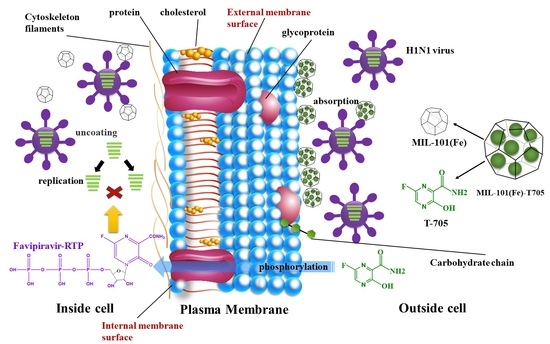
Anti-Influenza Virus Study of Composite Material with MIL-101(Fe)-Adsorbed Favipiravir
Abstract
1. Introduction
2. Experimental Section
2.1. Materials
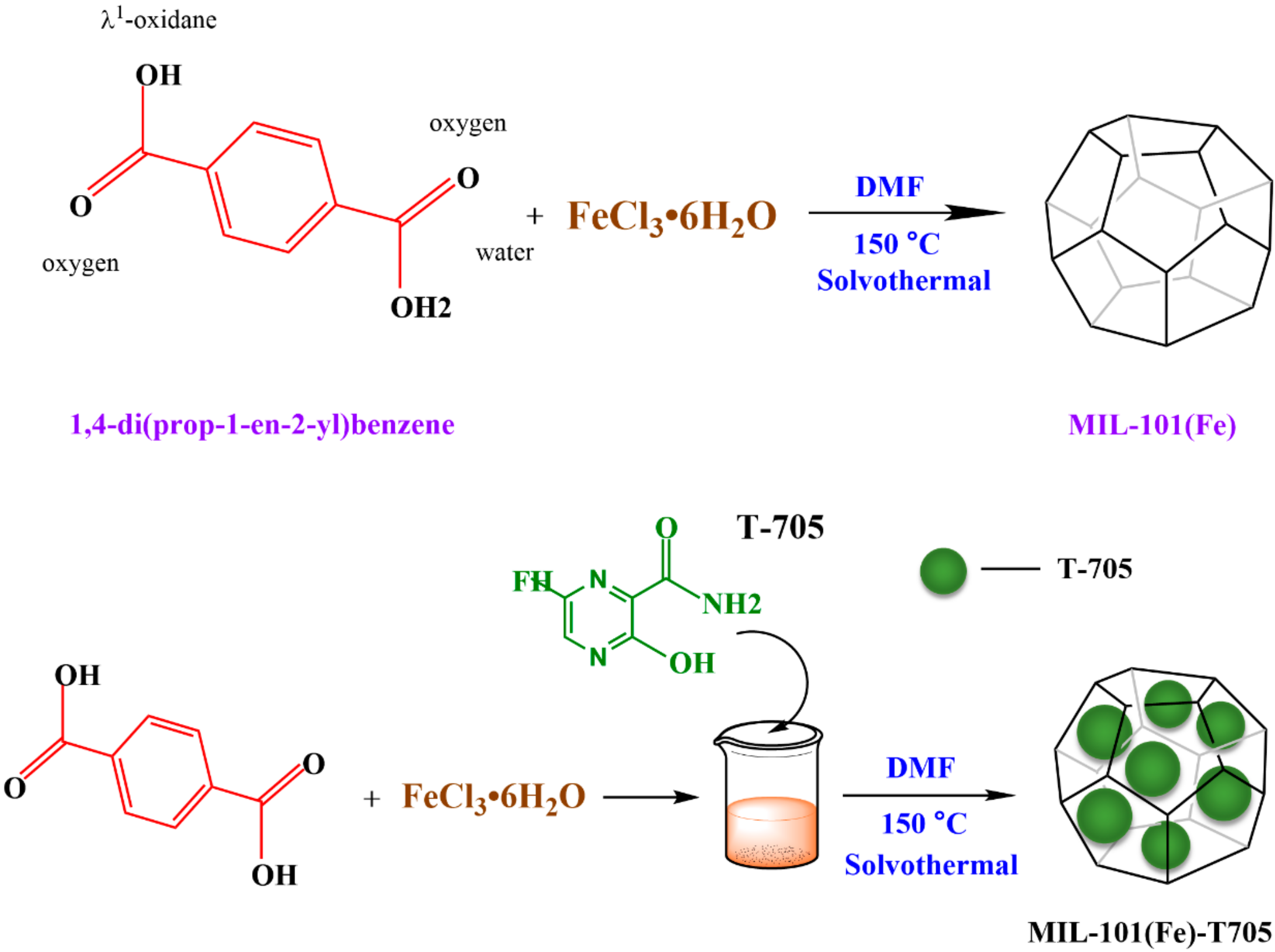
2.2. Synthesis of MIL-101(Fe)
2.3. Synthesis of MIL-101(Fe)-T705 Nanocomposites
2.4. Nanomaterial Characterization
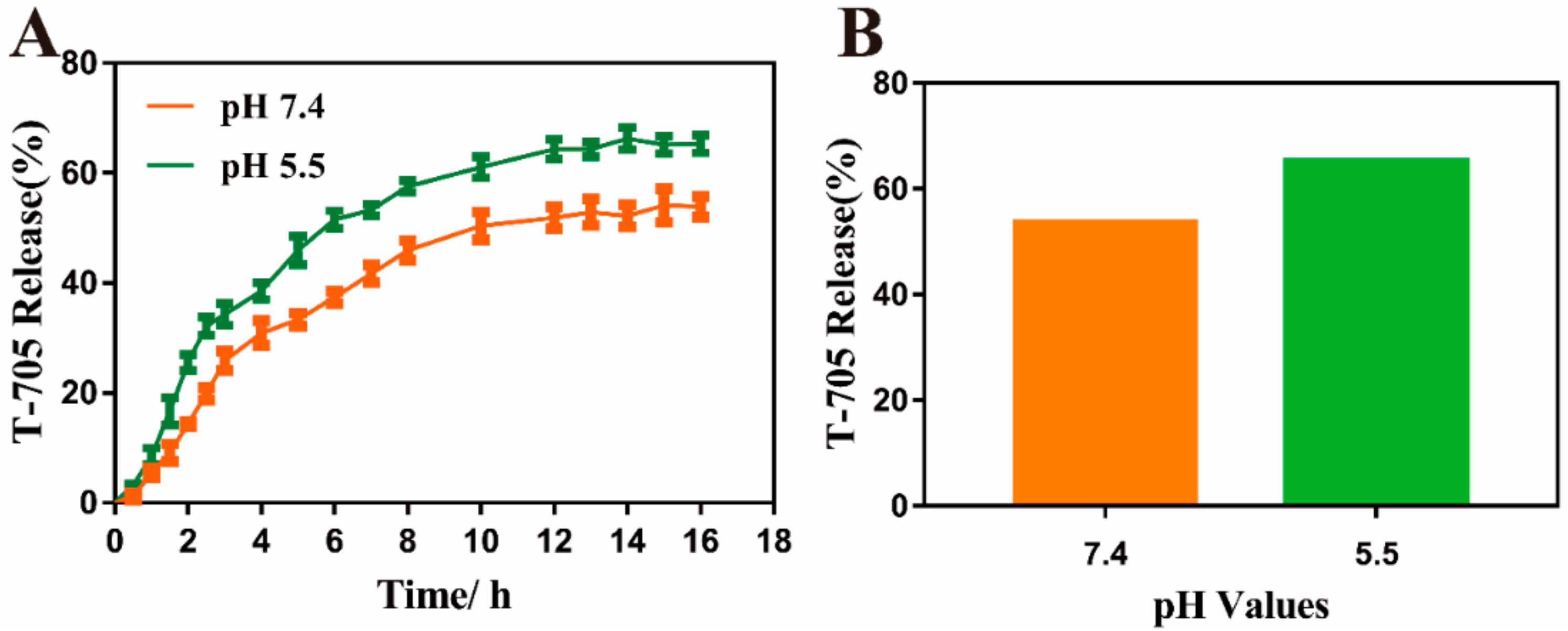
2.5. Loading and In Vitro Release of Drug
2.6. Bacterial Culture
2.7. Determination of the Bacterial Survival Rate
2.8. Determination of the Minimum Inhibitory Concentration
2.9. Determination of the Inhibitory Growth Curve
2.10. Cell Culture and Virus Amplification
2.11. Cytotoxicity Test of MIL-101(Fe)-T705
2.12. Virus Titer Determination
2.13. Determination of the Growth Curve of Influenza Virus
2.14. Determination of Fluorescence Quantitative PCR
2.15. Statistical Analysis
3. Results
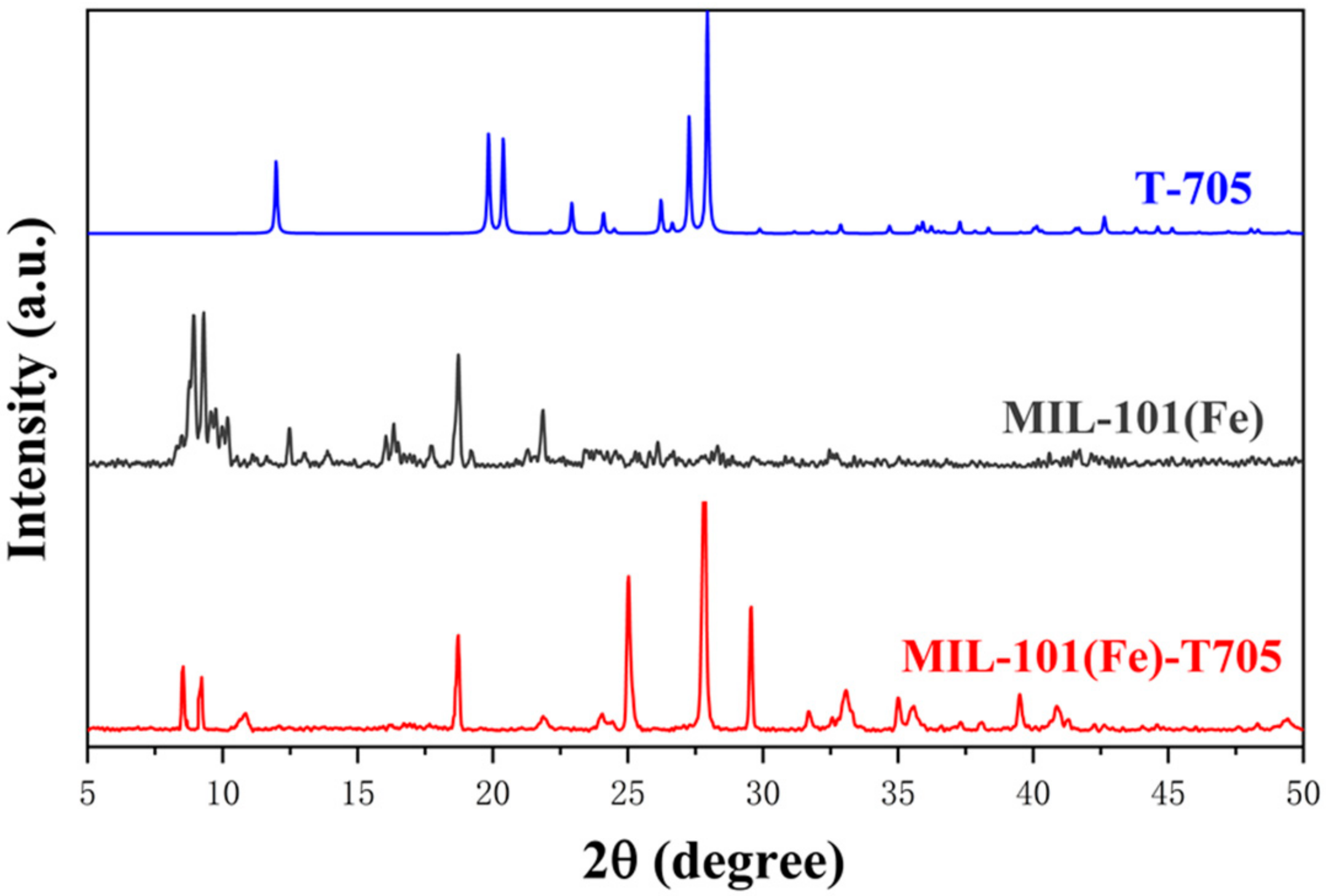
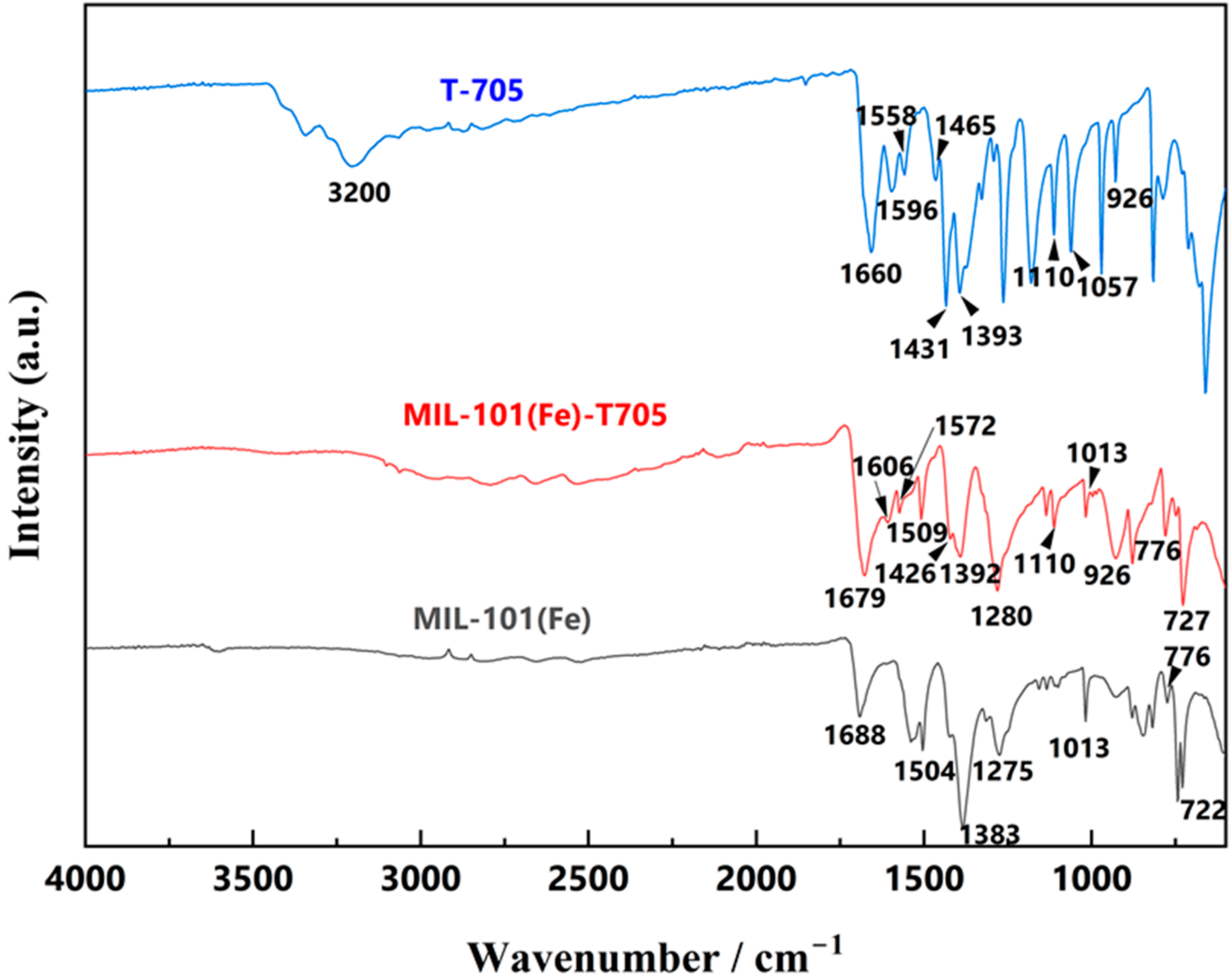
3.1. Characterization of the As-Made MIL-101(Fe)-T705
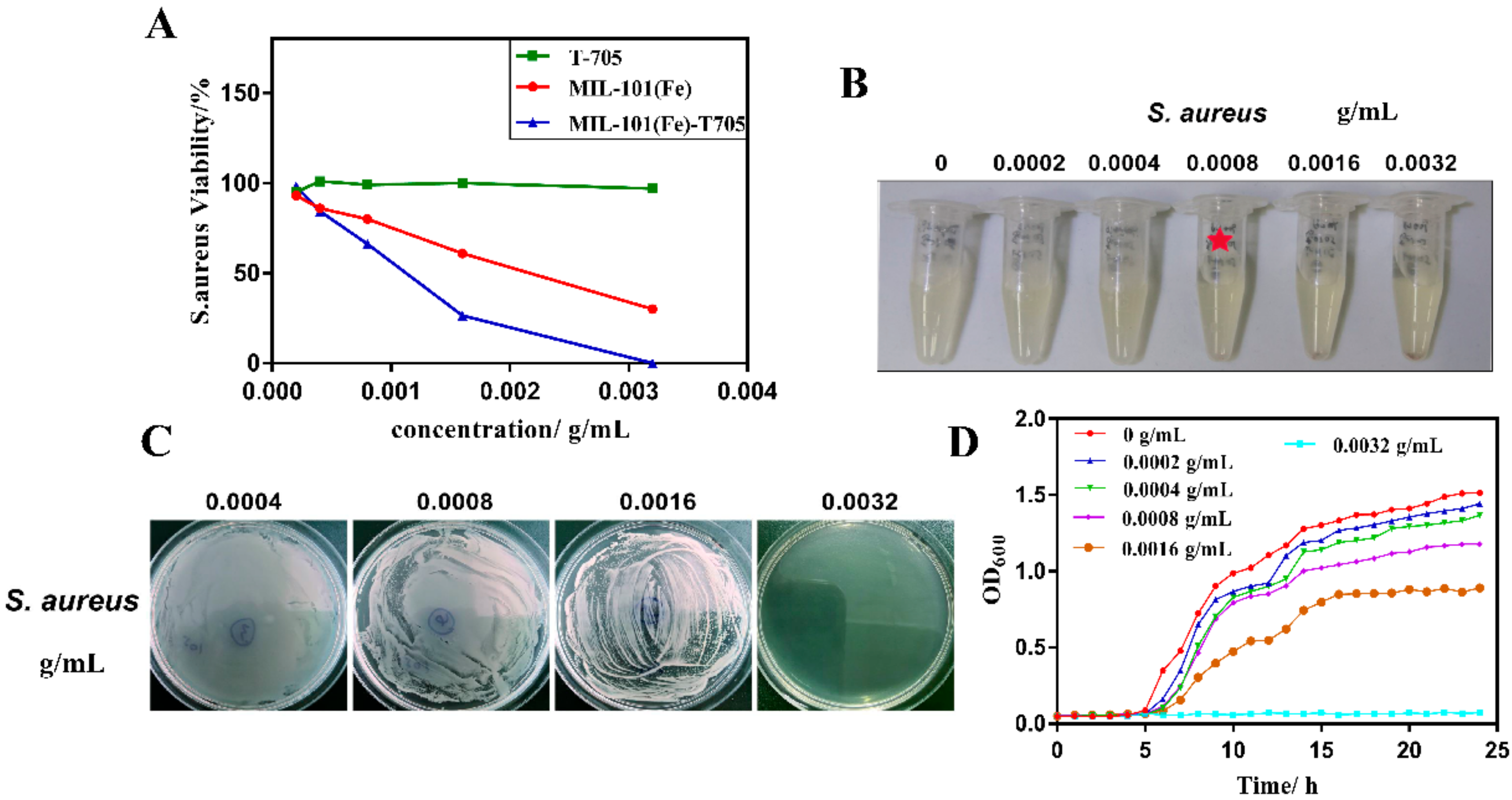
3.2. Antibacterial Properties of MIL-101(Fe)-T705
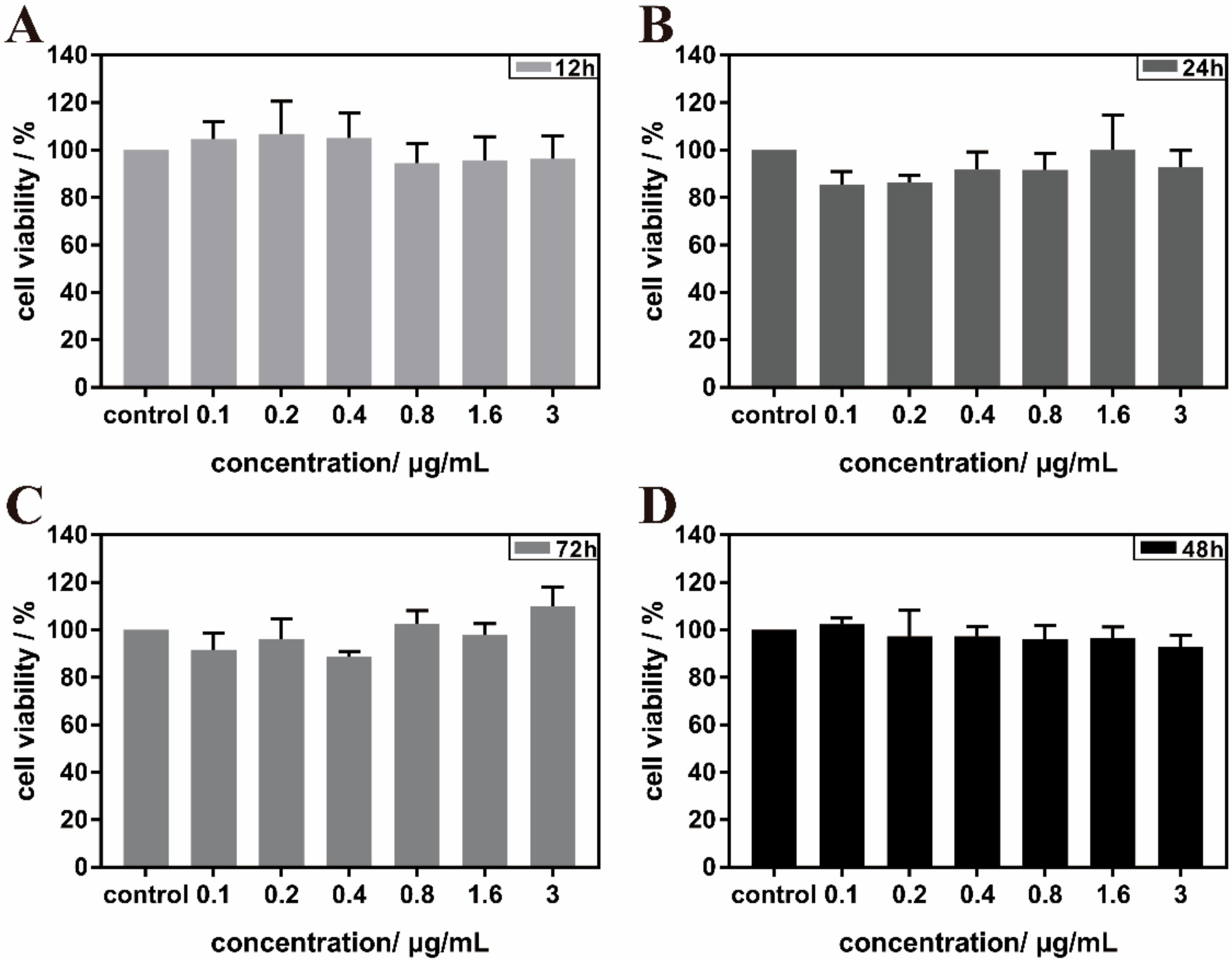
3.3. Biocompatibility Assessments
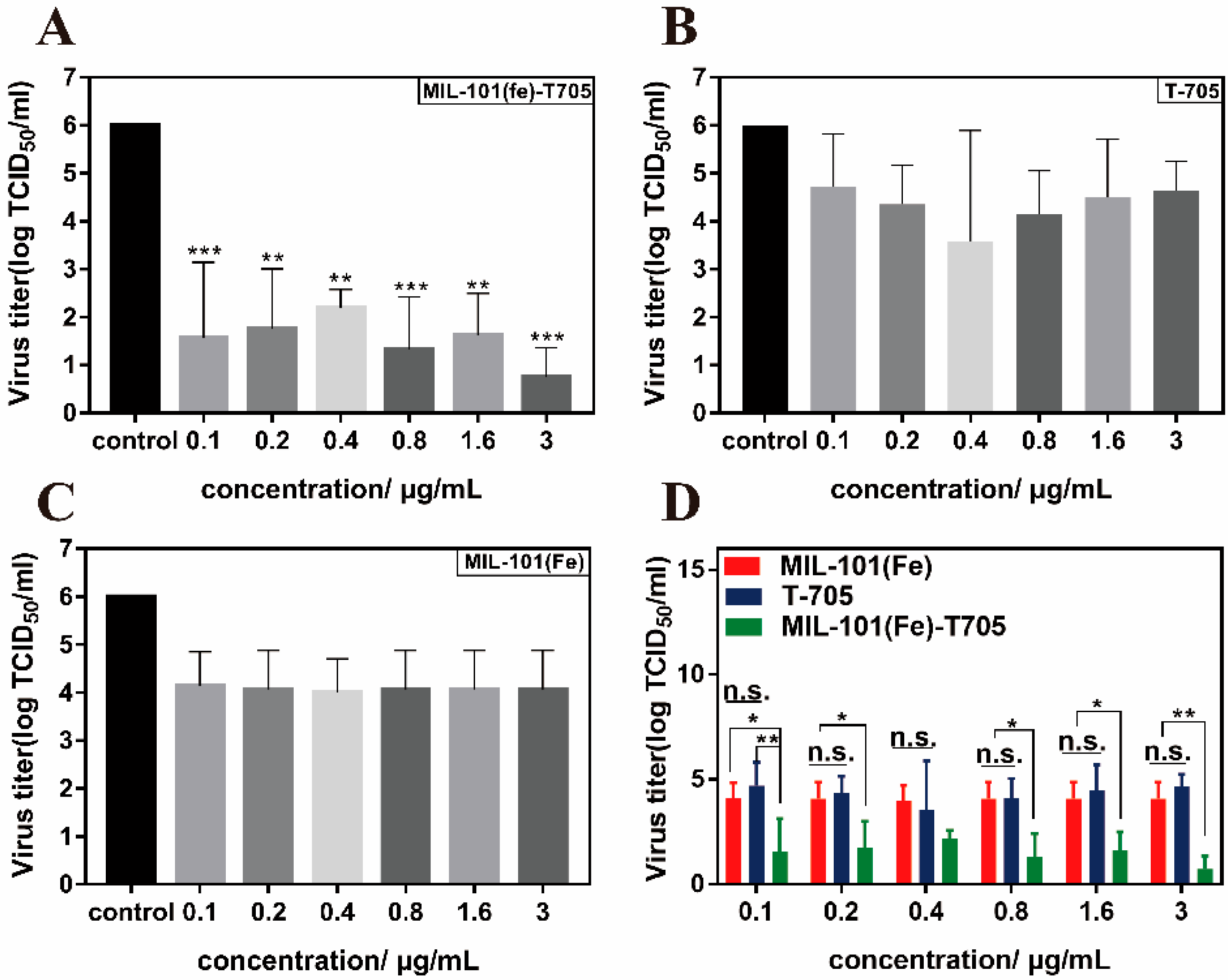
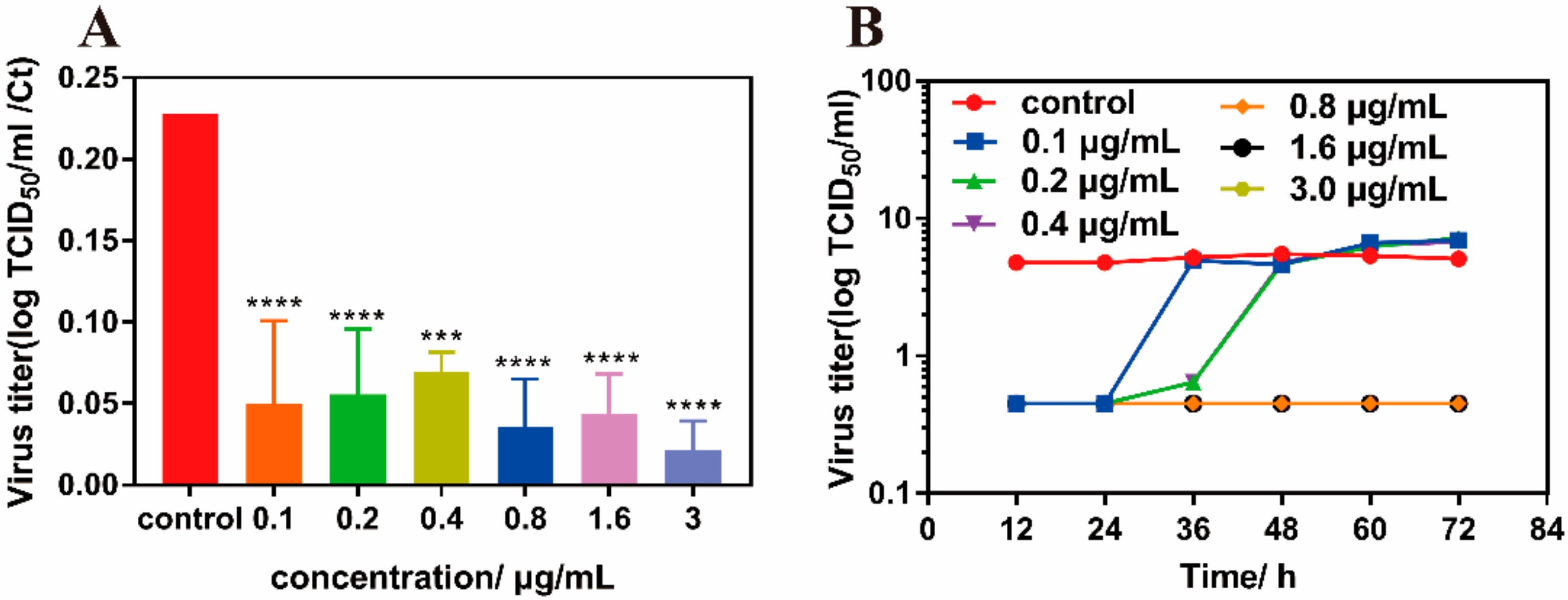
3.4. Antiviral Properties of MIL-101(Fe)-T705
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Avian influenza: Frequently asked questions. Relev. Epidemiol. Hebd. 2005, 80, 377–384. [Google Scholar]
- Vos, T.; Allen, C.; Arora, M.; Barber, R.; Bhutta, Z.; Brown, A.; Carter, A.; Casey, D.; Charlson, F.; Chen, A.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1545–1602. [Google Scholar] [CrossRef]
- Buchmeier, M.; Bowen, M.D.; Peters, C.J. Arenaviridae: The viruses and their replication. Fields Virol. 2001, 2, 1635–1668. [Google Scholar]
- Furuta, Y.; Takahashi, K.; Shiraki, K.; Sakamoto, K.; Smee, D.F.; Barnard, D.L.; Gowen, B.B.; Julander, J.G.; Morrey, J.D. T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antivir. Res. 2009, 82, 95–102. [Google Scholar] [CrossRef]
- Nogales, A.; DeDiego, M.L. Influenza Virus and Vaccination. Pathogens 2020, 9, 220. [Google Scholar] [CrossRef]
- Coates, B.M.; Staricha, K.L.; Wiese, K.M.; Ridge, K.M. Influenza A Virus Infection, Innate Immunity, and Childhood. JAMA Pediatrics 2015, 169, 956–963. [Google Scholar] [CrossRef]
- Hurt, A.C. The epidemiology and spread of drug resistant human influenza viruses. Curr. Opin. Virol. 2014, 8, 22–29. [Google Scholar] [CrossRef]
- Deyde, V.M.; Xu, X.; Bright, R.A.; Shaw, M.; Smith, C.B.; Zhang, Y.; Shu, Y.; Gubareva, L.V.; Cox, N.J.; Klimov, A.I. Surveillance of Resistance to Adamantanes among Influenza A(H3N2) and A(H1N1) Viruses Isolated Worldwide. J. Infect. Dis. 2007, 196, 249–257. [Google Scholar] [CrossRef]
- Fang, D.; Das, K.; Cao, W.; Malhotra, U.; Triadafilopoulos, G.; Najarian, R.; Hardie, L.; Lightdale, C.; Beales, I.; Felix, V.; et al. Barrett’s esophagus: Progression to adenocarcinoma and markers. Ann. N. Y. Acad. Sci. 2011, 1232, 210–229. [Google Scholar] [CrossRef]
- Boivin, G. Detection and management of antiviral resistance for influenza viruses. Influenza Other Respir. Viruses 2013, 7 (Suppl. 3), 18–23. [Google Scholar] [CrossRef] [PubMed]
- Sangawa, H.; Komeno, T.; Nishikawa, H.; Yoshida, A.; Takahashi, K.; Nomura, N.; Furuta, Y. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob. Agents Chemother. 2013, 57, 5202–5208. [Google Scholar] [CrossRef] [PubMed]
- Delang, L.; Abdelnabi, R.; Neyts, J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antivir. Res. 2018, 153, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, L.; Kuo, H.-C.D.; Shannar, A.; Peter, R.; Chou, P.J.; Li, S.; Hudlikar, R.; Liu, X.; Liu, Z.; et al. An Update on Current Therapeutic Drugs Treating COVID-19. Curr. Pharmacol. Rep. 2020, 6, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Baz, M.; Goyette, N.; Griffin, B.D.; Kobinger, G.P.; Boivin, G. In vitro susceptibility of geographically and temporally distinct Zika viruses to favipiravir and ribavirin. Antivir. Ther. 2017, 22, 613–618. [Google Scholar] [CrossRef]
- Delang, L.; Segura Guerrero, N.; Tas, A.; Quérat, G.; Pastorino, B.; Froeyen, M.; Dallmeier, K.; Jochmans, D.; Herdewijn, P.; Bello, F.; et al. Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J. Antimicrob. Chemother. 2014, 69, 2770–2784. [Google Scholar] [CrossRef]
- Tani, H.; Fukuma, A.; Fukushi, S.; Taniguchi, S.; Yoshikawa, T.; Iwata-Yoshikawa, N.; Sato, Y.; Suzuki, T.; Nagata, N.; Hasegawa, H.; et al. Efficacy of T-705 (Favipiravir) in the Treatment of Infections with Lethal Severe Fever with Thrombocytopenia Syndrome Virus. mSphere 2016, 1, e00061-15. [Google Scholar] [CrossRef]
- Zhang, T.; Zhai, M.; Ji, J.; Zhang, J.; Tian, Y.; Liu, X. Recent progress on the treatment of Ebola virus disease with Favipiravir and other related strategies. Bioorg. Med. Chem. Lett. 2017, 27, 2364–2368. [Google Scholar] [CrossRef]
- Sandre, R.M.; Shafran, S.D. Infective Endocarditis: Review of 135 Cases over 9 Years. Clin. Infect. Dis. 1996, 22, 276–286. [Google Scholar] [CrossRef]
- Lake, R.J.; Devleesschauwer, B.; Nasinyama, G.; Havelaar, A.H.; Kuchenmuller, T.; Haagsma, J.A.; Jensen, H.H.; Jessani, N.; Maertens de Noordhout, C.; Angulo, F.J.; et al. National Studies as a Component of the World Health Organization Initiative to Estimate the Global and Regional Burden of Foodborne Disease. PLoS ONE 2015, 10, e0140319. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Steinberg, J.P.; Clark, C.C.; Hackman, B.O. Nosocomial and Community-Acquired Staphylococcus aureus Bacteremias from 1980 to 1993: Impact of Intravascular Devices and Methicillin Resistance. Clin. Infect. Dis. 1996, 23, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Widmer, A.F.; Raad, P.I. Optimal duration of therapy for catheter-related Staphylococcus aureus bacteremia. Clin. Infect. Dis. 1992, 14, 1259–1260. [Google Scholar] [CrossRef] [PubMed]
- Tahmasebi, E.; Masoomi, M.Y.; Yamini, Y.; Morsali, A. Application of Mechanosynthesized Azine-Decorated Zinc(II) Metal–Organic Frameworks for Highly Efficient Removal and Extraction of Some Heavy-Metal Ions from Aqueous Samples: A Comparative Study. Inorg. Chem. 2015, 54, 425–433. [Google Scholar] [CrossRef]
- Kang, S.; Pinault, M.; Pfefferle, L.D.; Elimelech, M. Single-Walled Carbon Nanotubes Exhibit Strong Antimicrobial Activity. Langmuir 2007, 23, 8670–8673. [Google Scholar] [CrossRef]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.-H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.-Y.; et al. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 95–101. [Google Scholar] [CrossRef]
- Shrivastava, S.; Bera, T.; Singh, S.K.; Singh, G.; Ramachandrarao, P.; Dash, D. Characterization of Antiplatelet Properties of Silver Nanoparticles. ACS Nano 2009, 3, 1357–1364. [Google Scholar] [CrossRef]
- Muhsin, T.M.; Hachim, A.K. Mycosynthesis and characterization of silver nanoparticles and their activity against some human pathogenic bacteria. World J. Microbiol. Biotechnol. 2014, 30, 2081–2090. [Google Scholar] [CrossRef]
- Sondi, I.; Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: A case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef]
- Paredes, D.; Ortiz, C.; Torres, R. Synthesis, characterization, and evaluation of antibacterial effect of Ag nanoparticles against Escherichia coli O157:H7 and methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Nanomed. 2014, 9, 1717–1729. [Google Scholar] [CrossRef]
- Antoine, T.E.; Mishra, Y.K.; Trigilio, J.; Tiwari, V.; Adelung, R.; Shukla, D. Prophylactic, therapeutic and neutralizing effects of zinc oxide tetrapod structures against herpes simplex virus type-2 infection. Antivir. Res. 2012, 96, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Sametband, M.; Kalt, I.; Gedanken, A.; Sarid, R. Herpes Simplex Virus Type-1 Attachment Inhibition by Functionalized Graphene Oxide. ACS Appl. Mater. Interfaces 2014, 6, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wang, X.; Zhu, G.; Nian, Q.; Zhou, H.; Yang, D.; Qin, C.; Tang, R. Virus capture and destruction by label-free graphene oxide for detection and disinfection applications. Small 2015, 11, 1171–1176. [Google Scholar] [CrossRef]
- Meek, S.T.; Greathouse, J.A.; Allendorf, M.D. Metal-Organic Frameworks: A Rapidly Growing Class of Versatile Nanoporous Materials. Adv. Mater. 2011, 23, 249–267. [Google Scholar] [CrossRef]
- Gkiliopoulos, D.; Zamboulis, A.; Giannakoudakis, D.A.; Bikiaris, D.; Triantafyllidis, K. Polymer/Metal Organic Framework (MOF) Nanocomposites for Biomedical Applications. Molecules 2020, 25, 185. [Google Scholar] [CrossRef]
- Horcajada, P.; Serre, C.; Vallet-Regí, M.; Sebban, M.; Taulelle, F.; Férey, G. Metal–Organic Frameworks as Efficient Materials for Drug Delivery. Angew. Chem. Int. Ed. 2006, 45, 5974–5978. [Google Scholar] [CrossRef]
- Thallapally, P.K.; Tian, J.; Kishan, M.R.; Fernandez, C.A.; Dalgarno, S.J.; Mcgrail, P.B.; Warren, J.E.; Atwood, J.L. Flexible (breathing) interpenetrated metal-organic frameworks for CO2 separation applications. J. Am. Chem. Soc. 2008, 130, 16842–16843. [Google Scholar] [CrossRef]
- Keskin, S.; Kızılel, S. Biomedical Applications of Metal Organic Frameworks. Ind. Eng. Chem. Res. 2011, 50, 1799–1812. [Google Scholar] [CrossRef]
- Cabrera-García, A.; Checa-Chavarria, E.; Rivero-Buceta, E.; Moreno, V.; Fernández, E.; Botella, P. Amino modified metal-organic frameworks as pH-responsive nanoplatforms for safe delivery of camptothecin. J. Colloid Interface Sci. 2019, 541, 163–174. [Google Scholar] [CrossRef]
- Férey, G.; Mellot-Draznieks, C.; Serre, C.; Millange, F.; Dutour, J.; Surblé, S.; Margiolaki, I. A Chromium Terephthalate-Based Solid with Unusually Large Pore Volumes and Surface Area. Science 2005, 309, 2040–2042. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Pashow, K.M.L.; Della Rocca, J.; Xie, Z.; Tran, S.; Lin, W. Postsynthetic Modifications of Iron-Carboxylate Nanoscale Metal−Organic Frameworks for Imaging and Drug Delivery. J. Am. Chem. Soc. 2009, 131, 14261–14263. [Google Scholar] [CrossRef] [PubMed]
- Baati, T.; Njim, L.; Neffati, F.; Kerkeni, A.; Bouttemi, M.; Gref, R.; Najjar, M.F.; Zakhama, A.; Couvreur, P.; Serre, C.; et al. In depth analysis of the in vivo toxicity of nanoparticles of porous iron(iii) metal–organic frameworks. Chem. Sci. 2013, 4, 1597–1607. [Google Scholar] [CrossRef]
- Zhao, M.; Yuan, K.; Wang, Y.; Li, G.; Guo, J.; Gu, L.; Hu, W.; Zhao, H.; Tang, Z. Metal–organic frameworks as selectivity regulators for hydrogenation reactions. Nature 2016, 539, 76–80. [Google Scholar] [CrossRef]
- Deng, Y.; Zhang, R.; Li, D.; Sun, P.; Su, P.; Yang, Y. Preparation of iron-based MIL-101 functionalized polydopamine@Fe3O4 magnetic composites for extracting sulfonylurea herbicides from environmental water and vegetable samples. J. Sep. Sci. 2018, 41, 2046–2055. [Google Scholar] [CrossRef]
- He, L.; Dong, Y.; Zheng, Y.; Jia, Q.; Shan, S.; Zhang, Y. A novel magnetic MIL-101(Fe)/TiO2 composite for photo degradation of tetracycline under solar light. J. Hazard. Mater. 2019, 361, 85–94. [Google Scholar] [CrossRef]
- Denny, M.S., Jr.; Cohen, S.M. In Situ Modification of Metal-Organic Frameworks in Mixed-Matrix Membranes. Angew. Chem. Int. Ed. Engl. 2015, 54, 9029–9032. [Google Scholar] [CrossRef]
- Yan, D.; Hu, H.; Gao, N.; Ye, J.; Ou, H. Fabrication of carbon nanotube functionalized MIL-101(Fe) for enhanced visible-light photocatalysis of ciprofloxacin in aqueous solution. Appl. Surf. Sci. 2019, 498, 143836. [Google Scholar] [CrossRef]
- Zhang, C.; Ai, L.; Jiang, J. Graphene Hybridized Photoactive Iron Terephthalate with Enhanced Photocatalytic Activity for the Degradation of Rhodamine B under Visible Light. Ind. Eng. Chem. Res. 2015, 54, 153–163. [Google Scholar] [CrossRef]
- Wang, H.; Yuan, X.; Wu, Y.; Zeng, G.; Chen, X.; Leng, L.; Li, H. Synthesis and applications of novel graphitic carbon nitride/metal-organic frameworks mesoporous photocatalyst for dyes removal. Appl. Catal. B Environ. 2015, 174, 445–454. [Google Scholar] [CrossRef]
- Cui, F.; Deng, Q.; Sun, L. Prussian blue modified metal–organic framework MIL-101(Fe) with intrinsic peroxidase-like catalytic activity as a colorimetric biosensing platform. RSC Adv. 2015, 5, 98215–98221. [Google Scholar] [CrossRef]
- Du, J.-J.; Yuan, Y.-P.; Sun, J.-X.; Peng, F.-M.; Jiang, X.; Qiu, L.-G.; Xie, A.-J.; Shen, Y.-H.; Zhu, J.-F. New photocatalysts based on MIL-53 metal–organic frameworks for the decolorization of methylene blue dye. J. Hazard. Mater. 2011, 190, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Jing, F.; Shen, L.; Qin, N.; Wu, L. MIL-53(Fe) as a highly efficient bifunctional photocatalyst for the simultaneous reduction of Cr(VI) and oxidation of dyes. J. Hazard. Mater. 2015, 287, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Wyszogrodzka, G.; Marszalek, B.; Gil, B.; Dorozynski, P. Metal-organic frameworks: Mechanisms of antibacterial action and potential applications. Drug Discov. Today 2016, 21, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Szunerits, S.; Barras, A.; Khanal, M.; Pagneux, Q.; Boukherroub, R. Nanostructures for the Inhibition of Viral Infections. Molecules 2015, 20, 14051–14081. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26, D49–D53. [Google Scholar] [CrossRef]
- McKimm-Breschkin, J.L. Influenza neuraminidase inhibitors: Antiviral action and mechanisms of resistance. Influenza Other Respir. Viruses 2013, 7 (Suppl. 1), 25–36. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, H.; Wu, Y.; Liu, D.; Qi, X.; Shi, Y.; Gao, G.F. H7N9: A low pathogenic avian influenza A virus infecting humans. Curr. Opin. Virol. 2014, 5, 91–97. [Google Scholar] [CrossRef]
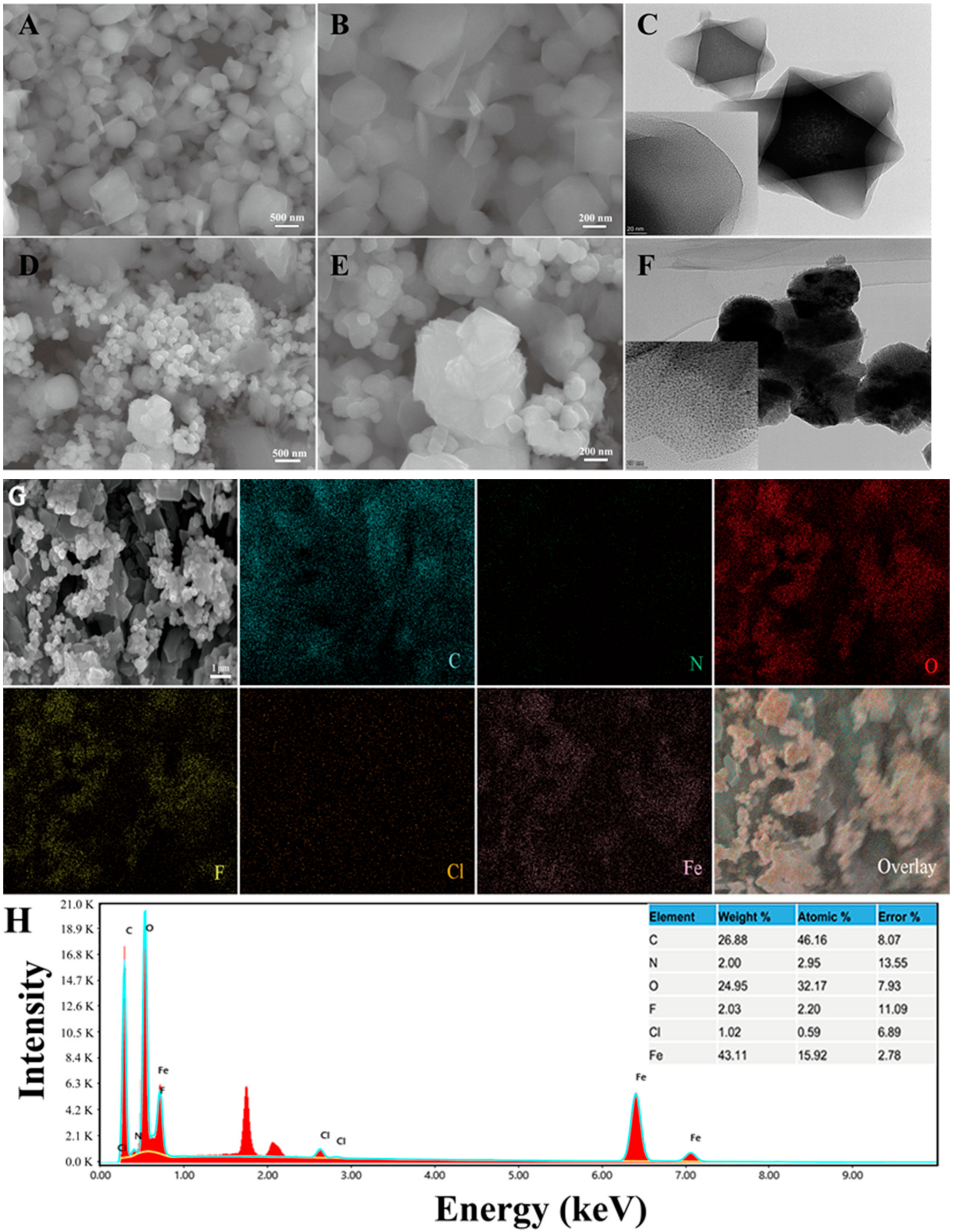
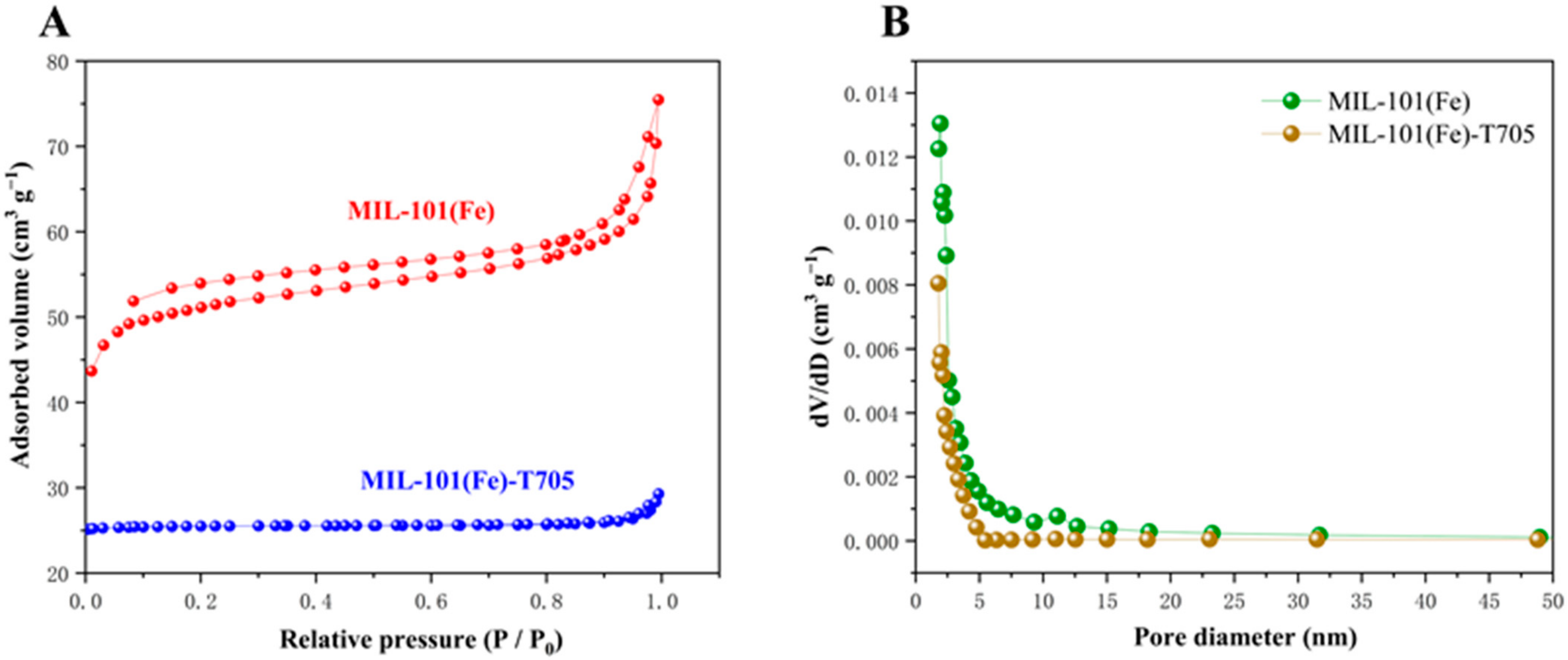

| Samples | Surface Area (m2 g−1) | Pore Diameter (nm) | Pore Volume (cm3 g−1) |
|---|---|---|---|
| MIL-101(Fe) | 199.7194 | 2.33771 | 0.116722 |
| MIL-101(Fe)-T705 | 116.7785 | 1.5159 | 0.008778 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Li, X.; Zheng, H.; Chen, J.; Ye, X.; Liu, T. Anti-Influenza Virus Study of Composite Material with MIL-101(Fe)-Adsorbed Favipiravir. Molecules 2022, 27, 2288. https://doi.org/10.3390/molecules27072288
Xu M, Li X, Zheng H, Chen J, Ye X, Liu T. Anti-Influenza Virus Study of Composite Material with MIL-101(Fe)-Adsorbed Favipiravir. Molecules. 2022; 27(7):2288. https://doi.org/10.3390/molecules27072288
Chicago/Turabian StyleXu, Mengyuan, Xi Li, Huiying Zheng, Jiehan Chen, Xiaohua Ye, and Tiantian Liu. 2022. "Anti-Influenza Virus Study of Composite Material with MIL-101(Fe)-Adsorbed Favipiravir" Molecules 27, no. 7: 2288. https://doi.org/10.3390/molecules27072288
APA StyleXu, M., Li, X., Zheng, H., Chen, J., Ye, X., & Liu, T. (2022). Anti-Influenza Virus Study of Composite Material with MIL-101(Fe)-Adsorbed Favipiravir. Molecules, 27(7), 2288. https://doi.org/10.3390/molecules27072288