Antioxidant Activity of Ruthenium Cyclopentadienyl Complexes Bearing Succinimidato and Phthalimidato Ligands
Abstract
:1. Introduction
2. Material and Methods
2.1. Chemicals
2.2. Cell Culture
2.3. Cell Viability
2.4. Cell Cycle
2.5. Effect of Ruthenium Complexes 1–3 on DNA Oxidative Damage
2.6. Comet Assay
2.7. Comet Analysis
2.8. Measurement of Reactive Oxygen Species
2.9. Measurement of SOD Activity
2.10. Statistical Analysis
3. Results
3.1. Cell Viability
3.2. Cell Cycle
3.3. DNA Oxidative Damage
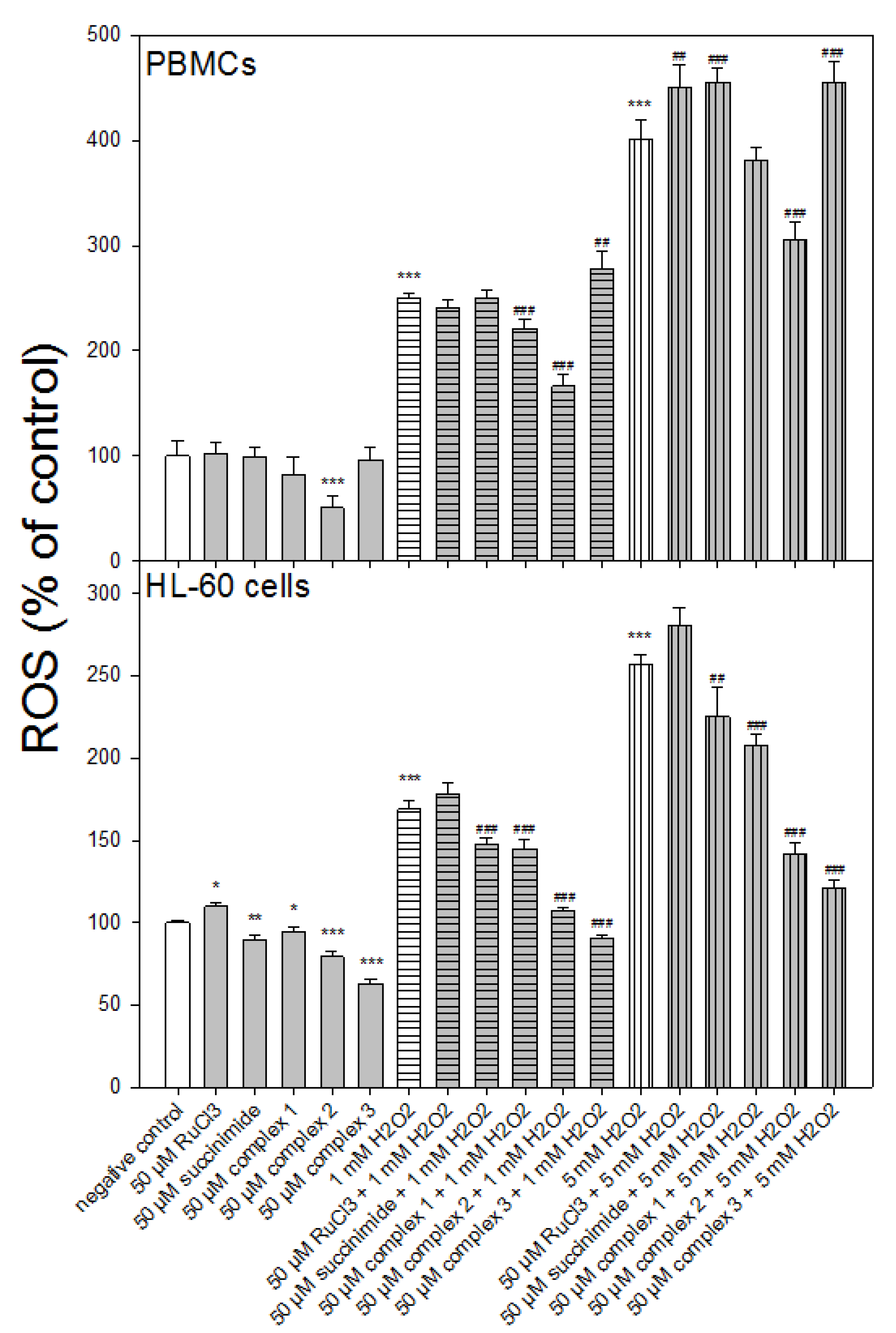
3.4. Reactive Oxygen Species Level
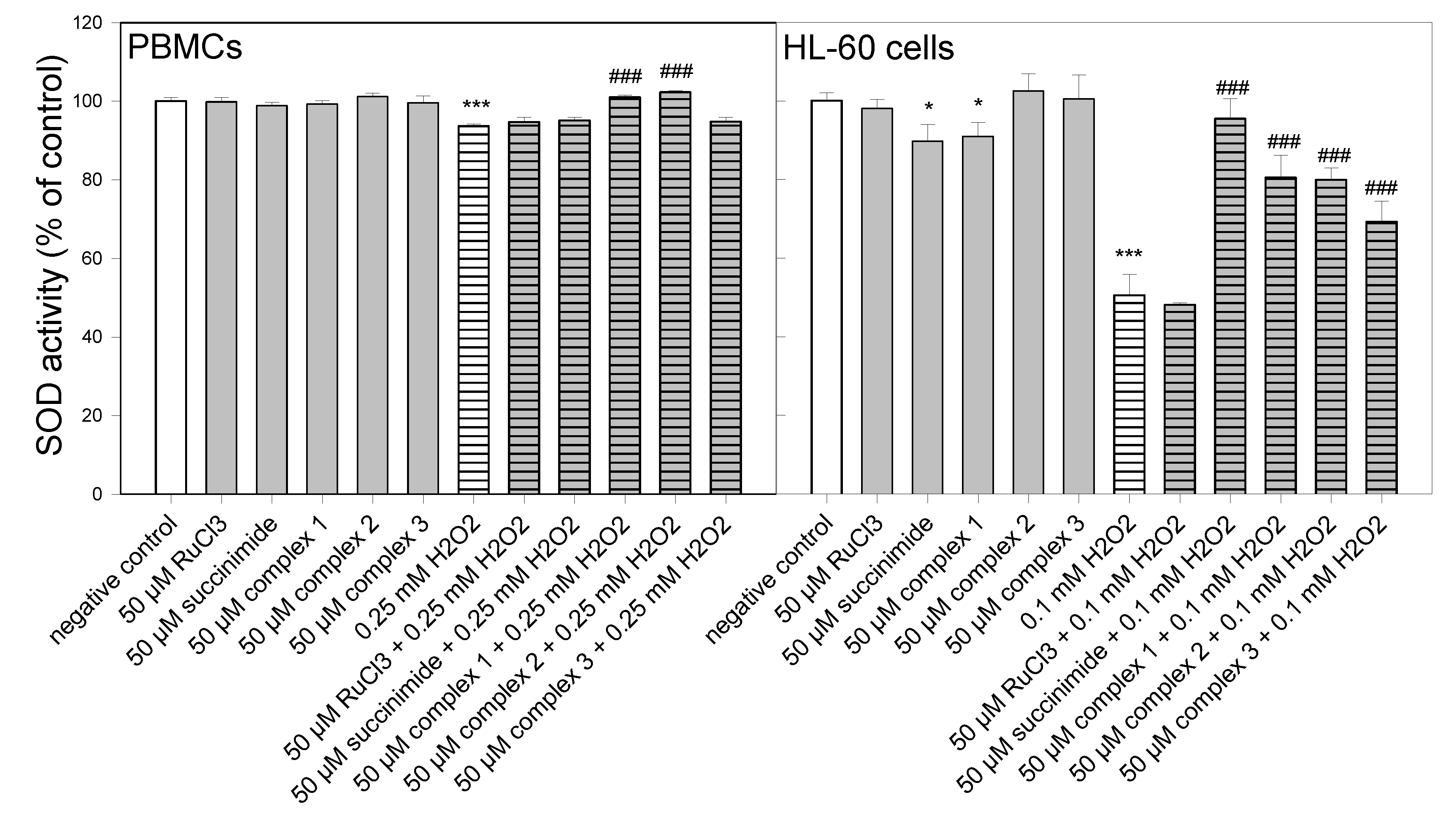
3.5. SOD Activity
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Singh, A.; Barman, P. Recent Advances in Schiff Base Ruthenium Metal Complexes: Synthesis and Applications. Top. Curr. Chem. 2021, 379, 29. [Google Scholar] [CrossRef] [PubMed]
- Kacsir, I.; Sipos, A.; Ujlaki, G.; Buglyó, P.; Somsák, L.; Bai, P.; Bokor, É. Ruthenium Half-Sandwich Type Complexes with Bidentate Monosaccharide Ligands Show Antineoplastic Activity in Ovarian Cancer Cell Models through Reactive Oxygen Species Production. Int. J. Mol. Sci. 2021, 22, 10454. [Google Scholar] [CrossRef]
- Mahmud, K.M.; Niloy, M.S.; Shakil, M.S.; Islam, M.A. Ruthenium Complexes: An Alternative to Platinum Drugs in Colorectal Cancer Treatment. Pharmaceutics 2021, 13, 1295. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Li, Y.; Shi, H.; Wang, Y.; Zhang, J.; Zhang, Q. Ruthenium Complexes as Promising Candidates against Lung Cancer. Molecules 2021, 26, 4389. [Google Scholar] [CrossRef] [PubMed]
- Juszczak, M.; Kluska, M.; Kosińska, A.; Palusiak, M.; Rybarczyk-Pirek, A.J.; Wzgarda-Raj, K.; Rudolf, B.; Woźniak, K. Cytotoxicity of piano-stool ruthenium cyclopentadienyl complexes bearing different imidato ligands. Appl. Organomet. Chem. 2022, 36, e6595. [Google Scholar] [CrossRef]
- Mu, C.; Prosser, K.E.; Harrypersad, S.; MacNeil, G.A.; Panchmatia, R.; Thompson, J.R.; Sinha, S.; Warren, J.J.; Walsby, C.J. Activation by Oxidation: Ferrocene-Functionalized Ru(II)-Arene Complexes with Anticancer, Antibacterial, and Antioxidant Properties. Inorg. Chem. 2018, 57, 15247–15261. [Google Scholar] [CrossRef]
- Buldurun, K.; Turan, N.; Aras, A.; Mantarcı, A.; Turkan, F.; Bursal, E. Spectroscopic and Structural Characterization, Enzyme Inhibitions, and Antioxidant Effects of New Ru(II) and Ni(II) Complexes of Schiff Base. Chem. Biodivers. 2019, 16, e1900243. [Google Scholar] [CrossRef]
- Mohankumar, A.; Devagi, G.; Shanmugam, G.; Nivitha, S.; Sundararaj, P.; Dallemer, F.; Kalaivani, P.; Prabhakaran, R. Organoruthenium(II) complexes attenuate stress in Caenorhabditis elegans through regulating antioxidant machinery. Eur. J. Med. Chem. 2019, 168, 123–133. [Google Scholar] [CrossRef]
- Sasahara, G.L.; Gouveia Júnior, F.S.; Rodrigues, R.O.; Zampieri, D.S.; Fonseca, S.; Gonçalves, R.C.R.; Athaydes, B.R.; Kitagawa, R.R.; Santos, F.A.; Sousa, E.H.S.; et al. Nitro-imidazole-based ruthenium complexes with antioxidant and anti-inflammatory activities. J. Inorg. Biochem. 2020, 206, 111048. [Google Scholar] [CrossRef]
- Nehru, S.; Veeralakshmi, S.; Kalaiselvam, S.; Subin David, S.P.; Sandhya, J.; Arunachalam, S. Protein binding and antioxidant studies of diimine based emissive surfactant-ruthenium(II) complexes. J. Biomol. Struct. Dyn. 2021, 39, 1535–1546. [Google Scholar] [CrossRef]
- Maikoo, S.; Chakraborty, A.; Vukea, N.; Dingle, L.M.K.; Samson, W.J.; de la Mare, J.A.; Edkins, A.L.; Booysen, I.N. Ruthenium complexes with mono- or bis-heterocyclic chelates: DNA/BSA binding, antioxidant and anticancer studies. J. Biomol. Struct. Dyn. 2021, 39, 4077–4088. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, S.A.; Badr, H.E.; di Biase, A.; El-Hendawy, A.M. Synthesis, characterization of ruthenium(II), nickel(II), palladium(II), and platinum(II) triphenylphosphine-based complexes bearing an ONS-donor chelating agent: Interaction with biomolecules, antioxidant, in vitro cytotoxic, apoptotic activity and cell cycle analysis. J. Inorg. Biochem. 2021, 223, 111549. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.E.; Braga, S.S. Redesigning Nature: Ruthenium Flavonoid Complexes with Antitumour, Antimicrobial and Cardioprotective Activities. Molecules 2021, 26, 4544. [Google Scholar] [CrossRef] [PubMed]
- Cuccioloni, M.; Bonfili, L.; Mozzicafreddo, M.; Cecarini, V.; Pettinari, R.; Condello, F.; Pettinari, C.; Marchetti, F.; Angeletti, M.; Eleuteri, A.M. A ruthenium derivative of quercetin with enhanced cholesterol-lowering activity. RSC Adv. 2016, 6, 39636–39641. [Google Scholar] [CrossRef]
- Ravishankar, D.; Salamah, M.; Attina, A.; Pothi, R.; Vallance, T.M.; Javed, M.; Williams, H.F.; Alzahrani, E.M.S.; Kabova, E.; Vaiyapuri, R.; et al. Ruthenium-conjugated chrysin analogues modulate platelet activity, thrombus formation and haemostasis with enhanced efficacy. Sci. Rep. 2017, 7, 5738. [Google Scholar] [CrossRef] [Green Version]
- Mabuza, L.P.; Gamede, M.W.; Maikoo, S.; Booysen, I.N.; Ngubane, P.S.; Khathi, A. Cardioprotective effects of a ruthenium (II) Schiff base complex in diet-induced prediabetic rats. Diabetes Metab. Syndr. Obes. 2019, 12, 217–223. [Google Scholar] [CrossRef] [Green Version]
- Sadiq, A.; Mahmood, F.; Ullah, F.; Ayaz, M.; Ahmad, S.; Haq, F.U.; Khan, G.; Jan, M.S. Synthesis, anticholinesterase and antioxidant potentials of ketoesters derivatives of succinimides: A possible role in the management of Alzheimer’s. Chem. Cent. J. 2015, 9, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, F.; Khan, Z.; Jan, M.S.; Ahmad, S.; Ahmad, A.; Rashid, U.; Ullah, F.; Ayaz, M.; Sadiq, A. Synthesis, in-vitro α-glucosidase inhibition, antioxidant, in-vivo antidiabetic and molecular docking studies of pyrrolidine-2,5-dione and thiazolidine-2,4-dione derivatives. Bioorg. Chem. 2019, 91, 103128. [Google Scholar] [CrossRef]
- Ahmad, A.; Ullah, F.; Sadiq, A.; Ayaz, M.; Saeed Jan, M.; Shahid, M.; Wadood, A.; Mahmood, F.; Rashid, U.; Ullah, R.; et al. Comparative Cholinesterase, α-Glucosidase Inhibitory, Antioxidant, Molecular Docking, and Kinetic Studies on Potent Succinimide Derivatives. Drug Des. Dev. Ther. 2020, 14, 2165–2178. [Google Scholar] [CrossRef]
- Waheed, B.; Mukarram Shah, S.M.; Hussain, F.; Khan, M.I.; Zeb, A.; Jan, M.S. Synthesis, Antioxidant, and Antidiabetic Activities of Ketone Derivatives of Succinimide. Evid. Based Complement. Altern. Med. 2022, 2022, 1445604. [Google Scholar] [CrossRef]
- Kubicka, A.; Fomal, E.; Olejniczak, A.B.; Rybarczyk-Pirek, A.J.; Wojtulewski, S.; Rudolf, B. Oxa-Michael reaction of metallocarbonyl complexes bearing the maleimidato ligand. Reactivity studies with selected hydroxy compounds. Polyhedron 2016, 107, 38–47. [Google Scholar] [CrossRef]
- Kosińska, A.; Wojtulewski, S.; Palusiak, M.; Tokarz, P.; Rudolf, B. A Useful Synthetic Route to N-Nonsubstituted Succinimides via Light-Induced Degradation of Metallocarbonyl Complexes. Organometallics 2021, 40, 663–673. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, P.; Piastowska-Ciesielska, A.W.; Kaarniranta, K.; Blasiak, J. All-Trans Retinoic Acid Modulates DNA Damage Response and the Expression of the VEGF-A and MKI67 Genes in ARPE-19 Cells Subjected to Oxidative Stress. Int. J. Mol. Sci. 2016, 17, 898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampson, J.B.; Beckman, J.S. Hydrogen peroxide damages the zinc-binding site of zinc-deficient Cu,Zn superoxide dismutase. Arch Biochem. Biophys. 2001, 392, 8–13. [Google Scholar] [CrossRef]
- Emamgholipour, S.; Hossein-Nezhad, A.; Ansari, M. Can Melatonin Act as an Antioxidant in Hydrogen Peroxide-Induced Oxidative Stress Model in Human Peripheral Blood Mononuclear Cells? Biochem. Res. Int. 2016, 2016, 5857940. [Google Scholar] [CrossRef] [Green Version]
- Bekeschus, S.; Liebelt, G.; Menz, J.; Singer, D.; Wende, K.; Schmidt, A. Cell cycle-related genes associate with sensitivity to hydrogen peroxide-induced toxicity. Redox Biol. 2022, 50, 102234. [Google Scholar] [CrossRef]
- Heo, S.; Kim, S.; Kang, D. The Role of Hydrogen Peroxide and Peroxiredoxins throughout the Cell Cycle. Antioxidants 2020, 9, 280. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Sohn, J.; Lee, J.H.; Lee, K.C.; Son, C.S.; Tockgo, Y.C. Regulation of bcl-2 family in hydrogen peroxide-induced apoptosis in human leukemia HL-60 cells. Exp. Mol. Med. 2000, 32, 42–46. [Google Scholar] [CrossRef] [Green Version]
- Bejarano, I.; Espino, J.; González-Flores, D.; Casado, J.G.; Redondo, P.C.; Rosado, J.A.; Barriga, C.; Pariente, J.A.; Rodríguez, A.B. Role of Calcium Signals on Hydrogen Peroxide-Induced Apoptosis in Human Myeloid HL-60 Cells. Int. J. Biomed. Sci. IJBS 2009, 5, 246–256. [Google Scholar]
- Barnes, J.L.; Zubair, M.; John, K.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA damage. Biochem. Soc. Trans. 2018, 46, 1213–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juszczak, M.; Kluska, M.; Wysokiński, D.; Woźniak, K. DNA damage and antioxidant properties of CORM-2 in normal and cancer cells. Sci. Rep. 2020, 10, 12200. [Google Scholar] [CrossRef] [PubMed]
- Wrona, M.; Patel, K.; Wardman, P. Reactivity of 2′,7′-dichlorodihydrofluorescein and dihydrorhodamine 123 and their oxidized forms toward carbonate, nitrogen dioxide, and hydroxyl radicals. Free Radic. Biol. Med. 2005, 38, 262–270. [Google Scholar] [CrossRef]
- Reiniers, M.J.; de Haan, L.R.; Reeskamp, L.F.; Broekgaarden, M.; van Golen, R.F.; Heger, M. Analysis and Optimization of Conditions for the Use of 2′,7′-Dichlorofluorescein Diacetate in Cultured Hepatocytes. Antioxidants 2021, 10, 674. [Google Scholar] [CrossRef] [PubMed]
- Gottfredsen, R.H.; Larsen, U.G.; Enghild, J.J.; Petersen, S.V. Hydrogen peroxide induce modifications of human extracellular superoxide dismutase that results in enzyme inhibition. Redox Biol. 2013, 1, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Lewandowski, Ł.; Kepinska, M.; Milnerowicz, H. Inhibition of copper-zinc superoxide dismutase activity by selected environmental xenobiotics. Environ. Toxicol. Pharmacol. 2018, 58, 105–113. [Google Scholar] [CrossRef]
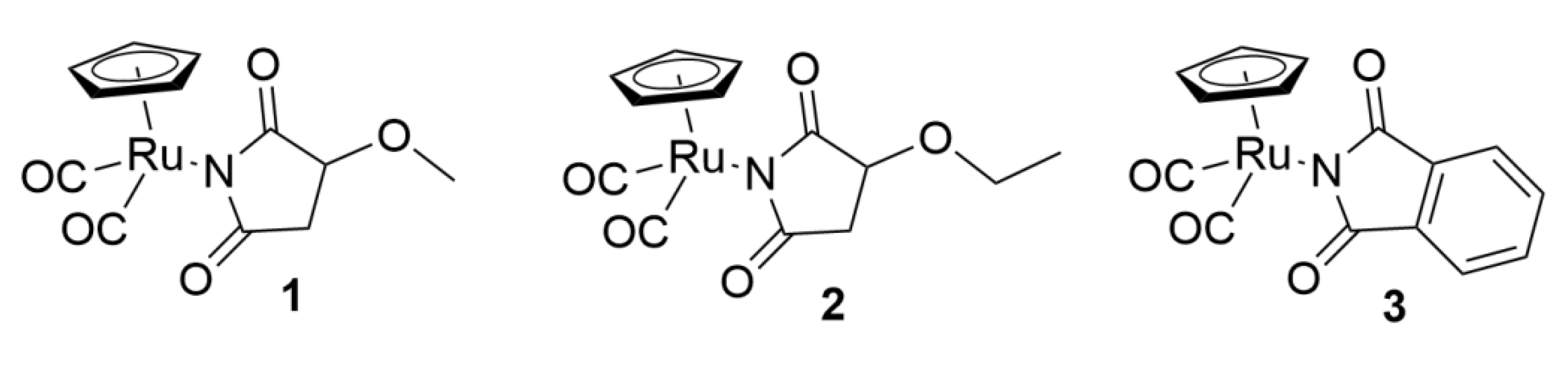


| Treatment | DNA Content % | |||
|---|---|---|---|---|
| Sub-G1 | G0/G1 | S | G2/M | |
| negative control | 1.19 ± 0.14 | 35.38 ± 1.64 | 25.66 ± 0.11 | 37.21 ± 1.72 |
| positive control (NOC 100 ng/mL) | 9.93 ± 2.46 ***↑ | 22.36 ± 6.91 *↓ | 21.09 ± 3.53 | 47.8 ± 4.22 *↑ |
| 0.1 mM H2O2 | 10.39 ± 0.37 ***↑ | 11.27 ± 0.88 ***↓ | 32.73 ± 1.27 ***↑ | 44.79 ± 0.42 **↑ |
| 0.2 mM H2O2 | 19.83 ± 0.76 ***↑ | 27.43 ± 0.62 **↓ | 23.82 ± 2.54 | 28.84 ± 2.38 **↓ |
| RuCl3 | 2.02 ± 0.43 | 35.28 ± 2.03 | 29.87 ± 0.17 | 32.5 ± 2.42 |
| RuCl3 + 0.1 mM H2O2 | 4.73 ± 0.55 ###↓ | 39.25 ± 0.4 ###↑ | 26.12 ± 0.35 ###↓ | 30.02 ± 0.79 ###↓ |
| RuCl3 + 0.2 mM H2O2 | 14.01 ± 0.98 ##↓ | 29.15 ± 1.48 | 11.6 ± 0.31 ###↓ | 45.37 ± 1.28 ###↑ |
| succinimide | 2.31 ± 0.33 | 44.42 ± 3.01 *↑ | 28.48 ± 2.02 | 24.17 ± 1.63 ***↓ |
| succinimide + 0.1 mM H2O2 | 2.07 ± 0.86 ###↓ | 41.95 ± 3.29 ###↑ | 22.63 ± 3.7 ###↓ | 33.17 ± 1.92 ###↓ |
| succinimide + 0.2 mM H2O2 | 5.12 ± 0.84 ###↓ | 27.34 ± 2.58 | 10.76 ± 0.15 ###↓ | 56.63 ± 1.66 ###↑ |
| complex 1 | 1.76 ± 0.13 | 48.81 ± 3.48 **↑ | 27.59 ± 3.97 | 21.48 ± 1.00 ***↓ |
| complex 1 + 0.1 mM H2O2 | 2.22 ± 0.36 ###↓ | 41.71 ± 1.21 ###↑ | 20.29 ± 2.95 ###↓ | 35.32 ± 1.31 ###↓ |
| complex 1 + 0.2 mM H2O2 | 5.34 ± 1.42 ###↓ | 29.82 ± 3.78 | 9.72 ± 0.29 ###↓ | 54.96 ± 2.42 ###↑ |
| complex 2 | 1.69 ± 0.13 | 49.3 ± 3.58 **↑ | 26.85 ± 3.54 | 21.24 ± 0.29 ***↓ |
| complex 2 + 0.1 mM H2O2 | 2.5 ± 0.56 ###↓ | 40.92 ± 1.8 ###↑ | 20.59 ± 2.28 ###↓ | 35.59 ± 0.53 ###↓ |
| complex 2 + 0.2 mM H2O2 | 6.64 ± 1.05 ###↓ | 27.92 ± 0.87 | 11.3 ± 0.62 ###↓ | 54.24 ± 0.92 ###↑ |
| complex 3 | 1.76 ± 0.17 | 51.4 ± 0.78 ***↑ | 25.21 ± 0.66 | 21.65 ± 0.38 ***↓ |
| complex 3 + 0.1 mM H2O2 | 2.25 ± 0.15 ###↓ | 43.67 ± 1.54 ###↑ | 18.11 ± 1.76 ###↓ | 35.86 ± 1.68 ###↓ |
| complex 3 + 0.2 mM H2O2 | 4.95 ± 1.23 ###↓ | 29.35 ± 4.14 | 9.5 ± 1.06 ###↓ | 56.07 ± 2.53 ###↑ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juszczak, M.; Kluska, M.; Kosińska, A.; Rudolf, B.; Woźniak, K. Antioxidant Activity of Ruthenium Cyclopentadienyl Complexes Bearing Succinimidato and Phthalimidato Ligands. Molecules 2022, 27, 2803. https://doi.org/10.3390/molecules27092803
Juszczak M, Kluska M, Kosińska A, Rudolf B, Woźniak K. Antioxidant Activity of Ruthenium Cyclopentadienyl Complexes Bearing Succinimidato and Phthalimidato Ligands. Molecules. 2022; 27(9):2803. https://doi.org/10.3390/molecules27092803
Chicago/Turabian StyleJuszczak, Michał, Magdalena Kluska, Aneta Kosińska, Bogna Rudolf, and Katarzyna Woźniak. 2022. "Antioxidant Activity of Ruthenium Cyclopentadienyl Complexes Bearing Succinimidato and Phthalimidato Ligands" Molecules 27, no. 9: 2803. https://doi.org/10.3390/molecules27092803
APA StyleJuszczak, M., Kluska, M., Kosińska, A., Rudolf, B., & Woźniak, K. (2022). Antioxidant Activity of Ruthenium Cyclopentadienyl Complexes Bearing Succinimidato and Phthalimidato Ligands. Molecules, 27(9), 2803. https://doi.org/10.3390/molecules27092803








