered N-benzyloxyethyl cyclic alpha-peptoids. Chem. Commun. 2008, 3927–3929. [Google Scholar] [CrossRef]
- Izzo, I.; Ianniello, G.; De Cola, C.; Nardone, B.; Erra, L.; Vaughan, G.; Tedesco, C.; De Riccardis, F. Structural effects of proline substitution and metal binding on hexameric cyclic peptoids. Org. Lett. 2013, 15, 598–601. [Google Scholar] [CrossRef]
- Meli, A.; Gambaro, S.; Costabile, C.; Talotta, C.; Della Sala, G.; Tecilla, P.; Milano, D.; Tosolini, M.; Izzo, I.; De Riccardis, F. Synthesis and complexing properties of cyclic benzylopeptoids—A new family of extended macrocyclic peptoids. Org. Biomol. Chem. 2016, 14, 9055–9062. [Google Scholar] [CrossRef]
- Schettini, R.; Costabile, C.; Della Sala, G.; Iuliano, V.; Tedesco, C.; Izzo, I.; De Riccardis, F. Cation-induced molecular switching based on reversible modulation of peptoid conformational states. J. Org. Chem. 2018, 83, 12648–12663. [Google Scholar] [CrossRef]
- Laussac, J.P.; Robert, A.; Haran, R.; Sarkar, B. Complexation of copper(II) with a macrocyclic peptide containing histidyl residues: Novel observation of NMR spectra of paramagnetic copper(II) compounds. Inorg. Chem. 1986, 25, 2760–2765. [Google Scholar] [CrossRef]
- Ozeki, E.; Kimura, S.; Imanishi, Y. Conformation and complexation with metal ions of cyclic hexapeptides: Cyclo(L-Leu-L-Phe-L-Pro)2 and cyclo[L-Cys(Acm)-L-Phe-L-Pro]2. Int. J. Peptide Protein Res. 1989, 34, 111–117. [Google Scholar] [CrossRef]
- Leipert, D.; Nopper, D.; Bauser, M.; Gauglitz, G.; Jung, G. Investigation of the molecular recognition of amino acids by cyclopeptides with reflectometric interference spectroscopy. Angew. Chem. Int. Ed. 1998, 37, 3308–3311. [Google Scholar] [CrossRef]
- Leipert, D.; Rathgeb, F.; Herold, M.; Mack, J.; Gauglitz, G.; Jung, G. Interaction between volatile organic compounds and cyclopeptides detected with reflectrometric interference spectroscopy. Anal. Chim. Acta 1999, 392, 213–221. [Google Scholar] [CrossRef]
- Ngu-Schwemlein, M.; Butko, P.; Cook, B.; Whigham, T. Interactions of an acidic cyclooctapeptide with metal ions: Microcalorimetric and fluorescence analyses. J. Peptide Res. 2006, 66 (Suppl. S1), 72–81. [Google Scholar] [CrossRef]
- Duléry, V.; Uhlich, N.A.; Maillard, N.; Fluxá, V.S.; Garcia, J.; Dumy, P.; Renaudet, O.; Reymond, J.-L.; Darbre, T. A cyclodecapeptide ligand to vitamin B12. Org. Biomol. Chem. 2008, 6, 4134–4141. [Google Scholar] [CrossRef] [PubMed]
- Ngu-Schwemlein, M.; Gilbert, W.; Askew, K.; Schwemlein, S. Thermodynamics and fluorescence studies of the interactions of cyclooctapeptides with Hg2+, Pb2+, and Cd2+. Bioorg. Med. Chem. 2008, 16, 5778–5787. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, D.; Haridas, V.; Karle, I.L. Cystinophanes, a novel family of aromatic-bridged cystine cyclic peptides: Synthesis, crystal structure, molecular recognition, and conformational studies. J. Am. Chem. Soc. 1998, 120, 2695–2702. [Google Scholar] [CrossRef]
- Chung, B.K.W.; Yudin, A.K. Disulfide-bridged peptide macrobicycles from nature. Org. Biomol. Chem. 2015, 13, 8768–8779. [Google Scholar] [CrossRef]
- Karle, I.L.; Ranganathan, D.; Haridas, V. Crystal structure of cyclo(Adm-Cyst)3: Example of a topologically defined double-helical cystine cyclic peptide. J. Am. Chem. Soc. 1996, 118, 10916–10917. [Google Scholar] [CrossRef]
- Ranganathan, D. Designer hybrid cyclopeptides for membrane ion transport and tubular structures. Acc. Chem. Res. 2001, 34, 919–930. [Google Scholar] [CrossRef]
- Ranganathan, D.; Lakshmi, C. Cystine-based cyclic oligoureas: A new class of hydrogen-bonding electroneutral anion receptors. Chem. Commun. 2001, 1250–1251. [Google Scholar] [CrossRef]
- Huang, H.; Mu, L.; He, J.; Cheng, J.-P. A cystine-bearing pseudo-cyclopeptide as a new amphi-receptor. Tetrahedron Lett. 2002, 43, 2255–2258. [Google Scholar] [CrossRef]
- Huang, H.; Mu, L.; He, J.; Cheng, J.-P. Ferrocenyl-bearing cyclopseudopeptides as redox-switchable cation receptors. J. Org. Chem. 2003, 68, 7605–7611. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, J.; He, J.Q.; Li, Z.C.; Cheng, J.-P. Polymethylene-bridged cystine–glycine-containing cyclopeptides as hydrogen-bonding electroneutral anion receptors: Design, synthesis, and halide ion recognition. Supramol. Chem. 2004, 16, 171–174. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, Z.; He, J.; Cheng, J.-P. Effective receptors for fluoride and acetate ions: Synthesis and binding study of pyrrole- and cystine-based cyclopeptido-mimetics. Tetrahedron Lett. 2007, 48, 6039–6043. [Google Scholar] [CrossRef]
- Corbett, P.T.; Leclaire, J.; Vial, L.; West, K.R.; Wietor, J.-L.; Sanders, J.K.M.; Otto, S. Dynamic combinatorial chemistry. Chem. Rev. 2006, 106, 3652–3711. [Google Scholar] [CrossRef]
- Atcher, J.; Moure, A.; Alfonso, I. The emergence of halophilic evolutionary patterns from a dynamic combinatorial library of macrocyclic pseudopeptides. Chem. Commun. 2013, 49, 487–489. [Google Scholar] [CrossRef]
- Miyake, H.; Kojima, Y. Macrocyclic pseudopeptides containing N,N′-ethylene-bridged-dipeptide units: Synthesis, binding properties toward metal and organic ammonium cations, and conformations. The first step in designing artificial metalloproteins. Coord. Chem. Rev. 1996, 148, 301–314. [Google Scholar] [CrossRef]
- Kojima, Y.; Yamashita, T.; Miyake, H. Structure of cyclic hexa-pseudopeptide constructed from N,N′-ethylene-bridged-(S)-alanyl-(S)-alanine and glycine. Chem. Lett. 1995, 24, 201–202. [Google Scholar] [CrossRef]
- Kojima, Y.; Ikeda, Y.; Miyake, H.; Iwadou, I.; Hirotsu, K.; Shibata, K.; Yamashita, T.; Ohsuka, A.; Sugihara, A. Macrocyclic peptides VI. Complex formations and conformations of an ionophorous cyclic octapeptide containing N,N′-ethylene-bridged (S)-leucyl-(S)-leucine and glycine in acetonitrile. Polym. J. 1991, 23, 1359–1363. [Google Scholar] [CrossRef][Green Version]
- Miyake, H.; Kato, N.; Kojima, Y.; Sugihara, A. Coordination abilities of amide oxygens in a 24-membered ring pseudopeptide toward several transition metal ions in acetonitrile. Inorg. Chim. Acta 1994, 223, 121–124. [Google Scholar] [CrossRef]
- Miyake, H.; Kojima, Y.; Yamashita, T.; Ohsuka, A. Macrocyclic peptides, 8. Enantiomeric differentiations of various (R)- and (S)- ammonium and (R)- and (S)-α-amino acid ester salts by macrocyclic pseudopeptides. Makromol. Chem. 1993, 194, 1925–1933. [Google Scholar] [CrossRef]
- Angelici, G.; Castellucci, N.; Tomasini, C. Conformational studies on a medium size cyclopseudopeptide containing the oxazolidin-2-one moiety. J. Phys. Org. Chem. 2012, 25, 965–970. [Google Scholar] [CrossRef]
- Tomasini, C.; Zanna, N. Oxazolidinone-containing pseudopeptides: Supramolecular materials, fibers, crystals, and gels. Pept. Sci. 2017, 108, e22898. [Google Scholar] [CrossRef]
- Ishida, H.; Suga, M.; Donowaki, K.; Ohkubo, K. Highly effective binding of phosphomonoester with neutral cyclic peptides which include a non-natural amino acid. J. Org. Chem. 1995, 60, 5374–5375. [Google Scholar] [CrossRef]
- Kubik, S. Large increase in cation binding affinity of artificial cyclopeptide receptors by an allosteric effect. J. Am. Chem. Soc. 1999, 121, 5846–5855. [Google Scholar] [CrossRef]
- Kubik, S.; Becker, S. Crystal Structure of cyclo(Ala-mAB)3 2022, CCDC 2165290: Experimental Crystal Structure Determination. Available online: https://www.ccdc.cam.ac.uk/structures/search?id=doi:10.5517/ccdc.csd.cc2bp529&sid=DataCite (accessed on 7 April 2022).
- Ishida, H.; Donowaki, K.; Suga, M.; Shimose, K.; Ohkubo, K. Serine proteases mimics: Hydrolytic activity of cyclic peptides which include a non-natural amino acid. Tetrahedron Lett. 1995, 36, 8987–8990. [Google Scholar] [CrossRef]
- Ishida, H.; Qi, Z.; Sokabe, M.; Donowaki, K.; Inoue, Y. Molecular design and synthesis of artificial ion channels based on cyclic peptides containing unnatural amino acids. J. Org. Chem. 2001, 66, 2978–2989. [Google Scholar] [CrossRef]
- Ma, J.C.; Dougherty, D.A. The cation-π interaction. Chem. Rev. 1997, 97, 1303–1324. [Google Scholar] [CrossRef]
- Dougherty, D.A. The cation-π interaction. Acc. Chem. Res. 2013, 46, 885–893. [Google Scholar] [CrossRef]
- Kubik, S.; Goddard, R. A new cyclic pseudopeptide composed of (L)-proline and 3-aminobenzoic acid subunits as a ditopic receptor for the simultaneous complexation of cations and anions. J. Org. Chem. 1999, 64, 9475–9486. [Google Scholar] [CrossRef]
- Kubik, S.; Goddard, R. Intramolecular conformational control in a cyclic peptide composed of alternating (L)-proline and substituted 3-aminobenzoic acid subunits. Chem. Commun. 2000, 633–634. [Google Scholar] [CrossRef]
- Kubik, S.; Goddard, R. Fine tuning of the cation affinity of artificial receptors based on cyclic peptides by intramolecular conformational control. Eur. J. Org. Chem. 2001, 311–322. [Google Scholar] [CrossRef]
- Cannizzaro, C.E.; Houk, K.N. Magnitudes and chemical consequences of R3N+-C-H···O=C hydrogen bonding. J. Am. Chem. Soc. 2002, 124, 7163–7169. [Google Scholar] [CrossRef] [PubMed]
- Zouatom Kamdam, M.A.; Kubik, S. unpublished results.
- Feigel, M.; Lugert, G. Anthranilsäure-Glycin-Sequenzen als Bauteil “schalenförmiger” Cyclopeptide. Liebigs Ann. Chem. 1989, 1989, 1089–1092. [Google Scholar] [CrossRef]
- Heinrichs, G.; Vial, L.; Lacour, J.; Kubik, S. Enantioselective recognition of a quaternary ammonium ion by C3 symmetric cyclic hexapeptides. Chem. Commun. 2003, 1252–1253. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, G.; Kubik, S.; Lacour, J.; Vial, L. Matched/mismatched interaction of a cyclic hexapeptide with ion pairs containing chiral cations and chiral anions. J. Org. Chem. 2005, 70, 4498–4501. [Google Scholar] [CrossRef]
- Bartoli, S.; Roelens, S. Binding of acetylcholine and tetramethylammonium to a cyclophane receptor: Anion’s contribution to the cation-π interaction. J. Am. Chem. Soc. 2002, 124, 8307–8315. [Google Scholar] [CrossRef]
- Böhmer, V.; Dalla Cort, A.; Mandolini, L. Counteranion effect on complexation of quats by a neutral calix[5]arene receptor. J. Org. Chem. 2001, 66, 1900–1902. [Google Scholar] [CrossRef]
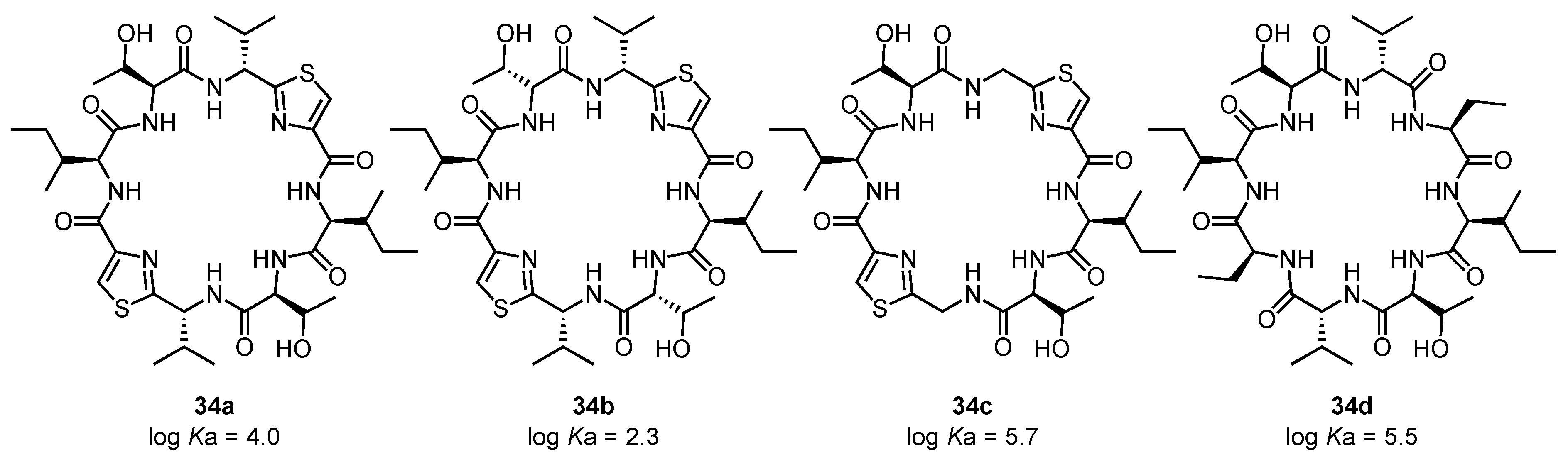
- Kubik, S.; Goddard, R.; Kirchner, R.; Nolting, D.; Seidel, J. A cyclic hexapeptide containing L-proline and 6-aminopicolinic acid subunits binds anions in water. Angew. Chem. Int. Ed. 2001, 40, 2648–2651. [Google Scholar] [CrossRef]
- Biedermann, F.; Nau, W.M.; Schneider, H.-J. The hydrophobic effect revisited—Studies with supramolecular complexes imply high-energy water as a noncovalent driving force. Angew. Chem. Int. Ed. 2014, 53, 11158–11171. [Google Scholar] [CrossRef]
- Kubik, S.; Goddard, R. Conformation and anion binding properties of cyclic hexapeptides containing L-4-hydroxyproline and 6-aminopicolinic acid subunits. Proc. Natl. Acad. Sci. USA 2002, 99, 5127–5132. [Google Scholar] [CrossRef]
- Rodriguez-Docampo, Z.; Pascu, S.I.; Kubik, S.; Otto, S. Noncovalent interactions within a synthetic receptor can reinforce guest binding. J. Am. Chem. Soc. 2006, 128, 11206–11210. [Google Scholar] [CrossRef] [PubMed]
- Kubik, S.; Kirchner, R.; Nolting, D.; Seidel, J. A molecular oyster: A neutral anion receptor containing two cyclopeptide subunits with a remarkable sulfate affinity in aqueous solution. J. Am. Chem. Soc. 2002, 124, 12752–12760. [Google Scholar] [CrossRef] [PubMed]
- Reyheller, C.; Hay, B.P.; Kubik, S. Influence of linker structure on the anion binding affinity of biscyclopeptides. New. J. Chem. 2007, 31, 2095–2102. [Google Scholar] [CrossRef]
- Otto, S.; Kubik, S. Dynamic combinatorial optimization of a neutral receptor that binds inorganic anions in aqueous solution. J. Am. Chem. Soc. 2003, 125, 7804–7805. [Google Scholar] [CrossRef]
- Fiehn, T.; Goddard, R.; Seidel, R.W.; Kubik, S. A cyclopeptide-derived molecular cage for sulfate that closes with a click. Chem. Eur. J. 2010, 16, 7241–7255. [Google Scholar] [CrossRef]
- Bartl, J.; Kubik, S. Anion binding of a cyclopeptide-derived molecular cage in aqueous solvent mixtures. ChemPlusChem 2020, 85, 963–969. [Google Scholar] [CrossRef]
- Rodriguez-Docampo, Z.; Eugenieva-Ilieva, E.; Reyheller, C.; Belenguer, A.M.; Kubik, S.; Otto, S. Dynamic combinatorial development of a neutral synthetic receptor that binds sulfate with nanomolar affinity in aqueous solution. Chem. Commun. 2011, 47, 9798–9800. [Google Scholar] [CrossRef]
- Sommer, F.; Kubik, S. Anion binding of a neutral bis(cyclopeptide) in water/methanol mixtures containing up to 95% of water. Org. Biomol. Chem. 2014, 12, 8851–8860. [Google Scholar] [CrossRef]
- Sommer, F.; Marcus, Y.; Kubik, S. Effects of solvent properties on the anion binding of neutral water-soluble bis(cyclopeptides) in water and aqueous solvent mixtures. ACS Omega 2017, 2, 3669–3680. [Google Scholar] [CrossRef]
- Reyheller, C.; Kubik, S. Selective sensing of sulfate in aqueous solution using a fluorescent bis(cyclopeptide). Org. Lett. 2007, 9, 5271–5274. [Google Scholar] [CrossRef]
- Kubik, S. Anion recognition in aqueous media by cyclopeptides and other synthetic receptors. Acc. Chem. Res. 2017, 50, 2870–2878. [Google Scholar] [CrossRef] [PubMed]
- Bitta, J.; Kubik, S. Cyclic hexapeptides with free carboxylate groups as new receptors for monosaccharides. Org. Lett. 2001, 3, 2637–2640. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, G.; Schellenträger, M.; Kubik, S. An enantioselective fluorescence sensor for glucose based on a cyclic tetrapeptide containing two boronic acid binding sites. Eur. J. Org. Chem. 2006, 4177–4186. [Google Scholar] [CrossRef]
- Bitta, J.; Kubik, S. Complexation of arginine with a cyclopeptide in polar solvents and water. J. Supramol. Chem. 2001, 1, 293–297. [Google Scholar] [CrossRef]
- Schaly, A.; Belda, R.; García-España, E.; Kubik, S. Selective recognition of sulfate anions by a cyclopeptide-derived receptor in aqueous phosphate buffer. Org. Lett. 2013, 15, 6238–6241. [Google Scholar] [CrossRef]
- Pedersen, D.S.; Abell, A. 1,2,3-Triazoles in peptidomimetic chemistry. Eur. J. Org. Chem. 2011, 2399–2411. [Google Scholar] [CrossRef]
- Krause, M.R.; Goddard, R.; Kubik, S. Anion-binding properties of a cyclic pseudohexapeptide containing 1,5-disubstituted 1,2,3-triazole subunits. J. Org. Chem. 2011, 76, 7084–7095. [Google Scholar] [CrossRef]
- Mungalpara, D.; Kelm, H.; Valkonen, A.; Rissanen, K.; Keller, S.; Kubik, S. Oxoanion binding to a cyclic pseudopeptide containing 1,4-disubstituted 1,2,3-triazole moieties. Org. Biomol. Chem. 2017, 15, 102–113. [Google Scholar] [CrossRef]
- Mungalpara, D.; Stegmüller, S.; Kubik, S. A neutral halogen bonding macrocyclic anion receptor based on a pseudocyclopeptide with three 5-iodo-1,2,3-triazole subunits. Chem. Commun. 2017, 53, 5095–5098. [Google Scholar] [CrossRef]
- Mungalpara, D.; Valkonen, A.; Rissanen, K.; Kubik, S. Efficient stabilisation of a dihydrogenphosphate tetramer and a dihydrogenpyrophosphate dimer by a cyclic pseudopeptide containing 1,4-disubstituted 1,2,3-triazole moieties. Chem. Sci. 2017, 8, 6005–6013. [Google Scholar] [CrossRef]
- He, Q.; Tu, P.; Sessler, J.L. Supramolecular chemistry of anionic dimers, trimers, tetramers, and clusters. Chem 2018, 4, 46–93. [Google Scholar] [CrossRef] [PubMed]
- Cousins, G.R.L.; Furlan, R.L.E.; Ng, Y.-F.; Redman, J.E.; Sanders, J.K.M. Identification and isolation of a receptor for N-methyl alkylammonium salts: Molecular amplification in a pseudo-peptide dynamic combinatorial library. Angew. Chem. Int. Ed. 2001, 40, 423–428. [Google Scholar] [CrossRef]
- Furlan, R.L.E.; Ng, Y.-F.; Otto, S.; Sanders, J.K.M. A new cyclic pseudopeptide receptor for Li+ from a dynamic combinatorial library. J. Am. Chem. Soc. 2001, 123, 8876–8877. [Google Scholar] [CrossRef] [PubMed]
- Bulos, F.; Roberts, S.L.; Furlan, R.L.E.; Sanders, J.K.M. Molecular amplification of two different receptors using diastereomeric templates. Chem. Commun. 2007, 3092–3093. [Google Scholar] [CrossRef]
- Lam, R.T.S.; Belenguer, A.; Roberts, S.L.; Naumann, C.; Jarrosson, T.; Otto, S.; Sanders, J.K. Amplification of acetylcholine-binding catenanes from dynamic combinatorial libraries. Science 2005, 308, 667–669. [Google Scholar] [CrossRef]
- Adrián, F.; Burguete, M.I.; Luis, S.V.; Miravet, J.F.; Querol, M.; García-España, E. An efficient β-turn directed cyclization of simple peptidomimetics. Tetrahedron Lett. 1999, 40, 1039–1040. [Google Scholar] [CrossRef]
- Becerril, J.; Bolte, M.; Burguete, M.I.; Galindo, F.; García-España, E.; Luis, S.V.; Miravet, J.F. Efficient macrocyclization of U-turn preorganized peptidomimetics: The role of intramolecular H-bond and solvophobic effects. J. Am. Chem. Soc. 2003, 125, 6677–6686. [Google Scholar] [CrossRef]
- Alfonso, I.; Burguete, M.I.; Luis, S.V. A hydrogen-bonding-modulated molecular rotor: Environmental effect in the conformational stability of peptidomimetic macrocyclic cyclophanes. J. Org. Chem. 2006, 71, 2242–2250. [Google Scholar] [CrossRef]
- Alfonso, I.; Burguete, M.I.; Galindo, F.; Luis, S.V.; Vigara, L. Molecular rotors as simple models to study amide NH-aromatic interactions and their role in the folding of peptide-like structures. J. Org. Chem. 2007, 72, 7947–7956. [Google Scholar] [CrossRef]
- Alfonso, I.; Burguete, M.I.; Luis, S.V.; Miravet, J.F.; Seliger, P.; Tomal, E. Silver complexes of peptidomimetic polyazapyridinophanes. The influence of the bonding cavity size and the nature of side chains. Org. Biomol. Chem. 2006, 4, 853–859. [Google Scholar] [CrossRef]
- Galindo, F.; Becerril, J.; Burguete, M.I.; Luis, S.V.; Vigara, L. Synthesis and study of a cyclophane displaying dual fluorescence emission: A novel ratiometric sensor for carboxylic acids in organic medium. Tetrahedron Lett. 2004, 45, 1659–1662. [Google Scholar] [CrossRef]
- Alfonso, I.; Burguete, M.I.; Galindo, F.; Luis, S.V.; Vigara, L. Unraveling the molecular recognition of amino acid derivatives by a pseudopeptidic macrocycle: ESI-MS, NMR, fluorescence, and modeling studies. J. Org. Chem. 2009, 74, 6130–6142. [Google Scholar] [CrossRef] [PubMed]
- Martí-Centelles, V.; Izquierdo, M.A.; Burguete, M.I.; Galindo, F.; Luis, S.V. Recognition of free tryptophan in water by synthetic pseudopeptides: Fluorescence and thermodynamic studies. Chem. Eur. J. 2014, 20, 7465–7478. [Google Scholar] [CrossRef] [PubMed]
- Martí-Centelles, V.; Burguete, M.I.; Galindo, F.; Izquierdo, M.A.; Kumar, D.K.; White, A.J.P.; Luis, S.V.; Vilar, R. Fluorescent acridine-based receptors for H2PO4–. J. Org. Chem. 2012, 77, 490–500. [Google Scholar] [CrossRef]
- Esteve, F.; Altava, B.; Bolte, M.; Burguete, M.I.; García-Verdugo, E.; Luis, S.V. Highly selective anion template effect in the synthesis of constrained pseudopeptidic macrocyclic cyclophanes. J. Org. Chem. 2020, 85, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Esteve, F.; Altava, B.; Burguete, M.I.; Bolte, M.; García-Verdugo, E.; Luis, S.V. Pseudopeptidic macrocycles as cooperative minimalistic synzyme systems for the remarkable activation and conversion of CO2 in the presence of the chloride anion. Green Chem. 2020, 22, 4697–4705. [Google Scholar] [CrossRef]
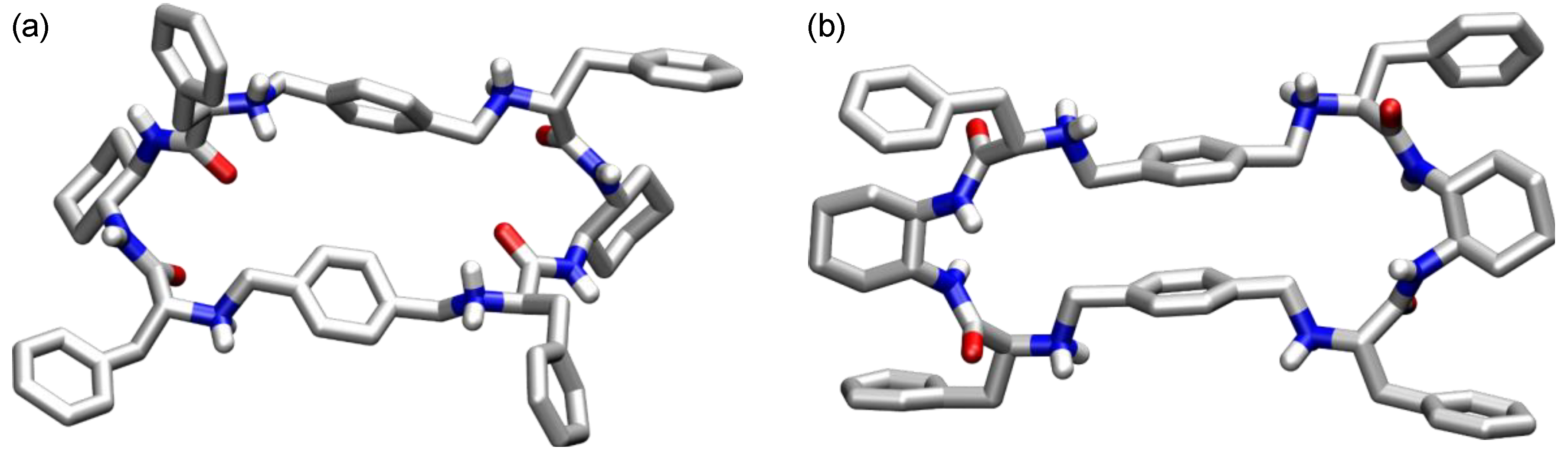
- Bru, M.; Alfonso, I.; Burguete, M.I.; Luis, S.V. Efficient syntheses of new chiral peptidomimetic macrocycles through a configurationally driven preorganization. Tetrahedron Lett. 2005, 46, 7781–7785. [Google Scholar] [CrossRef]
- Alfonso, I.; Bolte, M.; Bru, M.; Burguete, M.I.; Luis, S.V. Designed folding of pseudopeptides: The transformation of a configurationally driven preorganization into a stereoselective multicomponent macrocyclization reaction. Chem. Eur. J. 2008, 14, 8879–8891. [Google Scholar] [CrossRef]
- Bru, M.; Alfonso, I.; Bolte, M.; Burguete, M.I.; Luis, S.V. Structurally disfavoured pseudopeptidic macrocycles through anion templation. Chem. Commun. 2011, 47, 283–285. [Google Scholar] [CrossRef]
- Bru, M.; Alfonso, I.; Burguete, I.; Luis, S.V. Anion-templated syntheses of pseudopeptidic macrocycles. Angew. Chem. Int. Ed. 2006, 45, 6155–6159. [Google Scholar] [CrossRef]
- Alfonso, I.; Bolte, M.; Bru, M.; Burguete, M.I.; Luis, S.V.; Rubio, J. Supramolecular control for the modular synthesis of pseudopeptidic macrocycles through an anion-templated reaction. J. Am. Chem. Soc. 2008, 130, 6137–6144. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, I.; Bolte, M.; Bru, M.; Burguete, M.I.; Luis, S.V.; Vicent, C. Molecular recognition of N-protected dipeptides by pseudopeptidic macrocycles: A comparative study of the supramolecular complexes by ESI-MS and NMR. Org. Biomol. Chem. 2010, 8, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, I.; Bolte, M.; Bru, M.; Burguete, M.I.; Luis, S.V. Crystal structures of the HCl salts of pseudopeptidic macrocycles display “knobs into holes” hydrophobic interactions between aliphatic side chains. CrystEngComm 2009, 11, 735–738. [Google Scholar] [CrossRef]
- Alfonso, I.; Bru, M.; Burguete, M.I.; García-Verdugo, E.; Luis, S.V. Structural diversity in the self-assembly of pseudopeptidic macrocycles. Chem. Eur. J. 2010, 16, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Wagler, T.R.; Fang, Y.; Burrows, C.J. Optically active difunctionalized dioxocyclam macrocycles: Ligands for nickel-catalyzed oxidation of alkenes. J. Org. Chem. 1989, 54, 1584–1589. [Google Scholar] [CrossRef]
- Hopkins, R.B.; Albert, J.S.; Van Engen, D.; Hamilton, A.D. Synthesis and structure of chiral macrocycles containing 2,2’-bipyridine subunits. Bioorg. Med. Chem. 1996, 4, 1121–1128. [Google Scholar] [CrossRef]
- Hu, H.-Y.; Chen, C.-F. A new fluorescent chemosensor for anion based on an artificial cyclic tetrapeptide. Tetrahedron Lett. 2006, 47, 175–179. [Google Scholar] [CrossRef]
- Kataev, E.A.; Shumilova, T.A. Investigation of structural mimetics of natural phosphate ion binding motifs. Molecules 2015, 20, 3354–3370. [Google Scholar] [CrossRef]
- Smolyar, I.V.; Yudin, A.K.; Nenajdenko, V.G. Heteroaryl rings in peptide macrocycles. Chem. Rev. 2019, 119, 10032–10240. [Google Scholar] [CrossRef]
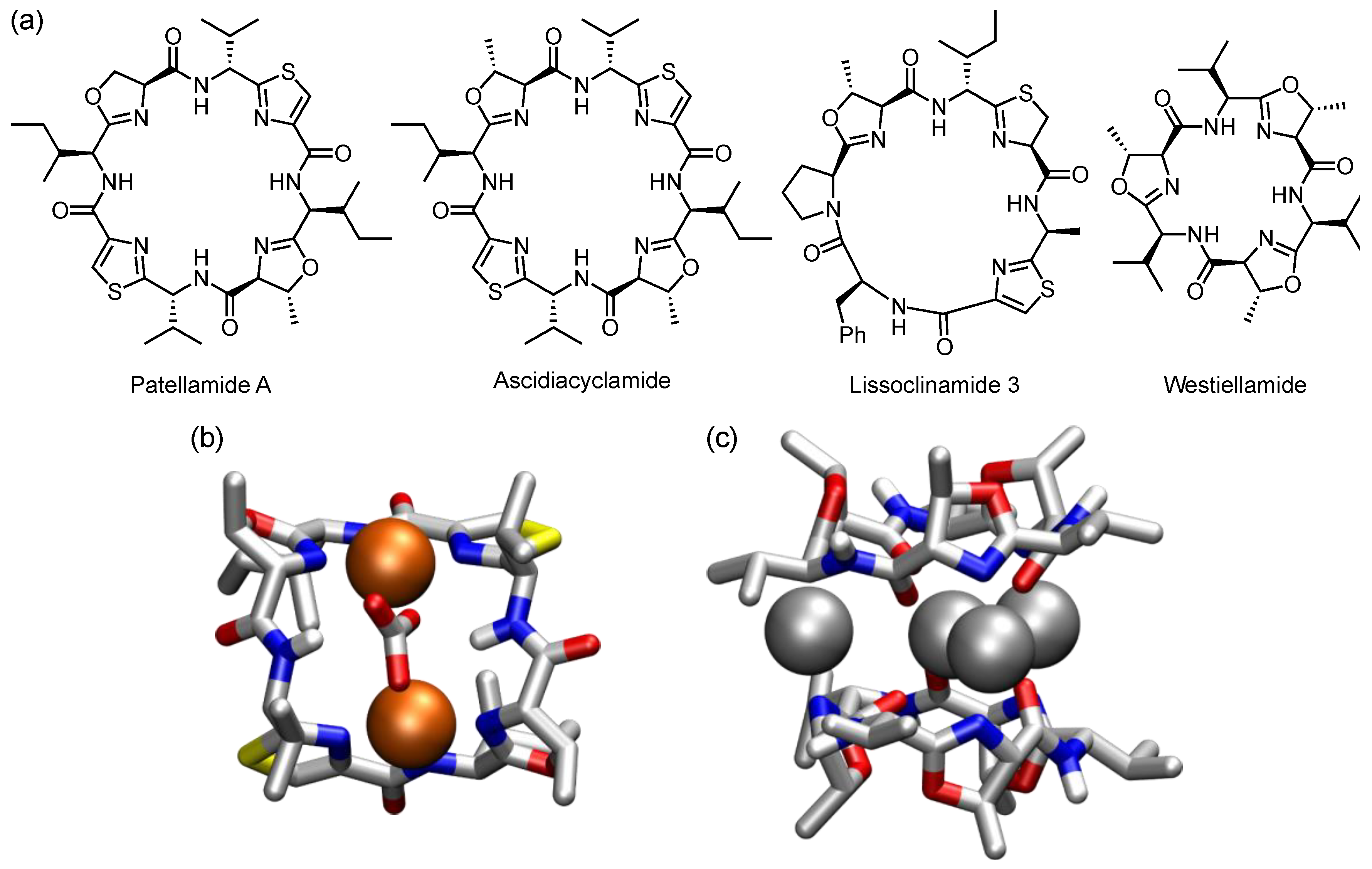
- Freeman, D.J.; Pattenden, G.; Drake, A.F.; Siligardi, G. Marine metabolites and metal ion chelation. Circular dichroism studies of metal binding to Lissoclinum cyclopeptides. J. Chem. Soc. Perkin Trans. 1998, 2, 129–135. [Google Scholar] [CrossRef]
- Bertram, A.; Pattenden, G. Marine metabolites: Metal binding and metal complexes of azole-based cyclic peptides of marine origin. Nat. Prod. Rep. 2007, 24, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Michael, J.P.; Pattenden, G. Marine metabolites and metal ion chelation: The facts and the fantasies. Angew. Chem. Int. Ed. Engl. 1993, 32, 1–23. [Google Scholar] [CrossRef]
- van den Brenk, A.L.; Byriel, K.A.; Fairlie, D.P.; Gahan, L.R.; Hanson, G.R.; Hawkins, C.J.; Jones, A.; Kennard, C.H.L.; Moubaraki, B.; Murray, K.S. Crystal structure and electrospray ionization mass spectrometry, electron paramagnetic resonance, and magnetic susceptibility study of [Cu2(ascidH2)(1,2-μ-CO3)(H2O)2]·2H2O, the bis(copper(II)) complex of ascidiacyclamide (ascidH4), a cyclic peptide isolated from the ascidian Lissoclinum patella. Inorg. Chem. 1994, 33, 3549–3557. [Google Scholar] [CrossRef]
- Wipf, P.; Venkatraman, S.; Miller, C.P.; Geib, S.J. Metal complexes of marine peptide metabolites: A novel Ag4 cluster. Angew. Chem. Int. Ed. 1994, 33, 1516–1518. [Google Scholar] [CrossRef]
- Comba, P.; Dovalil, N.; Gahan, L.R.; Hanson, G.R.; Westphal, M. Cyclic peptide marine metabolites and CuII. Dalton Trans. 2014, 43, 1935–1956. [Google Scholar] [CrossRef]
- Comba, P.; Gahan, L.R.; Haberhauer, G.; Hanson, G.R.; Noble, C.J.; Seibold, B.; van den Brenk, A.L. Copper(II) coordination chemistry of westiellamide and its imidazole, oxazole, and thiazole analogues. Chem. Eur. J. 2008, 14, 4393–4403. [Google Scholar] [CrossRef] [PubMed]
- Comba, P.; Dovalil, N.; Haberhauer, G.; Hanson, G.R.; Kato, Y.; Taura, T. Complex formation and stability of westiellamide derivatives with copper(II). J. Biol. Inorg. Chem. 2010, 15, 1129–1135. [Google Scholar] [CrossRef]
- Comba, P.; Dovalil, N.; Gahan, L.R.; Haberhauer, G.; Hanson, G.R.; Noble, C.J.; Seibold, B.; Vadivelu, P. Cu(II) coordination chemistry of patellamide derivatives: Possible biological functions of cyclic pseudopeptides. Chem. Eur. J. 2012, 18, 2578–2590. [Google Scholar] [CrossRef]
- Comba, P.; Dovalil, N.; Haberhauer, G.; Kowski, K.; Mehrkens, N.; Westphal, M. Copper solution chemistry of cyclic pseudo-octapeptides. Z. Anorg. Allg. Chem. 2013, 639, 1395–1400. [Google Scholar] [CrossRef]
- Cusack, R.M.; Grøndahl, L.; Abbenante, G.; Fairlie, D.P.; Gahan, L.R.; Hanson, G.R.; Hambley, T.W. Conformations of cyclic octapeptides and the influence of heterocyclic ring constraints upon calcium binding. J. Chem. Soc. Perkin Trans. 2000, 2, 323–331. [Google Scholar] [CrossRef]
- Cusack, R.M.; Grøndahl, L.; Fairlie, D.P.; Gahan, L.R.; Hanson, G.R. Cyclic octapeptides containing thiazole. Effect of stereochemistry and degree of flexibility on calcium binding properties. J. Chem. Soc. Perkin Trans. 2002, 2, 556–563. [Google Scholar] [CrossRef]
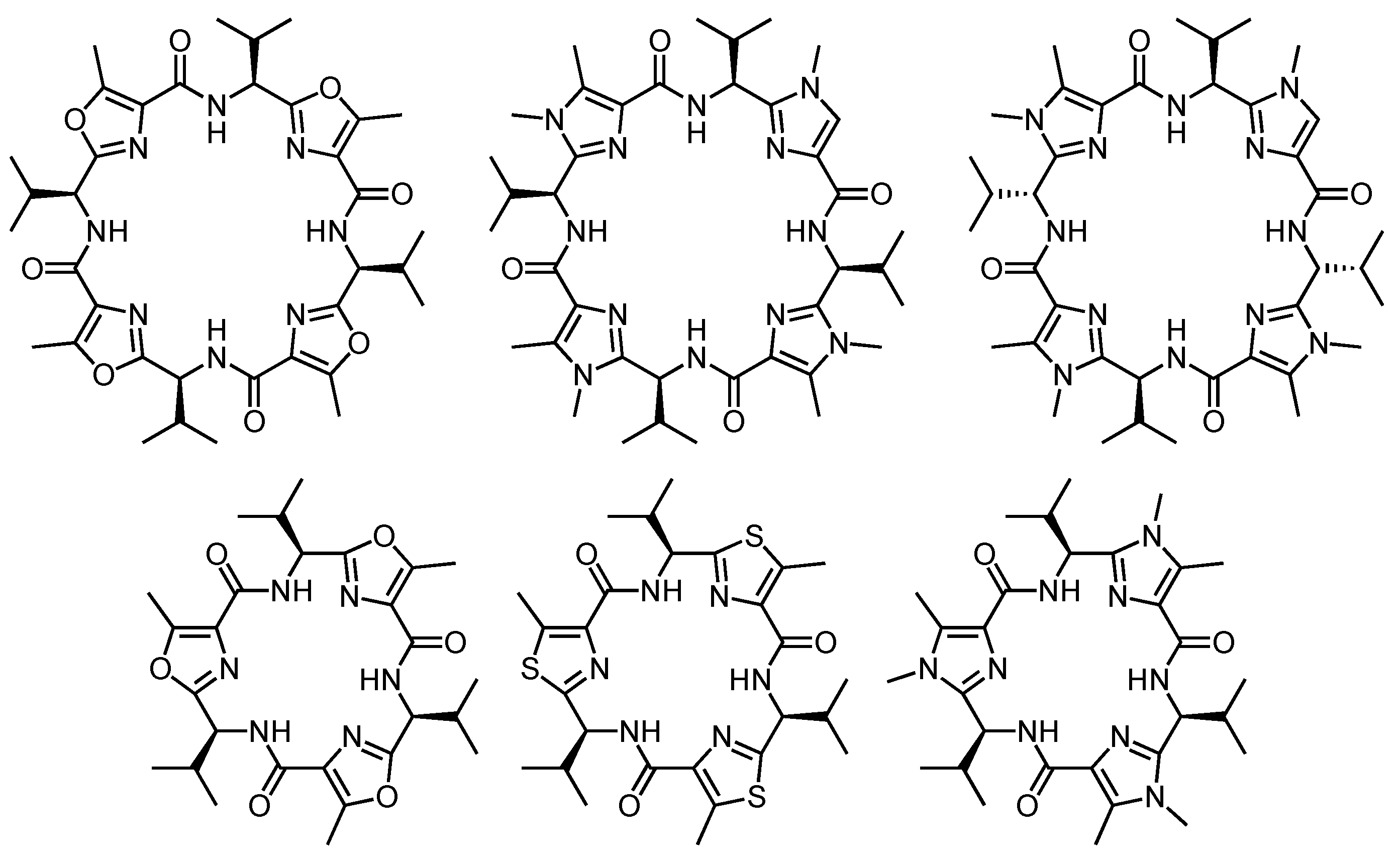
- Haberhauer, G.; Pintér, Á. Thieme Chemistry journal awardees—Where are they now? Macrocyclic peptide chemistry inspired by nature—From chiral artificial receptors toward molecular devices. Synlett 2009, 3082–3098. [Google Scholar] [CrossRef]
- Somogyi, L.; Haberhauer, G.; Rebek, J., Jr. Improved synthesis of functionalized molecular platforms related to marine cyclopeptides. Tetrahedron 2001, 57, 1699–1708. [Google Scholar] [CrossRef]
- Haberhauer, G.; Rominger, H. Synthesis of a new class of imidazole-based cyclic peptides. Tetrahedron Lett. 2002, 43, 6335–6338. [Google Scholar] [CrossRef]
- Haberhauer, G.; Rominger, F. Syntheses and structures of imidazole analogues of Lissoclinum cyclopeptides. Eur. J. Org. Chem. 2003, 3209–3218. [Google Scholar] [CrossRef]
- Haberhauer, G.; Pintér, Á.; Oeser, T.; Rominger, F. Synthesis and structural investigation of C4- and C2-symmetric molecular scaffolds based on imidazole peptides. Eur. J. Org. Chem. 2007, 1779–1792. [Google Scholar] [CrossRef]
- Haberhauer, G.; Drosdow, E.; Oeser, T.; Rominger, F. Structural investigation of westiellamide analogues. Tetrahedron 2008, 64, 1853–1859. [Google Scholar] [CrossRef]
- Pintér, Á.; Haberhauer, G. Synthesis of chiral threefold and sixfold functionalized macrocyclic imidazole peptides. Tetrahedron 2009, 65, 2217–2225. [Google Scholar] [CrossRef]
- Haberhauer, G.; Oeser, T.; Rominger, F. A C3-symmetric molecular scaffold for the construction of large receptors. Chem. Commun. 2004, 2044–2045. [Google Scholar] [CrossRef]
- Haberhauer, G.; Oeser, T.; Rominger, F. A widely applicable concept for predictable induction of preferred configuration in C3-symmetric systems. Chem. Commun. 2005, 2799–2801. [Google Scholar] [CrossRef]
- Haberhauer, G. Control of helicity in C3-symmetric systems by peptide-like β-turns. Tetrahedron Lett. 2008, 49, 2421–2424. [Google Scholar] [CrossRef]
- Pintér, À.; Haberhauer, G. Oxazole cyclopeptides for chirality transfer in C3-symmetric octahedral metal complexes. Eur. J. Org. Chem. 2008, 2375–2387. [Google Scholar] [CrossRef]
- Ziegler, E.; Haberhauer, G. Controlling the helicity of hydroxyquinoline metal complexes based on a macrocyclic peptide scaffold. Eur. J. Org. Chem. 2009, 3432–3438. [Google Scholar] [CrossRef]
- Haberhauer, G. Control of planar chirality: The construction of a copper-ion-controlled chiral molecular hinge. Angew. Chem. Int. Ed. 2008, 47, 3635–3638. [Google Scholar] [CrossRef] [PubMed]
- Ernst, S.; Haberhauer, G. A unidirectional open-close mechanism of metal-ion-driven molecular hinges with adjustable amplitude. Chem. Eur. J. 2009, 15, 13406–13416. [Google Scholar] [CrossRef] [PubMed]
- Haberhauer, G.; Kallweit, C. A bridged azobenzene derivative as a reversible, light-induced chirality switch. Angew. Chem. Int. Ed. 2010, 49, 2418–2421. [Google Scholar] [CrossRef]
- Tepper, C.; Haberhauer, G. Unidirectional redox-stimulated movement around a C-C single bond. Chem. Eur. J. 2011, 17, 8060–8065. [Google Scholar] [CrossRef]
- Kallweit, C.; Haberhauer, G.; Woitschetzki, S. 4,4′-Bipyridine as a unidirectional switching unit for a molecular pushing motor. Chem. Eur. J. 2014, 20, 6358–6365. [Google Scholar] [CrossRef]
- Haberhauer, G. A metal-ion-driven supramolecular chirality pendulum. Angew. Chem. Int. Ed. 2010, 49, 9286–9289. [Google Scholar] [CrossRef]
- Haberhauer, G. A molecular four-stroke motor. Angew. Chem. Int. Ed. 2011, 50, 6415–6418. [Google Scholar] [CrossRef]
- Schnopp, M.; Haberhauer, G. Highly selective recognition of α-chiral primary organoammonium ions by C3-symmetric peptide receptors. Eur. J. Org. Chem. 2009, 4458–4467. [Google Scholar] [CrossRef]
- Schnopp, M.; Ernst, S.; Haberhauer, G. Anion recognition by neutral macrocyclic azole amides. Eur. J. Org. Chem. 2009, 213–222. [Google Scholar] [CrossRef]
- Jolliffe, K.A. Pyrophosphate recognition and sensing in water using bis[zinc(iII)dipicolylamino]-functionalized peptides. Acc. Chem. Res. 2017, 50, 2254–2263. [Google Scholar] [CrossRef]
- Liu, X.; Ngo, H.T.; Ge, Z.; Butler, S.J.; Jolliffe, K.A. Tuning colourimetric indicator displacement assays for naked-eye sensing of pyrophosphate in aqueous media. Chem. Sci. 2013, 4, 1680–1686. [Google Scholar] [CrossRef]
- Butler, S.J.; Jolliffe, K.A. Selective pyrophosphate recognition by cyclic peptide receptors in physiological saline. Chem. Asian J. 2012, 7, 2621–2628. [Google Scholar] [CrossRef]
- Liu, X.; Smith, D.G.; Jolliffe, K.A. Are two better than one? Comparing intermolecular and intramolecular indicator displacement assays in pyrophosphate sensors. Chem. Commun. 2016, 52, 8463–8466. [Google Scholar] [CrossRef]
- Young, P.G.; Jolliffe, K.A. Selective recognition of sulfate ions by tripodal cyclic peptides functionalised with (thio)urea binding sites. Org. Biomol. Chem. 2012, 10, 2664–2672. [Google Scholar] [CrossRef]
- Dungan, V.J.; Ngo, H.T.; Young, P.G.; Jolliffe, K.A. High affinity sulfate binding in aqeous media by cyclic peptides with thiourea arms. Chem. Commun. 2013, 49, 264–266. [Google Scholar] [CrossRef]
- Chakraborty, T.K.; Tapadar, S.; Kumar, S.K. Cyclic trimer of 5-(aminomethyl)-2-furancarboxylic acid as a novel synthetic receptor for carboxylate recognition. Tetrahedron Lett. 2002, 43, 1317–1320. [Google Scholar] [CrossRef]
- Chakraborty, T.K.; Tapadar, S.; Raju, T.V.; Annapurna, J.; Singh, H. Cyclic trimers of chiral furan amino acids. Synlett 2004, 2484–2488. [Google Scholar] [CrossRef]
- Molina, L.; Moreno-Clavijo, E.; Moreno-Vargas, A.J.; Carmona, A.T.; Robina, I. Synthesis of a C3-symmetric furyl-cyclopeptide platform with anion recognition properties. Eur. J. Org. Chem. 2010, 4049–4055. [Google Scholar] [CrossRef]
- Albert, D.; Feigel, M. β-Loop, γ-loop, and helical peptide conformations in cyclopeptides containing a steroidal pseudo-amino acid. Helv. Chim. Acta. 1997, 80, 2168–2181. [Google Scholar] [CrossRef]
- Whitmarsh, S.D.; Redmond, A.P.; Sgarlata, V.; Davis, A.P. Cationic cyclocholamides; toroidal facial amphiphiles with potential for anionic transport. Chem. Commun. 2008, 3669–3671. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Widanapathirana, L.; Zhao, Y. Water-templated transmembrane nanopores from shape-persistent oligocholate macrocycles. J. Am. Chem. Soc. 2011, 133, 141–147. [Google Scholar] [CrossRef]
- Widanapathirana, L.; Zhao, Y. Aromatically functionalized cyclic tricholate macrocycles: Aggregation, transmembrane pore formation, flexibility, and cooperativity. J. Org. Chem. 2012, 77, 4679–4687. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kang, S.-W.; Gothard, C.M.; Maitra, S.; Atia-tul-Wahab; Nowick, J.S. A new class of macrocyclic receptors from iota-peptides. J. Am. Chem. Soc. 2007, 129, 1486–1487. [Google Scholar] [CrossRef] [PubMed]
- Gothard, C.M.; Nowick, J.S. Nanometer-scale water-soluble macrocycles from nanometer-sized amino acids. J. Org. Chem. 2010, 75, 1822–1830. [Google Scholar] [CrossRef][Green Version]
- Choi, K.; Hamilton, A.D. Elective anion binding by a macrocycle with convergent hydrogen bonding functionality. J. Am. Chem. Soc. 2001, 123, 2456–2457. [Google Scholar] [CrossRef]
- Choi, K.; Hamilton, A.D. Rigid macrocyclic triamides as anion receptors: Anion-dependent binding stoichiometries and 1H chemical shift changes. J. Am. Chem. Soc. 2003, 125, 10241–10249. [Google Scholar] [CrossRef]
- Jain, R.K.; Tsou, L.K.; Hamilton, A.D. Combined solid/solution phase synthesis of large surface area scaffolds derived from aminomethyl-benzoates. Tetrahedron Lett. 2009, 50, 2787–2789. [Google Scholar] [CrossRef]
- Meisel, J.W.; Hu, C.T.; Hamilton, A.D. Heterofunctionalized cavitands by macrocyclization of sequence-defined foldamers. Org. Lett. 2019, 21, 7763–7767. [Google Scholar] [CrossRef] [PubMed]
- Jwad, R.; Weissberger, D.; Hunter, L. Strategies for fine-tuning the conformations of cyclic peptides. Chem. Rev. 2020, 120, 9743–9789. [Google Scholar] [CrossRef] [PubMed]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubik, S. Synthetic Receptors Based on Abiotic Cyclo(pseudo)peptides. Molecules 2022, 27, 2821. https://doi.org/10.3390/molecules27092821
Kubik S. Synthetic Receptors Based on Abiotic Cyclo(pseudo)peptides. Molecules. 2022; 27(9):2821. https://doi.org/10.3390/molecules27092821
Chicago/Turabian StyleKubik, Stefan. 2022. "Synthetic Receptors Based on Abiotic Cyclo(pseudo)peptides" Molecules 27, no. 9: 2821. https://doi.org/10.3390/molecules27092821
APA StyleKubik, S. (2022). Synthetic Receptors Based on Abiotic Cyclo(pseudo)peptides. Molecules, 27(9), 2821. https://doi.org/10.3390/molecules27092821






