Predictive Models of Phytosterol Degradation in Rapeseeds Stored in Bulk Based on Artificial Neural Networks and Response Surface Regression
Abstract
:1. Introduction
2. Results
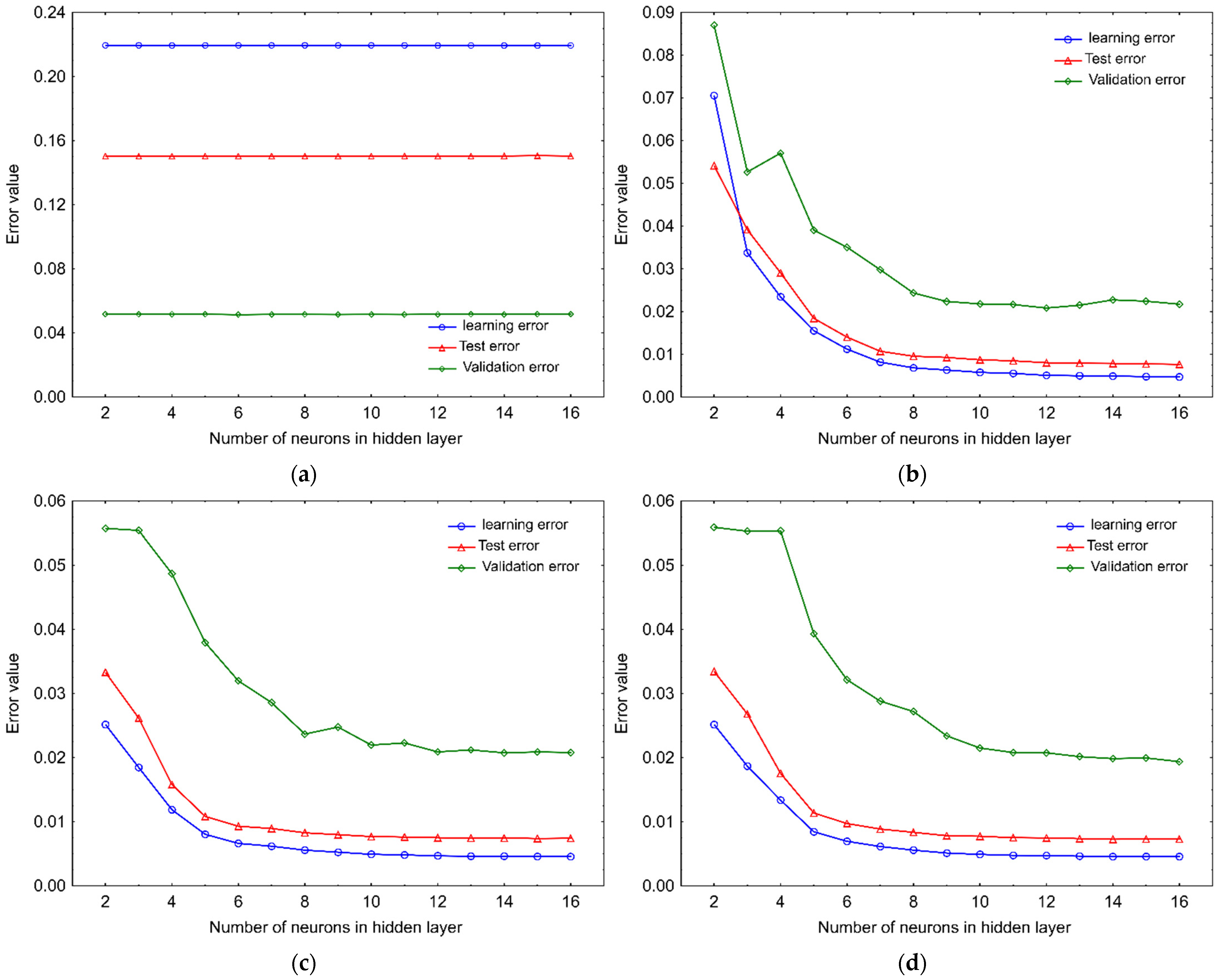
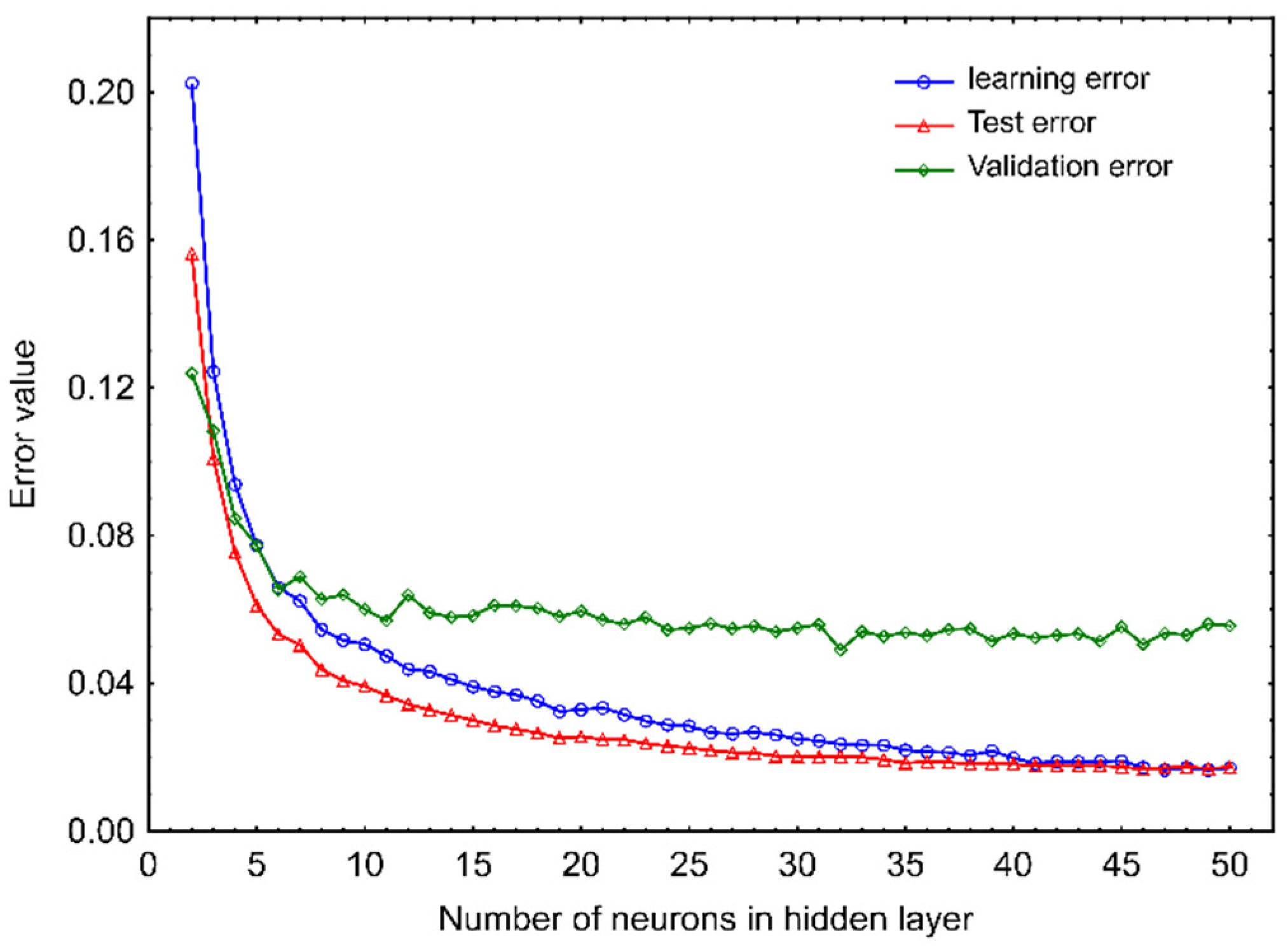
2.1. Artificial Neural Network Model
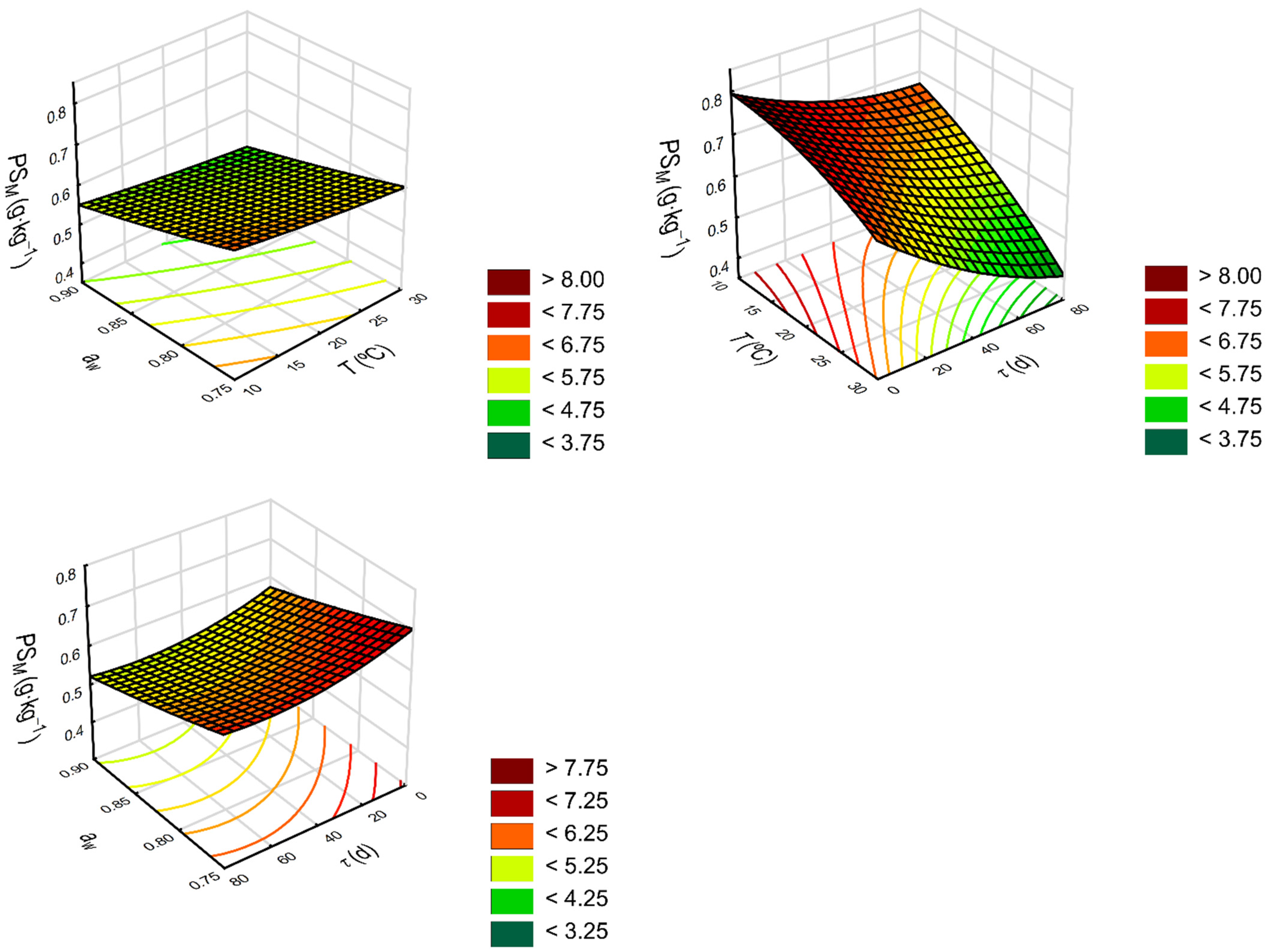
2.2. Response Surface Regression Model
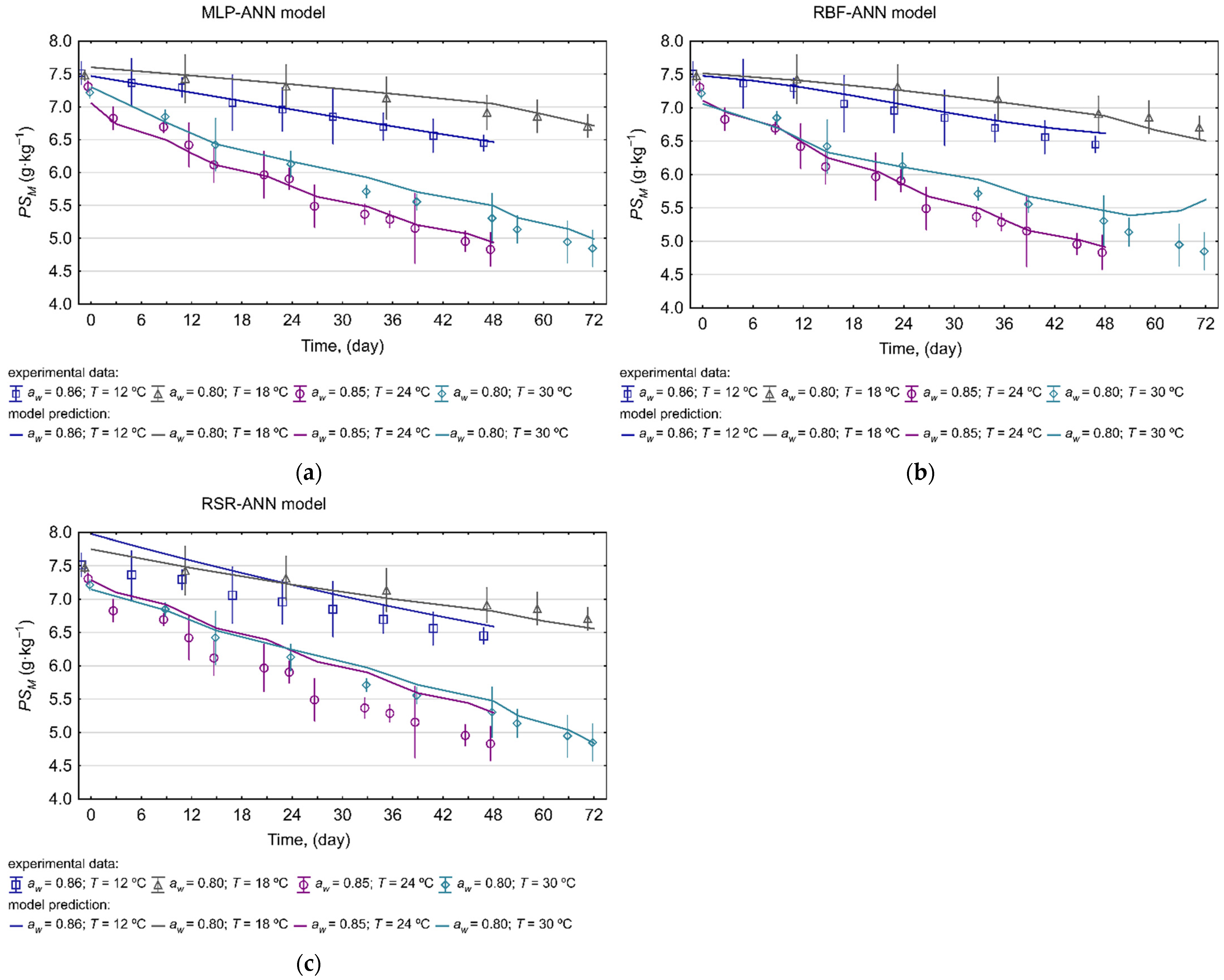
2.3. Model Performance Evaluation
3. Materials and Methods
3.1. Experimental Data Collection
Rapeseed Preparation and Experimental Design
3.2. Modeling Process
3.2.1. Data Sets
3.2.2. Artificial Neural Network Model Development
3.2.3. Response Surface Regression Modeling
3.2.4. Model Performance Assessment
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Lagarda, M.J.; García-Llatas, G.; Farré, R. Analysis of phytosterols in foods. J. Pharm. Biomed. Anal. 2006, 41, 1486–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Callaghan, J.; Coghlan, A.; Abbas, A.; García-Estrada, C.; Martín, J.F.; Dobson, A.D.W. Functional characterization of the polyketide synthase gene required for ochratoxin A biosynthesis in Penicillium verrucosum. Int. J. Food Microbiol. 2013, 161, 172–181. [Google Scholar] [CrossRef] [PubMed]
- De Smet, E.; Mensink, R.P.; Plat, J. Effects of plant sterols and stanols on intestinal cholesterol metabolism: Suggested mechanisms from past to present. Mol. Nutr. Food Res. 2012, 56, 1058–1072. [Google Scholar] [CrossRef] [PubMed]
- FAO Standard for Named Vegetable Oils CXS 210-1999. Available online: https://www.fao.org/3/y2774e/y2774e04.htm#bm4.1 (accessed on 1 February 2022).
- Moreau, R.A.; Nyström, L.; Whitaker, B.D.; Winkler-Moser, J.K.; Baer, D.J.; Gebauer, S.K.; Hicks, K.B. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and health-promoting uses. Prog. Lipid Res. 2018, 70, 35–61. [Google Scholar] [CrossRef]
- Moreau, R.A.; Whitaker, B.D.; Hicks, K.B. Phytosterols, phytostanols, and their conjugates in foods: Structural diversity, quantitative analysis, and health-promoting uses. Prog. Lipid Res. 2002, 41, 457–500. [Google Scholar] [CrossRef]
- Wawrzyniak, J.; Gawrysiak-Witulska, M.; Rudzińska, M. Dynamics of phytosterol degradation in a bulk of rapeseed stored under different temperature and humidity conditions. J. Stored Prod. Res. 2019, 83, 292–304. [Google Scholar] [CrossRef]
- García-Llatas, G.; Rodríguez-Estrada, M.T. Current and new insights on phytosterol oxides in plant sterol-enriched food. Chem. Phys. Lipids 2011, 164, 607–624. [Google Scholar] [CrossRef]
- Jaceldo-Siegl, K.; Lütjohann, D.; Sirirat, R.; Mashchak, A.; Fraser, G.E.; Haddad, E. Variations in dietary intake and plasma concentrations of plant sterols across plant-based diets among North American adults. Mol. Nutr. Food Res. 2017, 61, 1600828. [Google Scholar] [CrossRef]
- Ras, R.T.; Trautwein, E.A. Consumer purchase behaviour of foods with added phytosterols in six European countries: Data from a post-launch monitoring survey. Food Chem. Toxicol. 2017, 110, 42–48. [Google Scholar] [CrossRef]
- Botelho, P.B.; Galasso, M.; Dias, V.; Mandrioli, M.; Lobato, L.P.; Rodriguez-Estrada, M.T.; Castro, I.A. Oxidative stability of functional phytosterol-enriched dark chocolate. LWT Food Sci. Technol. 2014, 55, 444–451. [Google Scholar] [CrossRef] [Green Version]
- Sujith Kumar, M.S.; Mawlong, I.; Singh, D. Phytosterol recovery from oilseeds: Recent advances. J. Food Process Eng. 2017, 40, e12466. [Google Scholar] [CrossRef]
- Yang, R.; Xue, L.; Zhang, L.; Wang, X.; Qi, X.; Jiang, J.; Yu, L.; Wang, X.; Zhang, W.; Zhang, Q.; et al. Phytosterol contents of edible oils and their contributions to estimated phytosterol intake in the Chinese diet. Foods 2019, 8, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlahakis, C.; Hazebroek, J. Phytosterol accumulation in canola, sunflower, and soybean oils: Effects of genetics, planting location, and temperature. JAOCS J. Am. Oil Chem. Soc. 2000, 77, 49–53. [Google Scholar] [CrossRef]
- Maguire, L.; Konoplyannikov, M.; Ford, A.; Maguire, A.R.; O’Brien, N.M. Comparison of the cytotoxic effects of β-sitosterol oxides and a cholesterol oxide, 7β-hydroxycholesterol, in cultured mammalian cells. Br. J. Nutr. 2003, 90, 767–775. [Google Scholar] [CrossRef] [Green Version]
- Rudzinska, M.; Jelen, H.; Nogala-Kalucka, M.; Gawrysiak-Witulska, M. The influence of storage time and drying temperature on sterols content in seeds of rapeseed. Rośliny Oleiste Oilseed Crop. 2006, 27, 345–356. [Google Scholar]
- Gawrysiak-Witulska, M.; Rudzińska, M. Degradation of phytosterols during near-ambient drying of rapeseeds in a thick immobile layer. JAOCS J. Am. Oil Chem. Soc. 2012, 89, 1681–1689. [Google Scholar] [CrossRef] [Green Version]
- Gawrysiak-Witulska, M.; Rudzińska, M.; Siger, A.; Bartkowiak-Broda, I. A high drying temperature causes degradation of sterols and tocopherols in yellow-seeded Brassica napus oils. Eur. J. Lipid Sci. Technol. 2015, 117, 483–490. [Google Scholar] [CrossRef]
- Chelladurai, V.; Jian, F.; Jayas, D.S.; White, N.D.G.; Fields, P.G.; Manickavasagan, A. Feasibility of storing canola at different moisture contents in silo bags under Canadian Prairie conditions. Can. Biosyst. Eng. Genie Biosyst. Can. 2016, 58, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Gawrysiak-Witulska, M.; Wawrzyniak, J.; Ryniecki, A.; Perkowski, J. Relationship of ergosterol content and fungal contamination and assessment of technological quality of malting barley preserved in a metal silo using the near-ambient method. J. Stored Prod. Res. 2008, 44, 360–365. [Google Scholar] [CrossRef]
- Ryniecki, A.; Gawrysiak-Witulska, M.; Wawrzyniak, J. Correlation for the automatic identification of drying endpoint in near-ambient dryers: Application to malting barley. Biosyst. Eng. 2007, 98, 437–445. [Google Scholar] [CrossRef]
- Gawrysiak-Witulska, M.; Siger, A.; Wawrzyniak, J.; Nogala-Kalucka, M. Changes in tocochromanol content in seeds of Brassica napus L. during adverse conditions of storage. J. Am. Oil Chem. Soc. 2011, 88, 1379–1385. [Google Scholar] [CrossRef]
- Wawrzyniak, J.; Gawrysiak-Witulska, M.; Ryniecki, A. Management Control Points Related to the Lag Phase of Fungal Growth in a Stored Rapeseed Ecosystem. JAOCS J. Am. Oil Chem. Soc. 2018, 95, 1223–1235. [Google Scholar] [CrossRef]
- Wawrzyniak, J.; Kaczyński, Ł.K.; Cais-Sokolińska, D.; Wójtowski, J. Mathematical modelling of ethanol production as a function of temperature during lactic-alcoholic fermentation of goat’s milk after hydrolysis and transgalactosylation of lactose. Meas. J. Int. Meas. Confed. 2019, 135, 287–293. [Google Scholar] [CrossRef]
- Wawrzyniak, J. Prediction of fungal infestation in stored barley ecosystems using artificial neural networks. LWT 2021, 137, 110367. [Google Scholar] [CrossRef]
- Wawrzyniak, J. Model of Fungal Development in Stored Barley Ecosystems as a Prognostic Auxiliary Tool for Postharvest Preservation Systems. Food Bioprocess Technol. 2021, 14, 298–309. [Google Scholar] [CrossRef]
- Wawrzyniak, J. A Predictive Model for Assessment of the Risk of Mold Growth in Rapeseeds Stored in a bulk as a Decision Support Tool for Postharvest Management Systems. J. Am. Oil Chem. Soc. 2020, 97, 915–927. [Google Scholar] [CrossRef]
- Przybył, K.; Wawrzyniak, J.; Koszela, K.; Adamski, F.; Gawrysiak-Witulska, M. Application of deep and machine learning using image analysis to detect fungal contamination of rapeseed. Sensors 2020, 20, 7305. [Google Scholar] [CrossRef]
- Gomes, V.; Rendall, R.; Reis, M.S.; Mendes-Ferreira, A.; Melo-Pinto, P. Determination of sugar, pH, and anthocyanin contents in port wine grape berries through hyperspectral imaging: An extensive comparison of linear and non-linear predictive methods. Appl. Sci. 2021, 11, 10319. [Google Scholar] [CrossRef]
- Sharabiani, V.R.; Kaveh, M.; Taghinezhad, E.; Abbaszadeh, R.; Khalife, E.; Szymanek, M.; Dziwulska-Hunek, A. Application of Artificial Neural Networks, Support Vector, Adaptive Neuro-Fuzzy Inference Systems for the Moisture Ratio of Parboiled Hulls. Appl. Sci. 2022, 12, 1771. [Google Scholar] [CrossRef]
- Nosratimovafagh, A.; Fereidouni, A.E.; Krujatz, F. Modeling and Optimizing the Effect of Light Color, Sodium Chloride and Glucose Concentration on Biomass Production and the Quality of Arthrospira platensis Using Response Surface Methodology (RSM). Life 2022, 12, 371. [Google Scholar] [CrossRef]
- Nuapia, Y.; Cukrowska, E.; Tutu, H.; Chimuka, L. Statistical comparison of two modeling methods on pressurized hot water extraction of vitamin C and phenolic compounds from Moringa oleifera leaves. S. Afr. J. Bot. 2020, 129, 9–16. [Google Scholar] [CrossRef]
- Yu, H.C.; Huang, S.M.; Lin, W.M.; Kuo, C.H.; Shieh, C.J. Comparison of artificial neural networks and response surface methodology towards an efficient ultrasound-assisted extraction of chlorogenic acid from Lonicera japonica. Molecules 2019, 24, 2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghanta, S.; Bhaumik, C.; Manna, M.S. Process development for isolation of dietary eugenol from leaves of basil (Ocimum sanctum) in combination of optimization of process variables and modeling by artificial neural network. J. Indian Chem. Soc. 2022, 99, 100280. [Google Scholar] [CrossRef]
- Silva, S.F.; Anjos, C.A.R.; Cavalcanti, R.N.; Celeghini, R.M.D.S. Evaluation of extra virgin olive oil stability by artificial neural network. Food Chem. 2015, 179, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Kuvendziev, S.; Lisichkov, K.; Zeković, Z.; Marinkovski, M. Artificial neural network modelling of supercritical fluid CO2 extraction of polyunsaturated fatty acids from common carp (Cyprinus carpio L.) viscera. J. Supercrit. Fluids 2014, 92, 242–248. [Google Scholar] [CrossRef]
- Said, F.M.; Gan, J.Y.; Sulaiman, J. Correlation between response surface methodology and artificial neural network in the prediction of bioactive compounds of unripe Musa acuminata peel. Eng. Sci. Technol. Int. J. 2020, 23, 781–787. [Google Scholar] [CrossRef]
- Basheer, I.A.; Hajmeer, M. Artificial neural networks: Fundamentals, computing, design, and application. J. Microbiol. Methods 2000, 43, 3–31. [Google Scholar] [CrossRef]
- Panchal, F.S.; Panchal, M. Review on Methods of Selecting Number of Hidden Nodes in Artificial Neural Network. Int. J. Comput. Sci. Mob. Comput. 2014, 3, 455–464. [Google Scholar]
- Mirchandani, G.; Cao, W. On hidden nodes for neural nets. IEEE Trans. Circuits Syst. 1989, 36, 661–664. [Google Scholar] [CrossRef]
- Barron, A.R. Approximation and estimation bounds for artificial neural networks. Mach. Learn. 1994, 14, 115–133. [Google Scholar] [CrossRef]
- Gómez, I.; Franco, L.; Jerez, J.M. Neural network architecture selection: Can function complexity help? Neural Process. Lett. 2009, 30, 71–87. [Google Scholar] [CrossRef]
- Sheela, K.G.; Deepa, S.N. Selection of number of hidden neurons in neural networks in renewable energy systems. J. Sci. Ind. Res. 2014, 73, 686–688. [Google Scholar]
- Mittal, G.S. Chapter 18—Artificial Neural Network (ANN) Based Process Modeling. In Handbook of Farm, Dairy and Food Machinery Engineering, 2nd ed.; Kutz, M., Ed.; Academic Press: San Diego, CA, USA, 2013; pp. 467–473. ISBN 978-0-12-385881-8. [Google Scholar]
- Wawrzyniak, J. Application of artificial neural networks to assess the mycological state of bulk stored rapeseeds. Agriculture 2020, 10, 567. [Google Scholar] [CrossRef]
- Mateo, F.; Gadea, R.; Medina, Á.; Mateo, R.; Jiménez, M. Predictive assessment of ochratoxin A accumulation in grape juice based-medium by Aspergillus carbonarius using neural networks. J. Appl. Microbiol. 2009, 107, 915–927. [Google Scholar] [CrossRef]
- Mateo, F.; Gadea, R.; Mateo, E.M.; Jiménez, M. Multilayer perceptron neural networks and radial-basis function networks as tools to forecast accumulation of deoxynivalenol in barley seeds contaminated with Fusarium culmorum. Food Control 2011, 22, 88–95. [Google Scholar] [CrossRef]
- dos Santos, A.M.P.; Silva, E.F.R.; dos Santos, W.N.L.; da Silva, E.G.P.; dos Santos, L.O.; Bruna, B.R.; Maria, M.C.; dos Santos, W.P.C. Evaluation of minerals, toxic elements and bioactive compounds in rose petals (Rosa spp.) using chemometric tools and artificial neural networks. Microchem. J. 2018, 138, 98–108. [Google Scholar] [CrossRef]
- Jeyamkondan, S.; Jayas, D.S.; Holley, R.A. Microbial growth modelling with artificial neural networks. Int. J. Food Microbiol. 2001, 64, 343–354. [Google Scholar] [CrossRef]
- Te Giffel, M.C.; Zwietering, M.H. Validation of predictive models describing the growth of Listeria monocytogenes. Int. J. Food Microbiol. 1999, 46, 135–149. [Google Scholar] [CrossRef]
- Carbone, K.; Amoriello, T.; Iadecola, R. Exploitation of kiwi juice pomace for the recovery of natural antioxidants through microwave-assisted extraction. Agriculture 2020, 10, 435. [Google Scholar] [CrossRef]
- Yang, Q.-Q.; Gan, R.-Y.; Zhang, D.; Ge, Y.-Y.; Cheng, L.-Z.; Corke, H. Optimization of kidney bean antioxidants using RSM & ANN and characterization of antioxidant profile by UPLC-QTOF-MS. LWT 2019, 114, 108321. [Google Scholar] [CrossRef]
- Cheok, C.Y.; Chin, N.L.; Yusof, Y.A.; Talib, R.A.; Law, C.L. Optimization of total phenolic content extracted from Garcinia mangostana Linn. hull using response surface methodology versus artificial neural network. Ind. Crop. Prod. 2012, 40, 247–253. [Google Scholar] [CrossRef]
- Torrecilla, J.S.; Mena, M.L.; Yáñez-Sedeño, P.; García, J. Field determination of phenolic compounds in olive oil mill wastewater by artificial neural network. Biochem. Eng. J. 2008, 38, 171–179. [Google Scholar] [CrossRef]
- Dos Santos, D.A.; de Lima, K.P.; Consolin, M.F.B.; Filho, N.C.; Março, P.H.; Valderrama, P. Multi-product multivariate calibration: Determination of quality parameters in soybean industrialized juices. Acta Sci. Technol. 2019, 41, e37382. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.N.; Wu, H.L.; Qing, X.D.; Nie, C.C.; Li, S.F.; Yu, Y.J.; Zhang, S.R.; Yu, R.Q. The maintenance of the second-order advantage: Second-order calibration of excitation-emission matrix fluorescence for quantitative analysis of herbicide napropamide in various environmental samples. Talanta 2011, 85, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.S.; Gonzaga, F.B.; da Rocha, W.F.C.; Lima, I.C.A. A comparison of different strategies in multivariate regression models for the direct determination of Mn, Cr, and Ni in steel samples using laser-induced breakdown spectroscopy. Spectrochim. Acta Part B At. Spectrosc. 2018, 139, 20–26. [Google Scholar] [CrossRef]
- de Oliveira Ramos, R.; de Sousa Fernandes, D.D.; de Almeida, V.E.; Gonçalves Dias Diniz, P.H.; Lopes, W.S.; Leite, V.D.; de Araújo, M.C.U. A video processing and machine vision-based automatic analyzer to determine sequentially total suspended and settleable solids in wastewater. Anal. Chim. Acta 2022, 139, 20–26. [Google Scholar] [CrossRef]
- Olivieri, A.C. Practical guidelines for reporting results in single- and multi-component analytical calibration: A tutorial. Anal. Chim. Acta 2015, 868, 10–22. [Google Scholar] [CrossRef]
- Ryan, E.; McCarthy, F.O.; Maguire, A.R.; O’Brien, N.M. Phytosterol Oxidation Products: Their Formation, Occurrence, and Biological Effects. Food Rev. Int. 2009, 25, 157–174. [Google Scholar] [CrossRef]
- ASAE Standards, D245.5; Moisture Relationships of Plant-Based Agricultural Products. ASAE—The Society for Engineering in Agricultural, Food, and Biological Systems: St. Joseph, MI, USA, 2000.
- Wawrzyniak, J.; Ryniecki, A.; Gawrysiak-Witulska, M. Kinetics of mould growth in the stored barley ecosystem contaminated with Aspergillus westerdijkiae, Penicillium viridicatum and Fusarium poae at 23–30 °C. J. Sci. Food Agric. 2013, 93, 895–901. [Google Scholar] [CrossRef]
- Brazauskiene, I.; Petraitiene, E.; Mankeviciene, A. Effects of genotype and environmental factors on rape seed contamination with mycotoxins and mycotoxin-producing fungi. Ekologija 2006, 3, 14–20. [Google Scholar]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- AOCS Official Method Ch 2-91. Determination of the composition of the sterol fraction of animal and vegetable oils and fats by TLC and capillary GLC. In Methods and Recommended Practices of the AOCS, 6th ed.; Firestone, D., Ed.; AOCS Press: Champaign, IL, USA, 1997; pp. 1–5. [Google Scholar]
- Huang, Y. Advances in artificial neural networks—Methodological development and application. Algorithms 2009, 2, 973–1007. [Google Scholar] [CrossRef]
- Prashanth, D.S.; Mehta, R.V.K.; Sharma, N. Classification of Handwritten Devanagari Number—An analysis of Pattern Recognition Tool using Neural Network and CNN. Procedia Comput. Sci. 2020, 167, 2445–2457. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Kodogiannis, V.S. Application of neural networks as a non-linear modelling technique in food mycology. Expert Syst. Appl. 2009, 36, 121–131. [Google Scholar] [CrossRef]
- Keeratipibul, S.; Phewpan, A.; Lursinsap, C. Prediction of coliforms and Escherichia coli on tomato fruits and lettuce leaves after sanitizing by using Artificial Neural Networks. LWT Food Sci. Technol. 2011, 44, 130–138. [Google Scholar] [CrossRef]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Thibault, J.; Grandjean, B.P.A. Neural networks in process control—A survey. In IFAC Symposia Series, Advanced Control of Chemical Processes 1991; Najim, K., Dufour, E., Eds.; Pergamon: Toulouse, France, 1992; pp. 251–260. [Google Scholar]
- González, A.G.; Herrador, M.A.; Asuero, A.G. Intra-laboratory testing of method accuracy from recovery assays. Talanta 1999, 48, 729–736. [Google Scholar] [CrossRef]

| Network Parameters | Artificial Neural Network | |
|---|---|---|
| MLP 3-9-1 | RBF 3-27-1 | |
| Number of observation points (total) | 468 | |
| Learning | 243 | |
| Test | 108 | |
| Validation | 117 | |
| Activation functions in hidden layer | Log | Gau |
| Activation functions in output layer | Lin | Lin |
| Learning error | 0.0047 | 0.0147 |
| Test error | 0.0074 | 0.0153 |
| Validation error | 0.0098 | 0.0213 |
| Learning accuracy | 0.9969 | 0.9903 |
| Test accuracy | 0.9944 | 0.9886 |
| Validation accuracy | 0.9887 | 0.9769 |
| Equation Variable and Intercept | Regression Coefficients | Standard Error | p-Values |
|---|---|---|---|
| Intercept | −55.1677 | 5.6557 | <0.0001 |
| T | 0.9326 | 0.0438 | <0.0001 |
| T2 | −0.0037 | 0.0005 | <0.0001 |
| aw | 132.8619 | 13.3651 | <0.0001 |
| aw2 | −69.4989 | 7.9210 | <0.0001 |
| τ | 0.2277 | 0.0163 | <0.0001 |
| τ2 | 0.0001 | 0.0001 | 0.0129 |
| aw × T | −1.0046 | 0.0464 | <0.0001 |
| T × τ | −0.0013 | 0.0001 | <0.0001 |
| aw × τ | −0.2870 | 0.0176 | <0.0001 |
| Statistical Index | Model | |||||||
|---|---|---|---|---|---|---|---|---|
| MLP-ANN | RBF-ANN | RSR | ||||||
| Data set | B | V | B | V | B | V | ||
| L | T | L | T | |||||
| Coefficient of determination (R2) | 0.994 | 0.989 | 0.978 | 0.981 | 0.977 | 0.954 | 0.930 | 0.933 |
| Root mean square error (RMSE) | 0.097 | 0.122 | 0.140 | 0.172 | 0.175 | 0.206 | 0.320 | 0.302 |
| Mean absolute error (MAE) | 0.080 | 0.099 | 0.117 | 0.128 | 0.143 | 0.148 | 0.242 | 0.248 |
| Mean relative percentage error (MRPE, %) | −0.032 | 0.209 | −0.653 | −0.063 | 0.084 | −1.224 | −0.378 | −3.477 |
| Bias (Bf) | 1.000 | 0.998 | 1.006 | 1.000 | 0.999 | 1.012 | 1.002 | 1.034 |
| Mean absolute relative percentage error (MAPE, %) | 1.384 | 1.658 | 1.922 | 2.270 | 2.486 | 2.516 | 4.215 | 4.121 |
| Accuracy factor (Af) | 1.014 | 1.017 | 1.019 | 1.023 | 1.025 | 1.025 | 1.043 | 1.041 |
| Storage Conditions | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Temperature (T) | T = 12 °C | T = 18 °C | T = 24 °C | T = 30 °C | ||||||||||||
| Water activity (aw) | 0.76 | 0.80 | 0.86 | 0.90 | 0.76 | 0.80 | 0.86 | 0.90 | 0.75 | 0.81 | 0.85 | 0.90 | 0.75 | 0.80 | 0.84 | 0.90 |
| Experiment type | B | B | V | B | B | V | B | B | B | B | V | B | B | V | B | B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wawrzyniak, J.; Rudzińska, M.; Gawrysiak-Witulska, M.; Przybył, K. Predictive Models of Phytosterol Degradation in Rapeseeds Stored in Bulk Based on Artificial Neural Networks and Response Surface Regression. Molecules 2022, 27, 2445. https://doi.org/10.3390/molecules27082445
Wawrzyniak J, Rudzińska M, Gawrysiak-Witulska M, Przybył K. Predictive Models of Phytosterol Degradation in Rapeseeds Stored in Bulk Based on Artificial Neural Networks and Response Surface Regression. Molecules. 2022; 27(8):2445. https://doi.org/10.3390/molecules27082445
Chicago/Turabian StyleWawrzyniak, Jolanta, Magdalena Rudzińska, Marzena Gawrysiak-Witulska, and Krzysztof Przybył. 2022. "Predictive Models of Phytosterol Degradation in Rapeseeds Stored in Bulk Based on Artificial Neural Networks and Response Surface Regression" Molecules 27, no. 8: 2445. https://doi.org/10.3390/molecules27082445
APA StyleWawrzyniak, J., Rudzińska, M., Gawrysiak-Witulska, M., & Przybył, K. (2022). Predictive Models of Phytosterol Degradation in Rapeseeds Stored in Bulk Based on Artificial Neural Networks and Response Surface Regression. Molecules, 27(8), 2445. https://doi.org/10.3390/molecules27082445









