Lipid Droplets in Cancer: From Composition and Role to Imaging and Therapeutics
Abstract
1. Introduction
2. LDs Significance and Impact
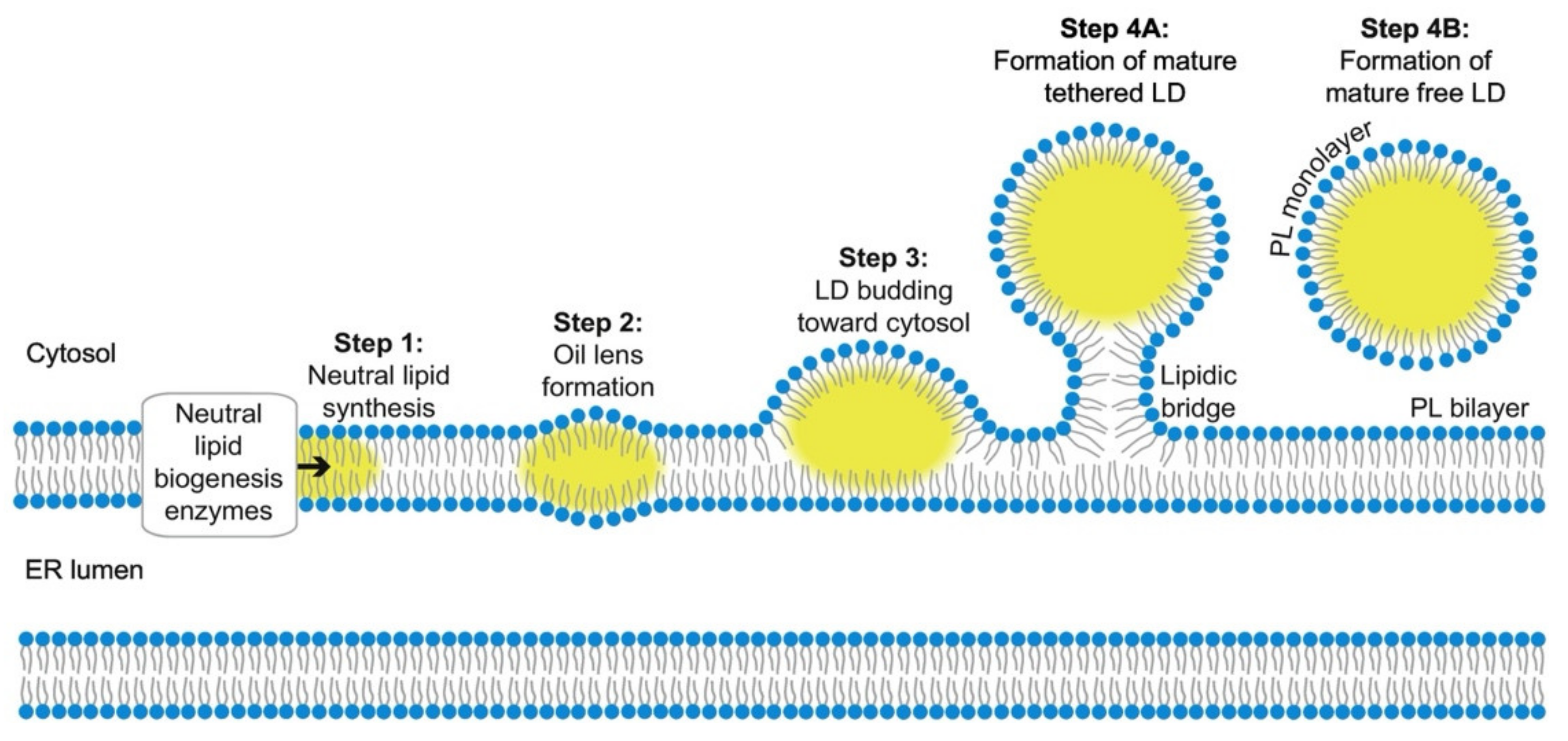
2.1. Biogenesis and Growth
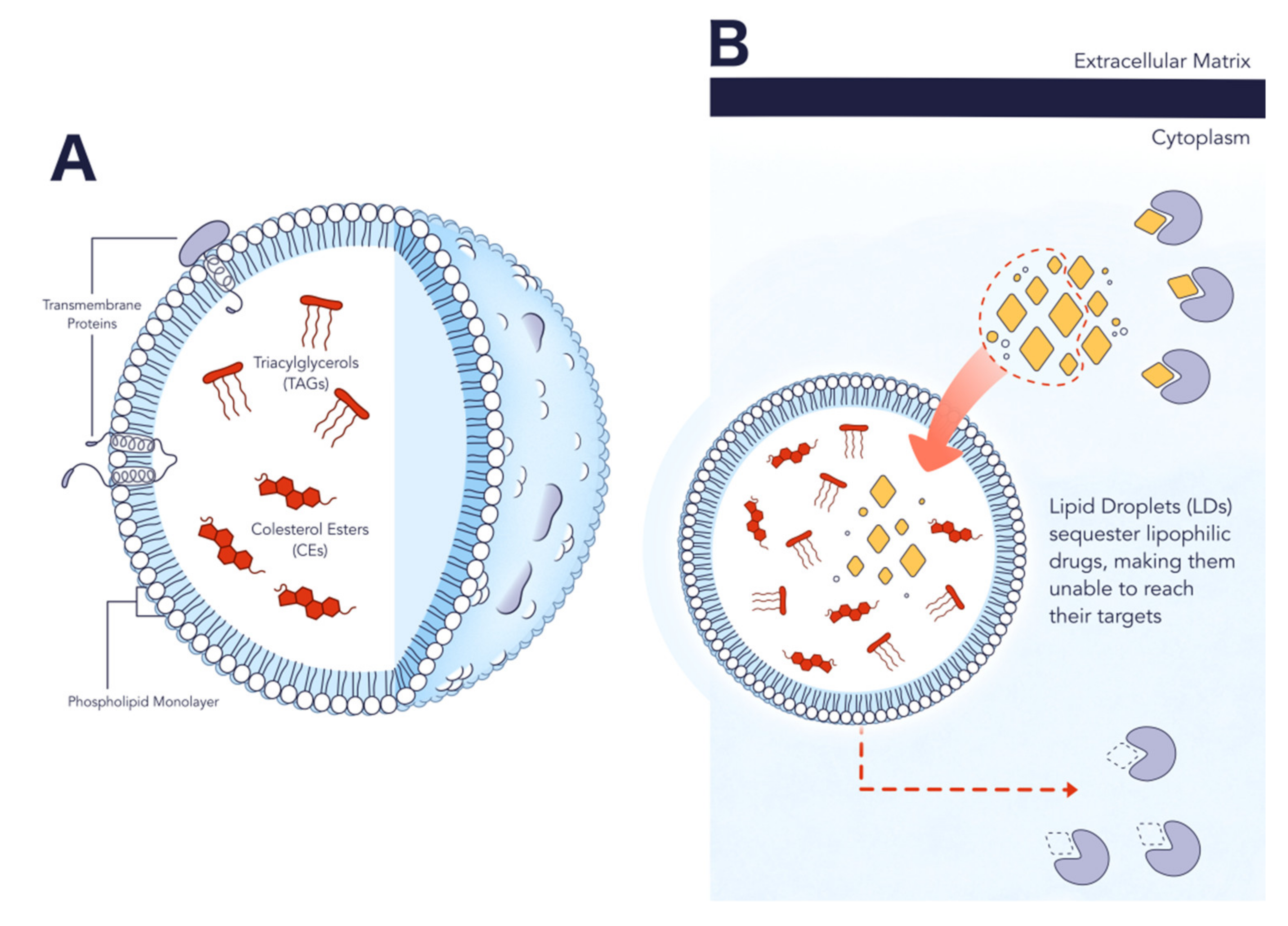
2.2. Structure and Composition
2.3. Role in Cancer
3. LDs in Lipid Metabolism-Related Disease Theranostics
3.1. Imaging Strategies
3.2. Targeting Novel Therapeutics
4. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cahill, D.P.; Kinzler, K.W.; Vogelstein, B.; Lengauer, C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999, 9, M57–M60. [Google Scholar] [CrossRef]
- Wishart, D.S. Is cancer a genetic disease or a metabolic disease? EBioMedicine 2015, 2, 478–479. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFbeta in cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A. Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 2005, 6, 611–622. [Google Scholar] [CrossRef]
- Lowe, S.W.; Cepero, E.; Evan, G. Intrinsic tumour suppression. Nature 2004, 432, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Taylor, N.M.I.; Manolaridis, I.; Jackson, S.M.; Kowal, J.; Stahlberg, H.; Locher, K.P. Structure of the human multidrug transporter ABCG2. Nature 2017, 546, 504–509. [Google Scholar] [CrossRef]
- Fodale, V.; Pierobon, M.; Liotta, L.; Petricoin, E. Mechanism of cell adaptation: When and how do cancer cells develop chemoresistance? Cancer J. 2011, 17, 89–95. [Google Scholar] [CrossRef]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Ameer, F.; Scandiuzzi, L.; Hasnain, S.; Kalbacher, H.; Zaidi, N. De novo lipogenesis in health and disease. Metabolism 2014, 63, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, Q.; Halim, A.; Song, G. Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett. 2017, 401, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Koizume, S.; Miyagi, Y. Lipid droplets: A key cellular organelle associated with cancer cell survival under normoxia and hypoxia. Int. J. Mol. Sci. 2016, 17, 1430. [Google Scholar] [CrossRef]
- Guo, Y.; Cordes, K.R.; Farese, R.V., Jr.; Walther, T.C. Lipid droplets at a glance. J. Cell Sci. 2009, 122, 749–752. [Google Scholar] [CrossRef]
- Bozza, P.T.; Viola, J.P. Lipid droplets in inflammation and cancer. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 243–250. [Google Scholar] [CrossRef]
- Ando, K.; Hu, Q.; Kasagi, Y.; Oki, E.; Mori, M. Recent developments in cancer research: Expectations for a new remedy. Ann. Gastroenterol. Surg. 2021, 5, 419–426. [Google Scholar] [CrossRef]
- Padma, V.V. An overview of targeted cancer therapy. Biomedicine 2015, 5, 19. [Google Scholar] [CrossRef]
- Masood, F. Polymeric nanoparticles for targeted drug delivery system for cancer therapy. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 60, 569–578. [Google Scholar] [CrossRef]
- Chen, F.; Ehlerding, E.B.; Cai, W. Theranostic nanoparticles. J. Nucl. Med. 2014, 55, 1919–1922. [Google Scholar] [CrossRef]
- Janib, S.M.; Moses, A.S.; MacKay, J.A. Imaging and drug delivery using theranostic nanoparticles. Adv. Drug Deliv. Rev. 2010, 62, 1052–1063. [Google Scholar] [CrossRef]
- Kim, B.Y.; Rutka, J.T.; Chan, W.C. Nanomedicine. N. Engl. J. Med. 2010, 363, 2434–2443. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Meyers, A.; Weiskittel, T.M.; Dalhaimer, P. Lipid droplets: Formation to breakdown. Lipids 2017, 52, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Sandager, L.; Gustavsson, M.H.; Ståhl, U.; Dahlqvist, A.; Wiberg, E.; Banas, A.; Lenman, M.; Ronne, H.; Stymne, S. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 2002, 277, 6478–6482. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, V.; Jacquier, N.; Schneiter, R. The topology of the triacylglycerol synthesizing enzyme Lro1 indicates that neutral lipids can be produced within the luminal compartment of the endoplasmatic reticulum: Implications for the biogenesis of lipid droplets. Commun. Integr. Biol. 2011, 4, 781–784. [Google Scholar] [CrossRef]
- Velázquez, A.P.; Tatsuta, T.; Ghillebert, R.; Drescher, I.; Graef, M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 2016, 212, 621–631. [Google Scholar] [CrossRef]
- Henne, M.; Goodman, J.M.; Hariri, H. Spatial compartmentalization of lipid droplet biogenesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158499. [Google Scholar] [CrossRef]
- Choudhary, V.; Ojha, N.; Golden, A.; Prinz, W.A. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 2015, 211, 261–271. [Google Scholar] [CrossRef]
- Choudhary, V.; Golden, A.; Prinz, W.A. Keeping FIT, storing fat: Lipid droplet biogenesis. Worm 2016, 5, e1170276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gross, D.A.; Zhan, C.; Silver, D.L. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. USA 2011, 108, 19581–19586. [Google Scholar] [CrossRef]
- Fei, W.; Du, X.; Yang, H. Seipin, adipogenesis and lipid droplets. Trends Endocrinol. Metab. 2011, 22, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Becuwe, M.; Housden, B.E.; Chitraju, C.; Porras, A.J.; Graham, M.M.; Liu, X.N.; Thiam, A.R.; Savage, D.B.; Agarwal, A.K.; et al. Seipin is required for converting nascent to mature lipid droplets. eLife 2016, 5, e35977. [Google Scholar] [CrossRef] [PubMed]
- Onal, G.; Kutlu, O.; Gozuacik, D.; Dokmeci Emre, S. Lipid droplets in health and disease. Lipids Health Dis. 2017, 16, 128. [Google Scholar] [CrossRef]
- Walther, T.C.; Chung, J.; Farese, R.V., Jr. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 2017, 33, 491–510. [Google Scholar] [CrossRef]
- Kory, N.; Farese, R.V., Jr.; Walther, T.C. Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol. 2016, 26, 535–546. [Google Scholar] [CrossRef]
- Wilfling, F.; Thiam, A.R.; Olarte, M.J.; Wang, J.; Beck, R.; Gould, T.J.; Allgeyer, E.S.; Pincet, F.; Bewersdorf, J.; Farese, R.V., Jr.; et al. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 2014, 3, e01607. [Google Scholar] [CrossRef]
- Boström, P.; Rutberg, M.; Ericsson, J.; Holmdahl, P.; Andersson, L.; Frohman, M.A.; Borén, J.; Olofsson, S.O. Cytosolic lipid droplets increase in size by microtubule-dependent complex formation. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 1945–1951. [Google Scholar] [CrossRef]
- Gong, J.; Sun, Z.; Wu, L.; Xu, W.; Schieber, N.; Xu, D.; Shui, G.; Yang, H.; Parton, R.G.; Li, P. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J. Cell Biol. 2011, 195, 953–963. [Google Scholar] [CrossRef]
- Nettebrock, N.T.; Bohnert, M. Born this way—Biogenesis of lipid droplets from specialized ER subdomains. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158448. [Google Scholar] [CrossRef]
- Dhiman, R.; Caesar, S.; Thiam, A.R.; Schrul, B. Mechanisms of protein targeting to lipid droplets: A unified cell biological and biophysical perspective. Semin. Cell Dev. Biol. 2020, 108, 4–13. [Google Scholar] [CrossRef]
- Tirinato, L.; Pagliari, F.; Limongi, T.; Marini, M.; Falqui, A.; Seco, J.; Candeloro, P.; Liberale, C.; Di Fabrizio, E. An overview of lipid droplets in cancer and cancer stem cells. Stem. Cells Int. 2017, 2017, 1656053. [Google Scholar] [CrossRef]
- Huang, J.; Li, L.; Lian, J.; Schauer, S.; Vesely, P.W.; Kratky, D.; Hoefler, G.; Lehner, R. Tumor-induced hyperlipidemia contributes to tumor growth. Cell Rep. 2016, 15, 336–348. [Google Scholar] [CrossRef]
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martín, M.; Castellanos, A.; Attolini, C.S.; Berenguer, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017, 541, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, J.B.; Chung, L.W.; Huang, W.C. Anti-cancer efficacy of SREBP inhibitor, alone or in combination with docetaxel, in prostate cancer harboring p53 mutations. Oncotarget 2015, 6, 41018–41032. [Google Scholar] [CrossRef]
- Zagani, R.; El-Assaad, W.; Gamache, I.; Teodoro, J.G. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumor suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget 2015, 6, 28282–28295. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Kalra, I.; Salunke, P.; Vasishta, R.K. Lipidized glioblastoma: A rare differentiation pattern. Neuropathology 2011, 31, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Almeida, L.Y.; Bastos, D.C.; Ortega, R.M.; Moreira, F.S.; Seguin, F.; Zecchin, K.G.; Raposo, H.F.; Oliveira, H.C.; Amoêdo, N.D.; et al. The fatty acid synthase inhibitor orlistat reduces the growth and metastasis of orthotopic tongue oral squamous cell carcinomas. Mol. Cancer Ther. 2014, 13, 585–595. [Google Scholar] [CrossRef]
- Baldo, S.; Antunes, P.; Felicidade, J.F.; Santos, F.M.F.; Arteaga, J.F.; Fernandes, F.; Pischel, U.; Pinto, S.N.; Gois, P.M.P. The BASHY platform enables the assembly of a fluorescent bortezomib-GV1001 conjugate. ACS Med. Chem. Lett. 2022, 13, 128–133. [Google Scholar] [CrossRef]
- Thiam, A.R.; Farese, R.V., Jr.; Walther, T.C. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 2013, 14, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, I.; Cui, Y.; Amiri, A.; Ding, Y.; Campbell, R.E.; Maysinger, D. Pharmacological inhibition of lipid droplet formation enhances the effectiveness of curcumin in glioblastoma. Eur. J. Pharm. Biopharm. 2016, 100, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Cotte, A.K.; Aires, V.; Fredon, M.; Limagne, E.; Derangère, V.; Thibaudin, M.; Humblin, E.; Scagliarini, A.; de Barros, J.P.; Hillon, P.; et al. Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat. Commun. 2018, 9, 322. [Google Scholar] [CrossRef] [PubMed]
- Nieva, C.; Marro, M.; Santana-Codina, N.; Rao, S.; Petrov, D.; Sierra, A. The lipid phenotype of breast cancer cells characterized by Raman microspectroscopy: Towards a stratification of malignancy. PLoS ONE 2012, 7, e46456. [Google Scholar] [CrossRef]
- Yu, W.; Cassara, J.; Weller, P.F. Phosphatidylinositide 3-kinase localizes to cytoplasmic lipid bodies in human polymorphonuclear leukocytes and other myeloid-derived cells. Blood 2000, 95, 1078–1085. [Google Scholar] [CrossRef]
- Bai, R.; Rebelo, A.; Kleeff, J.; Sunami, Y. Identification of prognostic lipid droplet-associated genes in pancreatic cancer patients via bioinformatics analysis. Lipids Health Dis. 2021, 20, 58. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Cheng, X.; Wu, X.; Yoo, J.Y.; Cheng, C.; Guo, J.Y.; Mo, X.; Ru, P.; Hurwitz, B.; Kim, S.H.; et al. Inhibition of SOAT1 suppresses glioblastoma growth via blocking SREBP-1-mediated lipogenesis. Clin. Cancer Res. 2016, 22, 5337–5348. [Google Scholar] [CrossRef]
- Rinia, H.A.; Burger, K.N.; Bonn, M.; Müller, M. Quantitative label-free imaging of lipid composition and packing of individual cellular lipid droplets using multiplex CARS microscopy. Biophys. J. 2008, 95, 4908–4914. [Google Scholar] [CrossRef]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef]
- Fam, T.K.; Klymchenko, A.S.; Collot, M. Recent advances in fluorescent probes for lipid droplets. Materials 2018, 11, 1768. [Google Scholar] [CrossRef]
- Melo, R.C.; D’Avila, H.; Wan, H.C.; Bozza, P.T.; Dvorak, A.M.; Weller, P.F. Lipid bodies in inflammatory cells: Structure, function, and current imaging techniques. J. Histochem. Cytochem. 2011, 59, 540–556. [Google Scholar] [CrossRef] [PubMed]
- Spandl, J.; White, D.J.; Peychl, J.; Thiele, C. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic 2009, 10, 1579–1584. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Lee, S.; Park, S.B. A Seoul-Fluor-based bioprobe for lipid droplets and its application in image-based high throughput screening. Chem. Commun. 2012, 48, 2331–2333. [Google Scholar] [CrossRef] [PubMed]
- Kuntam, S.; Puskás, L.G.; Ayaydin, F. Characterization of a new class of blue-fluorescent lipid droplet markers for live-cell imaging in plants. Plant Cell Rep. 2015, 34, 655–665. [Google Scholar] [CrossRef]
- Kikuchi, K. Design, synthesis and biological application of chemical probes for bio-imaging. Chem. Soc. Rev. 2010, 39, 2048–2053. [Google Scholar] [CrossRef] [PubMed]
- Ashoka, A.H.; Ashokkumar, P.; Kovtun, Y.P.; Klymchenko, A.S. Solvatochromic near-infrared probe for polarity mapping of biomembranes and lipid droplets in cells under stress. J. Phys. Chem. Lett. 2019, 10, 2414–2421. [Google Scholar] [CrossRef] [PubMed]
- Darragh, O.C.; Byrne, A.; Berselli, G.B.; Long, C.; Keyes, T.E. Mega-stokes pyrene ceramide conjugates for STED imaging of lipid droplets in live cells. Analyst 2019, 144, 1608–1621. [Google Scholar] [CrossRef]
- Öberg, E.; Appelqvist, H.; Nilsson, K.P.R. Non-fused phospholes as fluorescent probes for imaging of lipid droplets in living cells. Front. Chem. 2017, 5, 28. [Google Scholar] [CrossRef]
- Sharma, A.; Jha, A.K.; Mishra, S.; Jain, A.; Chauhan, B.S.; Kathuria, M.; Rawat, K.S.; Gupta, N.M.; Tripathi, R.; Mitra, K.; et al. Imaging and quantitative detection of lipid droplets by yellow fluorescent probes in liver sections of Plasmodium infected mice and third stage human cervical cancer tissues. Bioconjug. Chem. 2018, 29, 3606–3613. [Google Scholar] [CrossRef]
- Jana, P.; Siva, A.; Soppina, V.; Kanvah, S. Live-cell imaging of lipid droplets using solvatochromic coumarin derivatives. Org. Biomol. Chem. 2020, 18, 5608–5616. [Google Scholar] [CrossRef]
- Santos, F.M.; Rosa, J.N.; Candeias, N.R.; Carvalho, C.P.; Matos, A.I.; Ventura, A.E.; Florindo, H.F.; Silva, L.C.; Pischel, U.; Gois, P.M. A three-component assembly promoted by boronic acids delivers a modular fluorophore platform (BASHY dyes). Chemistry 2016, 22, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.X.; Ding, N.; Chen, S.; Yu, Y.L.; Wang, J.H. One-step synthesis of carbon nanoparticles capable of long-term tracking lipid droplet for real-time monitoring of lipid catabolism and pharmacodynamic evaluation of lipid-lowering drugs. Anal. Chem. 2021, 93, 5284–5290. [Google Scholar] [CrossRef]
- Klymchenko, A.S.; Roger, E.; Anton, N.; Anton, H.; Shulov, I.; Vermot, J.; Mely, Y.; Vandamme, T.F. Highly lipophilic fluorescent dyes in nano-emulsions: Towards bright non-leaking nano-droplets. RSC Adv. 2012, 2, 11876–11886. [Google Scholar] [CrossRef] [PubMed]
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005, 4, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Sapsford, K.E.; Algar, W.R.; Berti, L.; Gemmill, K.B.; Casey, B.J.; Oh, E.; Stewart, M.H.; Medintz, I.L. Functionalizing nanoparticles with biological molecules: Developing chemistries that facilitate nanotechnology. Chem. Rev. 2013, 113, 1904–2074. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Jana, N.R. Quantum dot-based designed nanoprobe for imaging lipid droplet. J. Phys. Chem. C 2017, 121, 23727–23735. [Google Scholar] [CrossRef]
- Jiang, G.; Li, C.; Liu, X.; Chen, Q.; Li, X.; Gu, X.; Zhang, P.; Lai, Q.; Wang, J. Lipid droplet-targetable fluorescence guided photodynamic therapy of cancer cells with an activatable AIE-active fluorescent probe for hydrogen peroxide. Adv. Opt. Mater. 2020, 8, 2001119. [Google Scholar] [CrossRef]
- Li, X.; Kwon, N.; Guo, T.; Liu, Z.; Yoon, J. Innovative strategies for hypoxic-tumor photodynamic therapy. Angew. Chem. Int. Ed. 2018, 57, 11522–11531. [Google Scholar] [CrossRef]
- Sun, X.; Zebibula, A.; Dong, X.; Zhang, G.; Zhang, D.; Qian, J.; He, S. Aggregation-induced emission nanoparticles encapsulated with PEGylated nano graphene oxide and their applications in two-photon fluorescence bioimaging and photodynamic therapy in vitro and in vivo. ACS Appl. Mater. Interfaces 2018, 10, 25037–25046. [Google Scholar] [CrossRef]
- Hong, Y.; Lam, J.W.; Tang, B.Z. Aggregation-induced emission. Chem. Soc. Rev. 2011, 40, 5361–5388. [Google Scholar] [CrossRef]
- Gu, X.; Kwok, R.T.K.; Lam, J.W.Y.; Tang, B.Z. AIEgens for biological process monitoring and disease theranostics. Biomaterials 2017, 146, 115–135. [Google Scholar] [CrossRef]
- Feng, H.T.; Yuan, Y.X.; Xiong, J.B.; Zheng, Y.S.; Tang, B.Z. Macrocycles and cages based on tetraphenylethylene with aggregation-induced emission effect. Chem. Soc. Rev. 2018, 47, 7452–7476. [Google Scholar] [CrossRef] [PubMed]
- Bennett, L.E.; Ghiggino, K.P.; Henderson, R.W. Singlet oxygen formation in monomeric and aggregated porphyrin c. J. Photochem. Photobiol. B 1989, 3, 81–89. [Google Scholar] [CrossRef]
- Park, S.Y.; Baik, H.J.; Oh, Y.T.; Oh, K.T.; Youn, Y.S.; Lee, E.S. A smart polysaccharide/drug conjugate for photodynamic therapy. Angew. Chem. Int. Ed. 2011, 50, 1644–1647. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Liu, B. Aggregation-induced emission (AIE) dots: Emerging theranostic nanolights. Acc. Chem. Res. 2018, 51, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Tang, B.Z. Aggregation-induced emission probes for cancer theranostics. Drug Discov. Today 2017, 22, 1288–1294. [Google Scholar] [CrossRef]
- Liang, J.; Tang, B.Z.; Liu, B. Specific light-up bioprobes based on AIEgen conjugates. Chem. Soc. Rev. 2015, 44, 2798–2811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mao, H.; Xu, W.; Shi, J.; Cai, Z.; Tong, B.; Dong, Y. Aggregation-induced emission of multiphenyl-substituted 1,3-butadiene derivatives: Synthesis, properties and application. Chemistry 2018, 24, 15965–15977. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Min, T.; Gong, J.; Du, L.; Phillips, D.L.; Liu, J.; Lam, J.W.Y.; Sung, H.H.Y.; Williams, I.D.; et al. Time-dependent photodynamic therapy for multiple targets: A highly efficient AIE-active photosensitizer for selective bacterial elimination and cancer cell ablation. Angew. Chem. Int. Ed. 2020, 59, 9470–9477. [Google Scholar] [CrossRef]
- Ni, X.; Zhang, X.; Duan, X.; Zheng, H.L.; Xue, X.S.; Ding, D. Near-infrared afterglow luminescent aggregation-induced emission dots with ultrahigh tumor-to-liver signal ratio for promoted image-guided cancer surgery. Nano Lett. 2019, 19, 318–330. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, X.; He, X.; He, Z.; Yang, X.; Tian, S.; Meng, F.; Ding, D.; Luo, L.; Tang, B.Z. A dual-functional photosensitizer for ultraefficient photodynamic therapy and synchronous anticancer efficacy monitoring. Adv. Funct. Mater. 2019, 29, 1902673. [Google Scholar] [CrossRef]
- Tabero, A.; García-Garrido, F.; Prieto-Castañeda, A.; Palao, E.; Agarrabeitia, A.R.; García-Moreno, I.; Villanueva, A.; de la Moya, S.; Ortiz, M.J. BODIPYs revealing lipid droplets as valuable targets for photodynamic theragnosis. Chem. Commun. 2020, 56, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, Y.; Yang, B.; Guan, P.; Chai, J.; Wen, G.; Liu, B. Tunable NIR AIE-active optical materials for lipid droplet imaging in typical model organisms and photodynamic therapy. J. Mater. Chem. B 2021, 9, 2417–2427. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Li, C.; Wang, Y.; Zhuang, W.; Chen, M.; Zhou, L.; Zhang, J.; Gong, Q.; Wei, Q.; You, J. A biheteroaryl-bridged fluorescence probe enables lipid droplets-specific bioimaging and photodynamic therapy in clinical clear cell renal cell carcinoma. Dye. Pigment. 2021, 188, 109215. [Google Scholar] [CrossRef]
- Tan, P.; Zhuang, W.; Li, S.; Zhang, J.; Xu, H.; Yang, L.; Liao, Y.; Chen, M.; Wei, Q. A lipid droplet targeted fluorescent probe for high-efficiency image-guided photodynamic therapy of renal cell carcinoma. Chem. Commun. 2021, 57, 1046–1049. [Google Scholar] [CrossRef]
- Frath, D.; Massue, J.; Ulrich, G.; Ziessel, R. Luminescent materials: Locking π-conjugated and heterocyclic ligands with boron(III). Angew. Chem. Int. Ed. 2014, 53, 2290–2310. [Google Scholar] [CrossRef]
- Liu, M.; Li, C. Recent advances in activatable organic photosensitizers for specific photodynamic therapy. ChemPlusChem 2020, 85, 948–957. [Google Scholar] [CrossRef]
- Xia, X.; Wang, R.; Hu, Y.; Liu, W.; Liu, T.; Sun, W.; Fan, J.; Peng, X. A novel photosensitizer for lipid droplet-location photodynamic therapy. Front. Chem. 2021, 9, 701771. [Google Scholar] [CrossRef]
- Feng, G.; Zhang, G.Q.; Ding, D. Design of superior phototheranostic agents guided by Jablonski diagrams. Chem. Soc. Rev. 2020, 49, 8179–8234. [Google Scholar] [CrossRef]
- Dai, Y.; Zhao, X.; Ji, H.; Zhang, D.; Zhang, P.; Xue, K.; Misal, S.; Zhu, H.; Qi, Z. Multifunctional aggregation-induced emission nanoparticle for high-fidelity imaging of lipid droplets in living cells and its application in photodynamic therapy. Chem. Eng. J. 2021, 410, 128186. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Bae, S.C.; Granick, S. Nanoparticle-induced surface reconstruction of phospholipid membranes. Proc. Natl. Acad. Sci. USA 2008, 105, 18171–18175. [Google Scholar] [CrossRef] [PubMed]
- Foreman-Ortiz, I.U.; Liang, D.; Laudadio, E.D.; Calderin, J.D.; Wu, M.; Keshri, P.; Zhang, X.; Schwartz, M.P.; Hamers, R.J.; Rotello, V.M.; et al. Anionic nanoparticle-induced perturbation to phospholipid membranes affects ion channel function. Proc. Natl. Acad. Sci. USA 2020, 117, 27854–27861. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antunes, P.; Cruz, A.; Barbosa, J.; Bonifácio, V.D.B.; Pinto, S.N. Lipid Droplets in Cancer: From Composition and Role to Imaging and Therapeutics. Molecules 2022, 27, 991. https://doi.org/10.3390/molecules27030991
Antunes P, Cruz A, Barbosa J, Bonifácio VDB, Pinto SN. Lipid Droplets in Cancer: From Composition and Role to Imaging and Therapeutics. Molecules. 2022; 27(3):991. https://doi.org/10.3390/molecules27030991
Chicago/Turabian StyleAntunes, Patrícia, Adriana Cruz, José Barbosa, Vasco D. B. Bonifácio, and Sandra N. Pinto. 2022. "Lipid Droplets in Cancer: From Composition and Role to Imaging and Therapeutics" Molecules 27, no. 3: 991. https://doi.org/10.3390/molecules27030991
APA StyleAntunes, P., Cruz, A., Barbosa, J., Bonifácio, V. D. B., & Pinto, S. N. (2022). Lipid Droplets in Cancer: From Composition and Role to Imaging and Therapeutics. Molecules, 27(3), 991. https://doi.org/10.3390/molecules27030991







