Potential Effects of Ibuprofen, Remdesivir and Omeprazole on Dexamethasone Metabolism in Control Sprague Dawley Male Rat Liver Microsomes (Drugs Often Used Together Alongside COVID-19 Treatment)
Abstract
:1. Introduction
2. Results and Discussion
2.1. HPLC Method Development and Validation
2.2. Optimisation of Substrate Concentration for Incubation System In Vitro
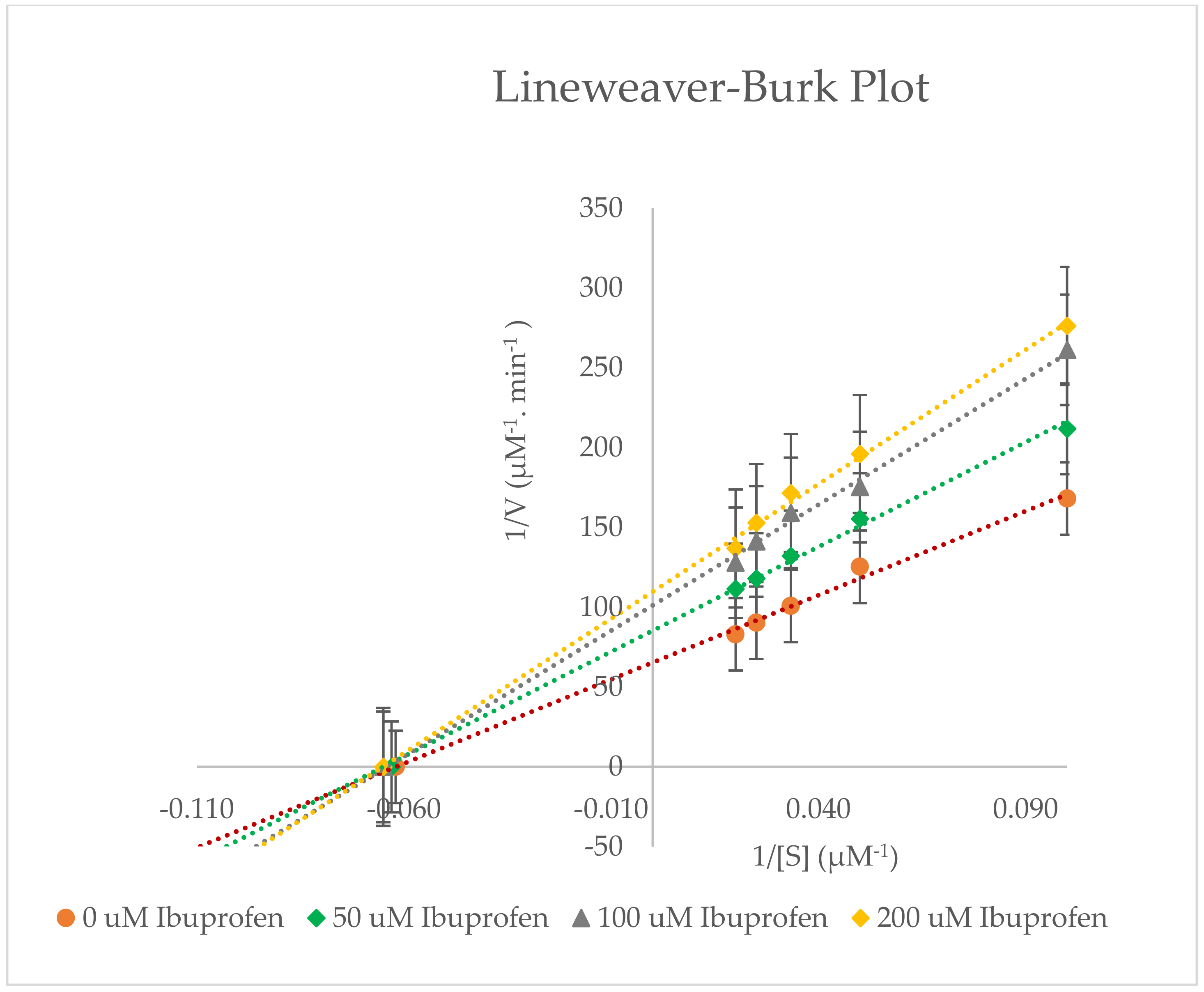
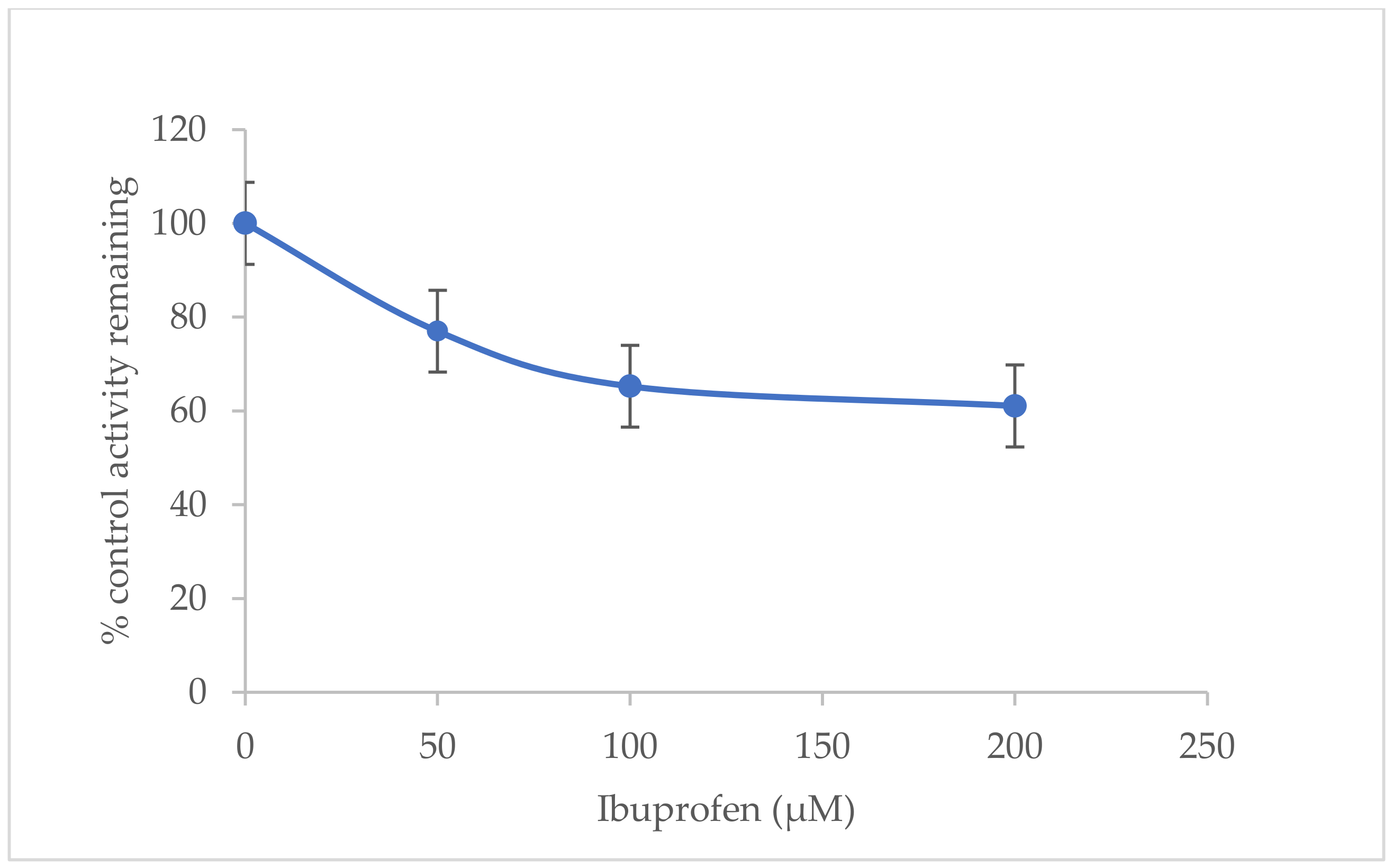
2.3. Inhibitory Effects of Ibuprofen on CYP3A2 Enzyme Activity in Rat Liver Microsomes (RLMs)
2.4. Inhibitory Effects of Remdesivir on CYP3A2 Enzyme Activity in RLMs
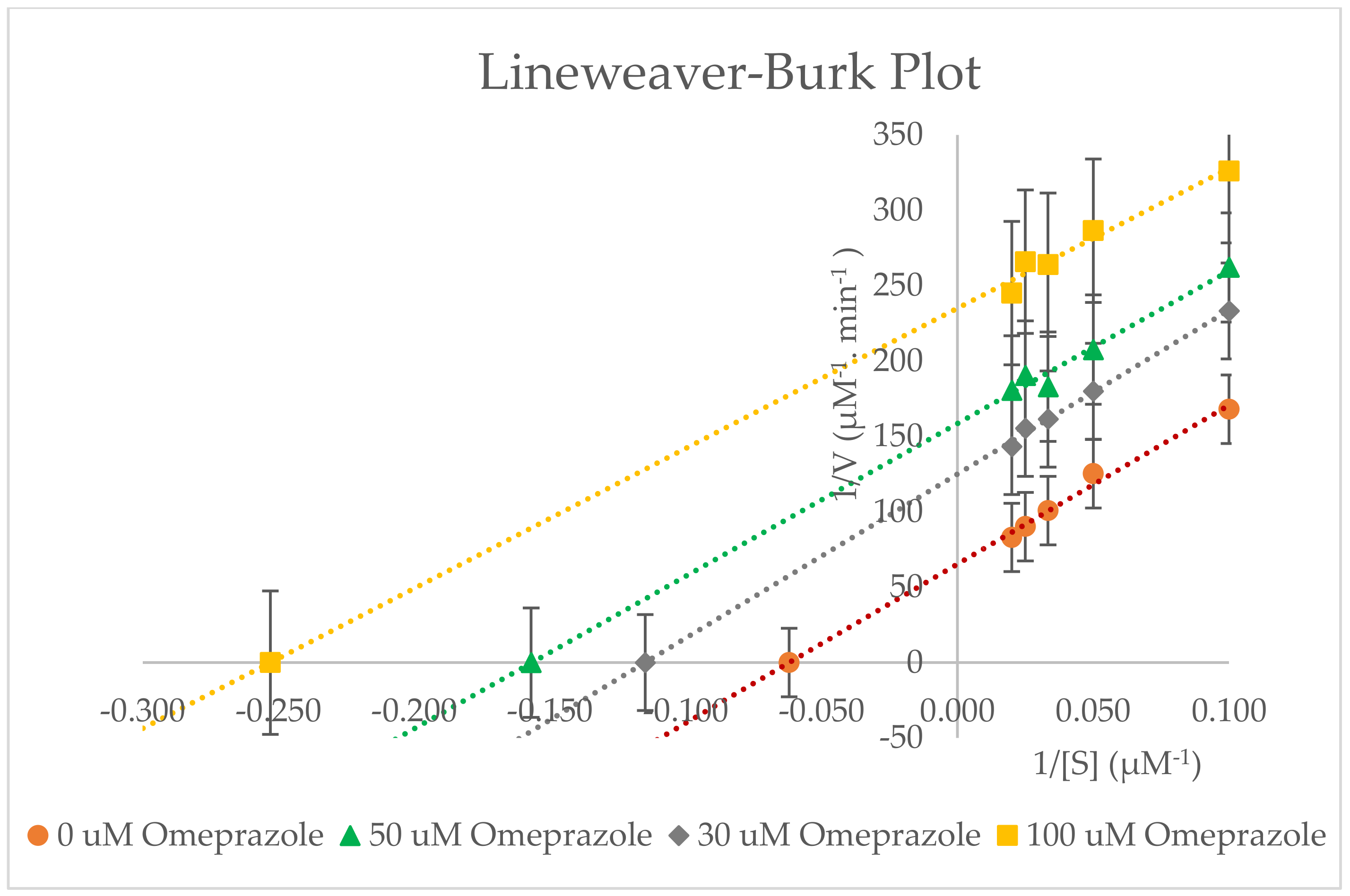
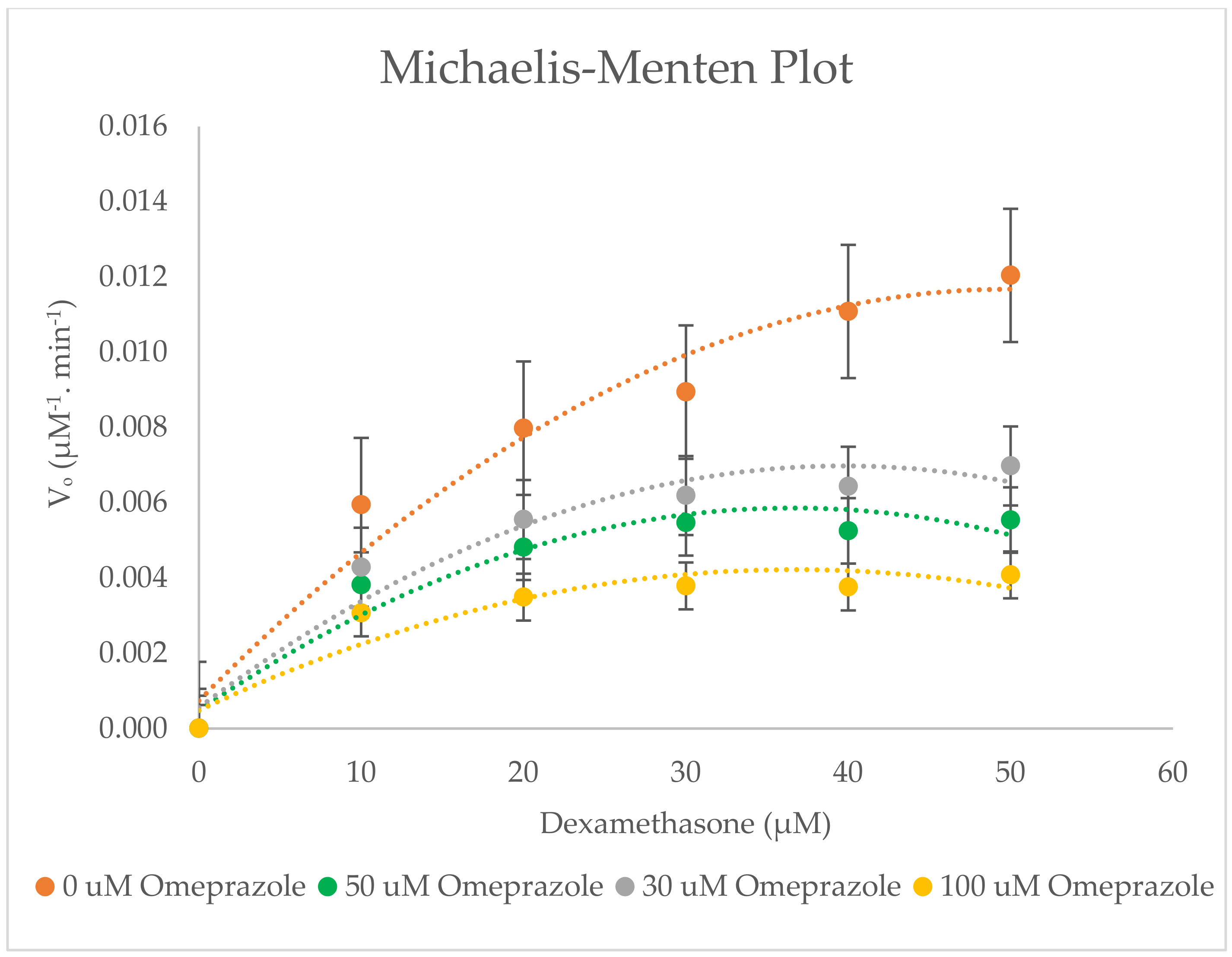
2.5. Inhibitory Effects of Omeprazole on CYP3A2 Enzyme Activity in RLMs
3. Materials and Methods
3.1. Chemicals
3.2. Rat Liver Microsomes
3.3. Instruments
3.4. Potential Effects of Inhibitors on CYP3A2 Activity In Vitro
3.5. Analytes Stock and Standard Solutions Preparation
3.6. Optimization of Substrate Concentration In Vitro
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, Y.; Wu, S.; Chen, Z.; Zhang, H.; Zhao, W. Inhibitory effects of cytochrome P450 enzymes CYP1A2, CYP2A6, CYP2E1 and CYP3A4 by extracts and alkaloids of Gelsemium elegans roots. J. Ethnopharmacol. 2015, 166, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zheng, L.; Qi, L.; Xu, Y.; Xu, L.; Yin, L.; Han, X.; Liu, K.; Peng, J. Inhibitory effects of dioscin on cytochrome P450 enzymes. RSC Adv. 2014, 4, 54026–54031. [Google Scholar] [CrossRef]
- Nebert, D.W.; Russell, D.W. Clinical importance of the cytochromes P450. Lancet 2002, 360, 1155–1162. [Google Scholar] [CrossRef]
- Sychev, D.A.; Ashraf, G.M.; Svistunov, A.A.; Maksimov, M.L.; Tarasov, V.V.; Chubarev, V.N.; Otdelenov, V.A.; Denisenko, N.P.; Barreto, G.E.; Aliev, G. The cytochrome P450 isoenzyme and some new opportunities for the prediction of negative drug interaction in vivo. Drug Des. Dev. Ther. 2018, 12, 1147–1156. [Google Scholar] [CrossRef] [Green Version]
- Gentile, D.M.; Maggs, J.L.; Park, B.K.; Back, D.J. Dexamethasone metabolism by human liver in vitro. Metabolite identification and inhibition of 6-hydroxylation. J. Pharmacol. Exp. Ther. 1996, 277, 105–112. [Google Scholar]
- Sun, M.; Tang, Y.; Ding, T.; Liu, M.; Wang, X. Inhibitory effects of celastrol on rat liver cytochrome P450 1A2, 2C11, 2D6, 2E1 and 3A2 activity. Fitoterapia 2014, 92, 1–8. [Google Scholar] [CrossRef]
- Shayeganpour, A.; El-Kadi, A.O.S.; Brocks, D.R. Determination of enzymes(s) involved in the metabolism of amiodarone in liver and intestine of rat: The contribution of cytochrome P450 3A isoforms. Drug Metab. Dispos. 2006, 34, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Zhou, X.; Tang, S.; Liu, M.; Wang, X. Evaluation of the inhibition potential of plumbagin against cytochrome P450 using LC-MS/MS and cocktail approach. Sci. Rep. 2016, 6, 28482. [Google Scholar] [CrossRef] [Green Version]
- Puisset, F.; Chatelut, E.; Sparreboom, A.; Delord, J.-P.; Berchery, D.; Lochon, T.; Lafont, T.; Roche, H. Dexamethasone as a probe for CYP3A4 metabolism: Evidence of gender effect. Cancer Chemother. Pharmacol. 2007, 60, 305–308. [Google Scholar] [CrossRef]
- Desjardins, J.P.; Iversen, P.L. Inhibition of the rat cytochrome P450 3A2 by an antisense phosphorothioate oligodeoxynucleotide in vivo. J. Pharmacol. Exp. Ther. 1995, 275, 1608–1613. [Google Scholar]
- Tomlinson, E.S.; Maggs, J.L.; Park, B.K.; Back, D.J. Dexamethasone Metabolism in Species Differences. J. Steroid Biochem. Mol. Biol. 1997, 62, 345–352. [Google Scholar] [CrossRef]
- Li, L.; Li, Z.; Deng, C.; Ning, M.; Li, H.; Bi, S.; Zhou, T.; Lu, W. A mechanism-based pharmacokinetic/pharmacodynamic model for CYP3A1/2 induction by dexamethasone in rats. Acta Pharmacol. Sin. 2012, 33, 127–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehrer, S.; Rheinstein, P. Common drugs, vitamins, nutritional supplements and COVID-19 mortality. Int. J. Funct. Nutr. 2021, 2, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Aguila, E.J.T.; Cua, I.H.Y. Repurposed GI Drugs in the Treatment of COVID-19. Dig. Dis. Sci. 2020, 65, 2452–2453. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Jamieson, L.; Edoka, I.; Long, L.; Silal, S.; Pulliam, J.R.C.; Moultire, H.; Sanne, I.; Meyer-Rath, G.; Nichols, B.E. Cost-effectiveness of remdesivir and dexamethasone for COVID-19 treatment in South Africa. Open Forum Infect. Dis. 2021, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Drake, T.M.; Fairfield, C.J.; Pius, R.; Knight, S.R.; Norman, L.; Girvan, M.; Hardwick, H.E.; Docherty, A.B.; Thwaites, R.S.; Openshaw, P.J.M.; et al. Non-steroidal anti-inflammatory drug use and outcomes of COVID-19 in the ISARIC Clinical Characterisation Protocol UK cohort: A matched, prospective cohort study. Lancet Rheumatol. 2021, 3, e498–e506. [Google Scholar] [CrossRef]
- Hussain, A.; Naughton, D.P.; Barker, J. Development and Validation of a Novel HPLC Method to Analyse Metabolic Reaction Products Catalysed by the CYP3A2 Isoform: In Vitro Inhibition of CYP3A2 Enzyme Activity by Aspirin (Drugs Often Used Together in COVID-19 Treatment). Molecules 2022, 27, 927. [Google Scholar] [CrossRef]
- Burenheide, A.; Kunze, T.; Clement, B. Inhibitory effects on cytochrome P450 enzymes of pentamidine and its amidoxime pro-drug. Basic Clin. Pharmacol. Toxicol. 2008, 103, 61–65. [Google Scholar] [CrossRef]
- Russell, B.; Moss, C.; George, G.; Santaolalla, A.; Cope, A.; Papa, S.; Hemelrijck, M.V. Associations between immune-suppressive and stimulating drugs and novel COVID-19—A systematic review of current evidence. Ecancermedicalscience 2020, 14, 1022. [Google Scholar] [CrossRef] [Green Version]
- Rinott, E.; Kozer, E.; Shapira, Y.; Bar-Haim, A.; Youngster, I. Ibuprofen use and clinical outcomes in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, e1259. [Google Scholar] [CrossRef]
- Mazaleuskaya, L.L.; Theken, K.N.; Gong, L.; Thorn, C.F.; FitzGerald, G.A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Ibuprofen pathways. Pharm. Genom. 2015, 25, 96–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Chen, S.; Hu, A.; Zhang, L.; Sun, W.; Chen, J.; Tang, W.; Zhang, H.; Liu, C.; Ke, C.; et al. The Establishment and Validation of the Human U937 Cell Line as a Cellular Model to Screen Immunomodulatory Agents Regulating Cytokine Release Induced by Influenza Virus Infection. Virol. Sin. 2019, 34, 648–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, J.; Fowle, C.J. Inhibition of Endocannabinoid Metabolism by the Metabolites of Ibuprofen and Flurbiprofen. PLoS ONE 2014, 9, e103589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karttunen, P.; Saano, V.; Paronen, P.; Peura, P.; Vidgren, M. Pharmacokinetics of ibuprofen in man: A single-dose comparison of two over-the-counter, 200 mg preparation. Int. J. Clin. Pharmacol. Ther. Toxicol. 1990, 28, 251–255. [Google Scholar]
- Tanaka, E.; Kobayashi, S.; Oguchi, K.; Kuroiwa, Y.; Yasuhara, H. The effects of short-term administration of ibuprofen on trimethadione metabolism and antipyrine metabolite formation in the rat. Res. Commun. Chem. Pathol. Pharmacol. 1985, 48, 317–320. [Google Scholar]
- Deb, S.; Reeves, A.A.; Hopefl, R.; Bejusca, R. ADME and Pharmacokinetic Properties of Remdesivir: Its Drug Interaction Potential. Pharmaceuticals 2021, 14, 655. [Google Scholar] [CrossRef]
- Yang, K. What Do We Know About Remdesivir Drug Interactions? Clin. Transl. Sci. 2020, 13, 842–844. [Google Scholar] [CrossRef]
- Keeling, D.J.; Fallowfield, C.; Milliner, K.J.; Tingley, S.K.; Ife, R.J.; Anderwood, A.H. Studies on the mechanism of action of omeprazole. Biochem. Pharmacol. 1985, 34, 2967–2973. [Google Scholar] [CrossRef]
- Meent, D.G.D.; Adel, M.D.; Dijk, J.V.; Barroso-Fernandez, B.; Galta, R.E.; Golor, G.; Schaddelee, M. Effect of Multiple Doses of Omeprazole on the Pharmacokinetics, Safety, and Tolerability of Roxadustat in Healthy Subjects. Eur. J. Drug Metab. Pharmacokinet. 2018, 43, 685–692. [Google Scholar] [CrossRef] [Green Version]
- Li, X.Q.; Andersson, T.B.; Ahlstrom, M.; Weidolf, L. Comparison of inhibitory effects of the proton pump-inhibiting drugs omeprazole, esomeprazole, lansoprazole, pantoprazole, and rabeprazole on human cytochrome P450 activities. Drug Metab. Dispos. 2004, 32, 821–827. [Google Scholar] [CrossRef]
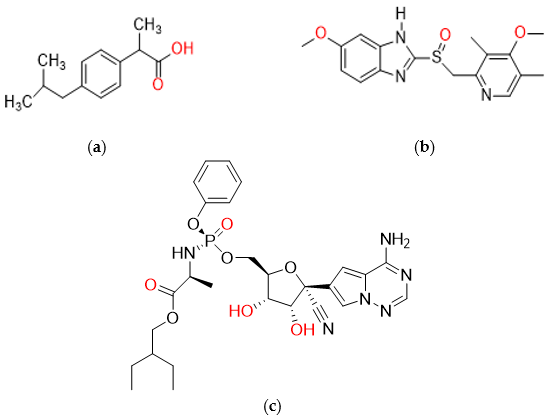





| Investigated Parameters | Characteristic |
|---|---|
| Mobile phase | 70% acetonitrile and 30% H2O, v/v |
| Column temperature | 25 °C |
| Wavelength | 243 nm |
| Flow rate | 0.6 mL/min |
| Internal standard | 4-hydroxyoctanophenone |
| Injection volume | 10 µL |
| Run time | 8 min |
| Column | Waters C18 column, 15 mm × 4.6 mm, 3.5 µm |
| Ibuprofen Concentration | Pharmacokinetic Parameters for 6β-Hydroxylase | |||
|---|---|---|---|---|
| Km (µM) | Vmax (µM−1∙min−1) | Clint (µM−2∙min−1) | % Inhibition | |
| 0 µM Ibuprofen | 15.967 ± 1.582 | 0.0151 ± 0.058 × 10−3 | 0.0009 ± 0.093 × 10−3 | - |
| 50 µM Ibuprofen | 15.873 ± 1.680 | 0.0116 ± 0.608 × 10−3 | 0.0007 ± 0.054 × 10−3 | 22.998 ± 4.341 |
| 100 µM Ibuprofen | 15.443 ± 1.034 | 0.0099 ± 0.040 × 10−3 | 0.0006 ± 0.043 × 10−3 | 34.734 ± 0.336 |
| 200 µM Ibuprofen | 15.153 ± 0.446 | 0.0092 ± 0.100 × 10−3 | 0.0006 ± 0.024 × 10−3 | 38.939 ± 0.476 |
| Remdesivir Concentration | Pharmacokinetic Parameters for 6β-Hydroxylase | ||||
|---|---|---|---|---|---|
| Km (µM) | Vmax (µM−1∙min−1) | Clint (µM−2∙min−1) | IC50 (µM) | Ki (µM) | |
| 0 µM Remdesivir | 9.637 ± 0.550 | 0.0119 ± 0.700 × 10−3 | 0.0001 ± 0.010 × 10−3 | 45.007 ± 0.016 | 22.504 ± 0.008 |
| 30 µM Remdesivir | 22.097 ± 0.922 | 0.0122 ± 0.306 × 10−3 | 0.0006 ± 0.011 × 10−3 | - | - |
| 50 µM Remdesivir | 23.167 ± 1.002 | 0.0146 ± 0.586 × 10−3 | 0.0006 ± 0.018 × 10−3 | - | - |
| 100 µM Remdesivir | 13.300 ± 0.436 | 0.0132 ± 0.153 × 10−3 | 0.0010 ± 0.025 × 10−3 | - | - |
| Omeprazole Concentration | Pharmacokinetic Parameters for 6β-Hydroxylase | ||||
|---|---|---|---|---|---|
| Km (µM) | Vmax (µM−1∙min−1) | Clint (µM−2∙min−1) | IC50 (µM) | Ki (µM) | |
| 0 µM Omeprazole | 16.033 ± 1.498 | 0.0152 ± 0.208 × 10−3 | 0.0010 ± 0.093 × 10−3 | 78.351 ± 0.460 | 39.175 ± 0.230 |
| 30 µM Omeprazole | 8.800 ± 1.307 | 0.0080 ± 0.346 × 10−3 | 0.0009 ± 0.091 × 10−3 | ||
| 50 µM Omeprazole | 6.423 ± 1.050 | 0.0062 ± 0.473 × 10−3 | 0.0010 ± 0.093 × 10−3 | ||
| 100 µM Omeprazole | 3.867 ± 0.230 | 0.0042 ± 0.058 × 10−3 | 0.0011 ± 0.066 × 10−3 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, A.; Naughton, D.P.; Barker, J. Potential Effects of Ibuprofen, Remdesivir and Omeprazole on Dexamethasone Metabolism in Control Sprague Dawley Male Rat Liver Microsomes (Drugs Often Used Together Alongside COVID-19 Treatment). Molecules 2022, 27, 2238. https://doi.org/10.3390/molecules27072238
Hussain A, Naughton DP, Barker J. Potential Effects of Ibuprofen, Remdesivir and Omeprazole on Dexamethasone Metabolism in Control Sprague Dawley Male Rat Liver Microsomes (Drugs Often Used Together Alongside COVID-19 Treatment). Molecules. 2022; 27(7):2238. https://doi.org/10.3390/molecules27072238
Chicago/Turabian StyleHussain, Amira, Declan P. Naughton, and James Barker. 2022. "Potential Effects of Ibuprofen, Remdesivir and Omeprazole on Dexamethasone Metabolism in Control Sprague Dawley Male Rat Liver Microsomes (Drugs Often Used Together Alongside COVID-19 Treatment)" Molecules 27, no. 7: 2238. https://doi.org/10.3390/molecules27072238
APA StyleHussain, A., Naughton, D. P., & Barker, J. (2022). Potential Effects of Ibuprofen, Remdesivir and Omeprazole on Dexamethasone Metabolism in Control Sprague Dawley Male Rat Liver Microsomes (Drugs Often Used Together Alongside COVID-19 Treatment). Molecules, 27(7), 2238. https://doi.org/10.3390/molecules27072238







