A Helquat-like Compound as a Potent Inhibitor of Flaviviral and Coronaviral Polymerases
Abstract
:1. Introduction
2. Results
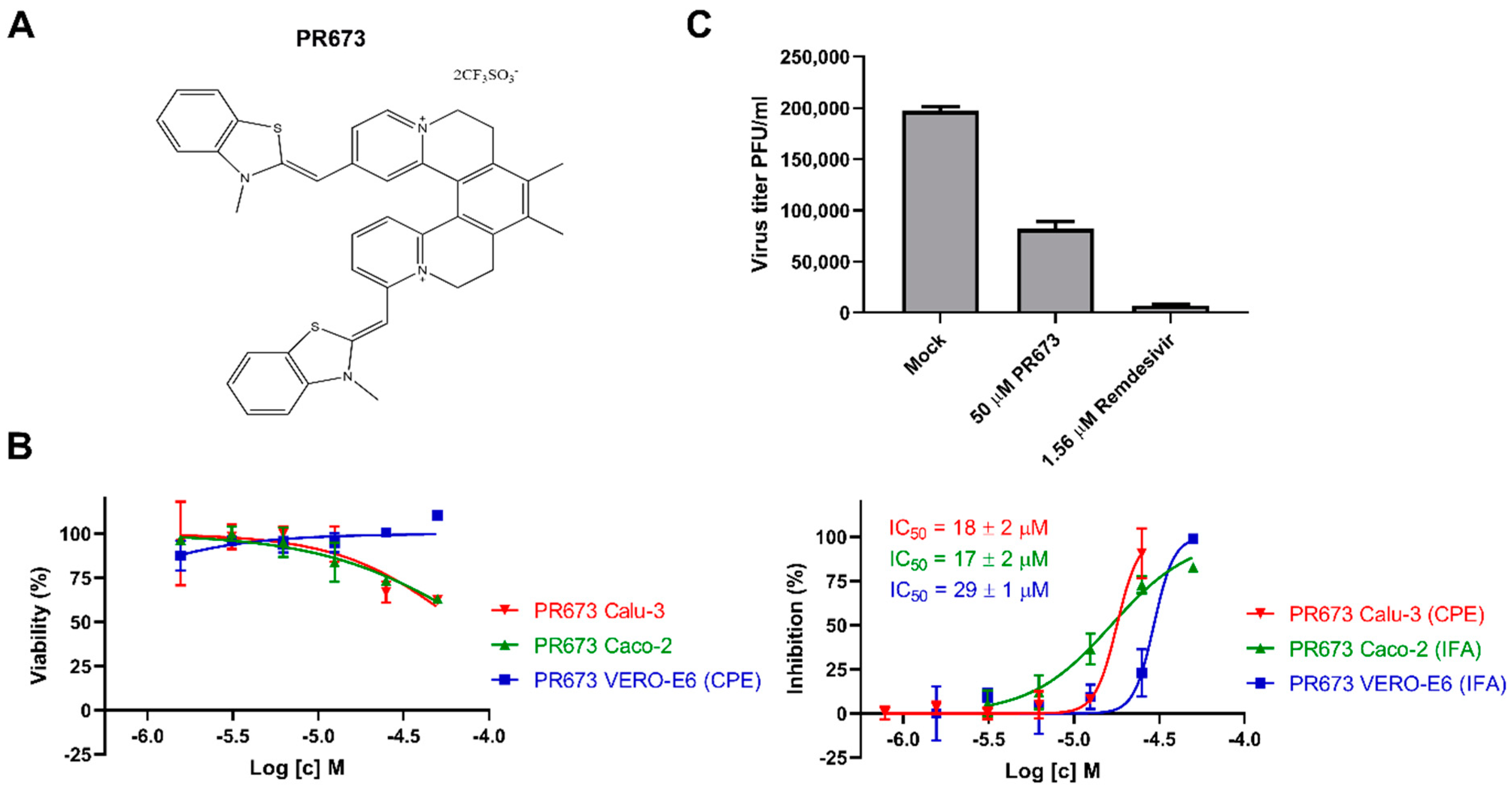
2.1. Identification of PR673
2.2. PR673 Inhibits the Coronaviral RdRp
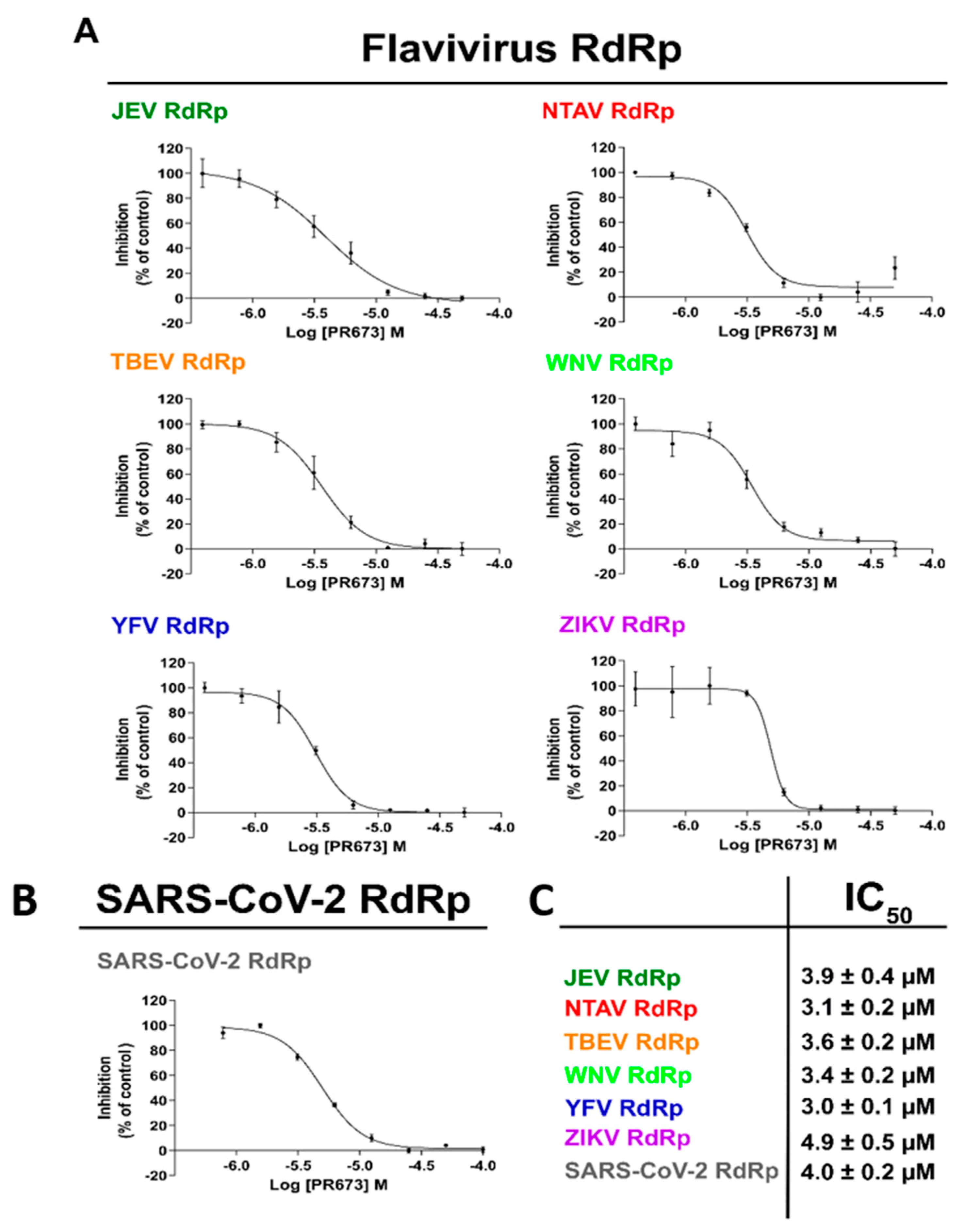
2.3. PR673 Inhibitory Activity against Flaviviral RdRps
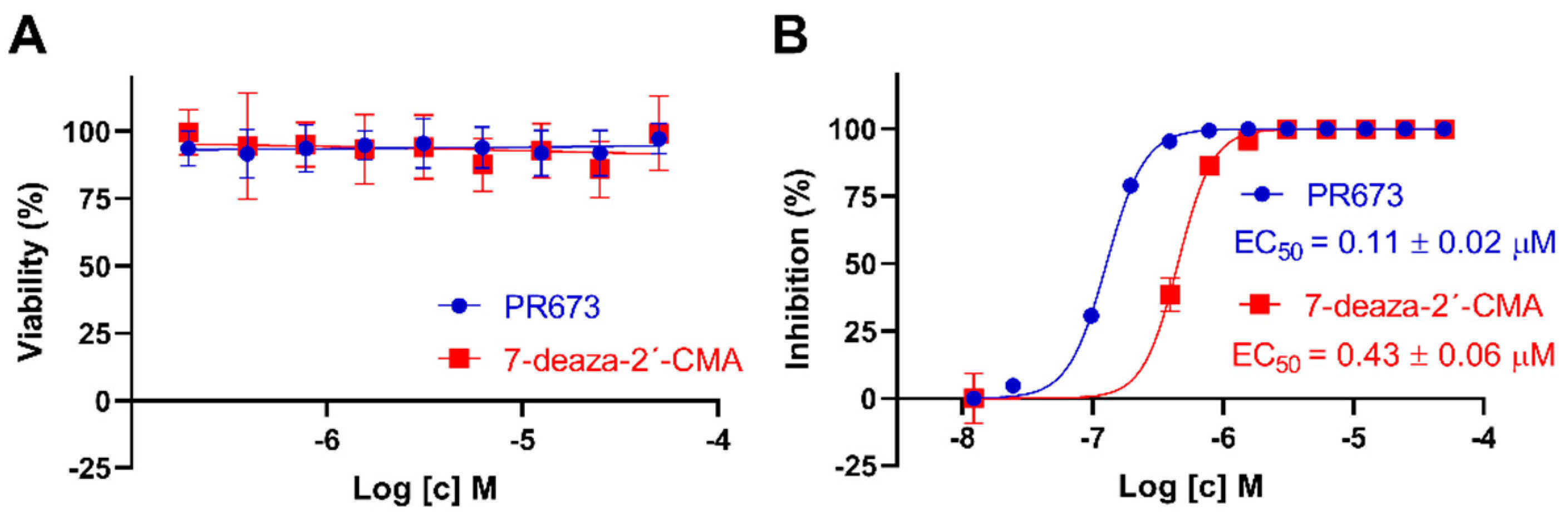
2.4. PR673 Inhibits Replication of the TBEV in Cell Culture
3. Discussion
4. Material and Methods
4.1. Anti-SARS-CoV-2 Activity Determination Using Immunofluorescence Assay
4.2. Anti-SARS-CoV-2 Activity Determination Using Cytopathic Effect-Based Assay
4.3. Cytotoxicity Determination in SARS-CoV-2 Assays
4.4. SARS-CoV-2 Yield Reduction Assay
4.5. Anti-TBEV Studies
4.6. Protein Expression and Purification
4.7. Primer Extension Polymerase Activity Assay
4.8. In Vitro Determination of IC50
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Moureau, G.; Cook, S.; Lemey, P.; Nougairede, A.; Forrester, N.L.; Khasnatinov, M.; Charrel, R.N.; Firth, A.E.; Gould, E.A.; de Lamballerie, X. New Insights into Flavivirus Evolution, Taxonomy and Biogeographic History, Extended by Analysis of Canonical and Alternative Coding Sequences. PLoS ONE 2015, 10, e0117849. [Google Scholar]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, O.S.; Waffa, Z. Neuropathogenic human coronaviruses: A review. Rev. Med. Virol. 2020, 30, e2118. [Google Scholar] [CrossRef] [PubMed]
- Shchelkanov, M.Y.; Popova, A.Y.; Dedkov, V.G.; Akimkin, V.G.; Maleev, V.V. History of Investigation and Current Classification of Coronaviruses (Nidovirales: Coronaviridae). Russ. J. Infect. Immun. 2020, 10, 221–246. [Google Scholar] [CrossRef] [Green Version]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Donaldson, E.F.; Baric, R.S. A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013, 11, 836–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Paixao, E.S.; Barreto, F.; Teixeira Mda, G.; Costa Mda, C.; Rodrigues, L.C. History, Epidemiology, and Clinical Manifestations of Zika: A Systematic Review. Am. J. Public Health 2016, 106, 606–612. [Google Scholar] [CrossRef]
- Rubin, D.; Chan-Tack, K.; Farley, J.; Sherwat, A. FDA Approval of Remdesivir—A Step in the Right Direction. N. Engl. J. Med. 2020, 383, 2598–2600. [Google Scholar] [CrossRef]
- Fischer, W.; Eron, J.J.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.; Mollan, K.R.; Wolfe, C.R.; et al. Molnupiravir, an Oral Antiviral Treatment for COVID-19. medRxiv 2021. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Li, P.F.; Wang, Y.N.; Lavrijsen, M.; Lamers, M.M.; de Vries, A.C.; Rottier, R.J.; Bruno, M.J.; Peppelenbosch, M.P.; Haagmans, B.L.; Pan, Q.W. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022, 32, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Frazier, M.N.; Dillard, L.B.; Krahn, J.M.; Perera, L.; Williams, J.G.; Wilson, I.M.; Stewart, Z.D.; Pillon, M.C.; Deterding, L.J.; Borgnia, M.J.; et al. Characterization of SARS2 Nsp15 nuclease activity reveals it’s mad about U. Nucleic Acids Res. 2021, 49, 10136–10149. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.A.; Douangamath, A.; Yadzani, S.; Yosaatmadja, Y.; Aimon, A.; Brandao-Neto, J.; Dunnett, L.; Gorrie-stone, T.; Skyner, R.; Fearon, D.; et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021, 12, 4848. [Google Scholar] [CrossRef] [PubMed]
- Cihlova, B.; Huskova, A.; Boserle, J.; Nencka, R.; Boura, E.; Silhan, J. High-Throughput Fluorescent Assay for Inhibitor Screening of Proteases from RNA Viruses. Molecules 2021, 26, 3792. [Google Scholar] [CrossRef] [PubMed]
- Nencka, R.; Silhan, J.; Klima, M.; Otava, T.; Kocek, H.; Krafcikova, P.; Boura, E. Coronaviral RNA-methyltransferases: Function, structure and inhibition. Nucleic Acids Res. 2022, 50, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Konkolova, E.; Klima, M.; Nencka, R.; Boura, E. Structural analysis of the putative SARS-CoV-2 primase complex. J. Struct. Biol. 2020, 211, 107548. [Google Scholar] [CrossRef] [PubMed]
- Otava, T.; Sala, M.; Li, F.; Fanfrlik, J.; Devkota, K.; Perveen, S.; Chau, I.; Pakarian, P.; Hobza, P.; Vedadi, M.; et al. The Structure-Based Design of SARS-CoV-2 nsp14 Methyltransferase Ligands Yields Nanomolar Inhibitors. ACS Infect. Dis. 2021, 7, 2214–2220. [Google Scholar] [CrossRef]
- Devkota, K.; Schapira, M.; Perveen, S.; Khalili Yazdi, A.; Li, F.; Chau, I.; Ghiabi, P.; Hajian, T.; Loppnau, P.; Bolotokova, A.; et al. Probing the SAM Binding Site of SARS-CoV-2 Nsp14 In Vitro Using SAM Competitive Inhibitors Guides Developing Selective Bisubstrate Inhibitors. SLAS Discov. 2021, 26, 1200–1211. [Google Scholar] [CrossRef]
- Perveen, S.; Khalili Yazdi, A.; Devkota, K.; Li, F.; Ghiabi, P.; Hajian, T.; Loppnau, P.; Bolotokova, A.; Vedadi, M. A High-Throughput RNA Displacement Assay for Screening SARS-CoV-2 nsp10-nsp16 Complex toward Developing Therapeutics for COVID-19. SLAS Discov. 2021, 26, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Dejmek, M.; Konkolova, E.; Eyer, L.; Strakova, P.; Svoboda, P.; Sala, M.; Krejcova, K.; Ruzek, D.; Boura, E.; Nencka, R. Non-Nucleotide RNA-Dependent RNA Polymerase Inhibitor That Blocks SARS-CoV-2 Replication. Viruses 2021, 13, 1585. [Google Scholar] [CrossRef] [PubMed]
- Eggleton, J.S.; Nagalli, S. Highly Active Antiretroviral Therapy (HAART); StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Tykvart, J.; Navratil, V.; Kugler, M.; Sacha, P.; Schimer, J.; Hlavackova, A.; Tenora, L.; Zemanova, J.; Dejmek, M.; Kral, V.; et al. Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA). SLAS Discov. 2020, 25, 1026–1037. [Google Scholar] [CrossRef] [PubMed]
- Eyer, L.; Smidkova, M.; Nencka, R.; Neca, J.; Kastl, T.; Palus, M.; De Clercq, E.; Ruzek, D. Structure-activity relationships of nucleoside analogues for inhibition of tick-borne encephalitis virus. Antivir. Res. 2016, 133, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Eyer, L.; Valdes, J.J.; Gil, V.A.; Nencka, R.; Hrebabecky, H.; Sala, M.; Salat, J.; Cerny, J.; Palus, M.; De Clercq, E.; et al. Nucleoside inhibitors of tick-borne encephalitis virus. Antimicrob. Agents Chemother. 2015, 59, 5483–5493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozuch, O.; Mayer, V. Pig kidney epithelial (PS) cells: A perfect tool for the study of flaviviruses and some other arboviruses. Acta Virol. 1975, 19, 498. [Google Scholar]
- Zeng, J.; Weissmann, F.; Bertolin, A.P.; Posse, V.; Canal, B.; Ulferts, R.; Wu, M.; Harvey, R.; Hussain, S.; Milligan, J.C.; et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase. Biochem. J. 2021, 478, 2405–2423. [Google Scholar] [CrossRef] [PubMed]
- Perez-Lemus, G.R.; Menendez, C.A.; Alvarado, W.; Bylehn, F.; de Pablo, J.J. Toward wide-spectrum antivirals against coronaviruses: Molecular characterization of SARS-CoV-2 NSP13 helicase inhibitors. Sci. Adv. 2022, 8, eabj4526. [Google Scholar] [CrossRef] [PubMed]
- Rona, G.; Zeke, A.; Miwatani-Minter, B.; de Vries, M.; Kaur, R.; Schinlever, A.; Garcia, S.F.; Goldberg, H.V.; Wang, H.; Hinds, T.R.; et al. The NSP14/NSP10 RNA repair complex as a Pan-coronavirus therapeutic target. Cell Death Differ. 2022, 29, 285–292. [Google Scholar] [CrossRef]
- Canal, B.; Fujisawa, R.; McClure, A.W.; Deegan, T.D.; Wu, M.; Ulferts, R.; Weissmann, F.; Drury, L.S.; Bertolin, A.P.; Zeng, J.K.; et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp15 endoribonuclease. Biochem. J. 2021, 478, 2465–2479. [Google Scholar] [CrossRef]
- Khalili Yazdi, A.; Li, F.; Devkota, K.; Perveen, S.; Ghiabi, P.; Hajian, T.; Bolotokova, A.; Vedadi, M. A High-Throughput Radioactivity-Based Assay for Screening SARS-CoV-2 nsp10-nsp16 Complex. SLAS Discov. 2021, 26, 757–765. [Google Scholar] [CrossRef]
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Gotte, M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020, 295, 4773–4779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Gotte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konkolova, E.; Dejmek, M.; Hrebabecky, H.; Sala, M.; Boserle, J.; Nencka, R.; Boura, E. Remdesivir triphosphate can efficiently inhibit the RNA-dependent RNA polymerase from various flaviviruses. Antivir. Res. 2020, 182, 104899. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Daifuku, R.; Loeb, L.A. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 2004, 58, 183–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seley-Radtke, K.L.; Yates, M.K. The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antivir. Res. 2018, 154, 66–86. [Google Scholar] [CrossRef] [PubMed]
- Eastman, R.T.; Roth, J.S.; Brimacombe, K.R.; Simeonov, A.; Shen, M.; Patnaik, S.; Hall, M.D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Central Sci. 2020, 6, 672–683. [Google Scholar] [CrossRef]
- De Clercq, E.; Neyts, J. Antiviral agents acting as DNA or RNA chain terminators. In The Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 53–84. [Google Scholar]
- Menendez-Arias, L.; Andino, R. Viral polymerases. Virus Res. 2017, 234, 1–3. [Google Scholar] [CrossRef]
- Dubankova, A.; Boura, E. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antivir. Res. 2019, 169, 104536. [Google Scholar] [CrossRef]
- Dubankova, A.; Humpolickova, J.; Klima, M.; Boura, E. Negative charge and membrane-tethered viral 3B cooperate to recruit viral RNA dependent RNA polymerase 3D (pol). Sci. Rep. 2017, 7, 17309. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konkolova, E.; Krejčová, K.; Eyer, L.; Hodek, J.; Zgarbová, M.; Fořtová, A.; Jirasek, M.; Teply, F.; Reyes-Gutierrez, P.E.; Růžek, D.; et al. A Helquat-like Compound as a Potent Inhibitor of Flaviviral and Coronaviral Polymerases. Molecules 2022, 27, 1894. https://doi.org/10.3390/molecules27061894
Konkolova E, Krejčová K, Eyer L, Hodek J, Zgarbová M, Fořtová A, Jirasek M, Teply F, Reyes-Gutierrez PE, Růžek D, et al. A Helquat-like Compound as a Potent Inhibitor of Flaviviral and Coronaviral Polymerases. Molecules. 2022; 27(6):1894. https://doi.org/10.3390/molecules27061894
Chicago/Turabian StyleKonkolova, Eva, Kateřina Krejčová, Luděk Eyer, Jan Hodek, Michala Zgarbová, Andrea Fořtová, Michael Jirasek, Filip Teply, Paul E. Reyes-Gutierrez, Daniel Růžek, and et al. 2022. "A Helquat-like Compound as a Potent Inhibitor of Flaviviral and Coronaviral Polymerases" Molecules 27, no. 6: 1894. https://doi.org/10.3390/molecules27061894
APA StyleKonkolova, E., Krejčová, K., Eyer, L., Hodek, J., Zgarbová, M., Fořtová, A., Jirasek, M., Teply, F., Reyes-Gutierrez, P. E., Růžek, D., Weber, J., & Boura, E. (2022). A Helquat-like Compound as a Potent Inhibitor of Flaviviral and Coronaviral Polymerases. Molecules, 27(6), 1894. https://doi.org/10.3390/molecules27061894






