A Photosensitized Singlet Oxygen (1O2) Toolbox for Bio-Organic Applications: Tailoring 1O2 Generation for DNA and Protein Labelling, Targeting and Biosensing
Abstract
1. Introduction
2. Reaction of Singlet Oxygen with Biomolecules
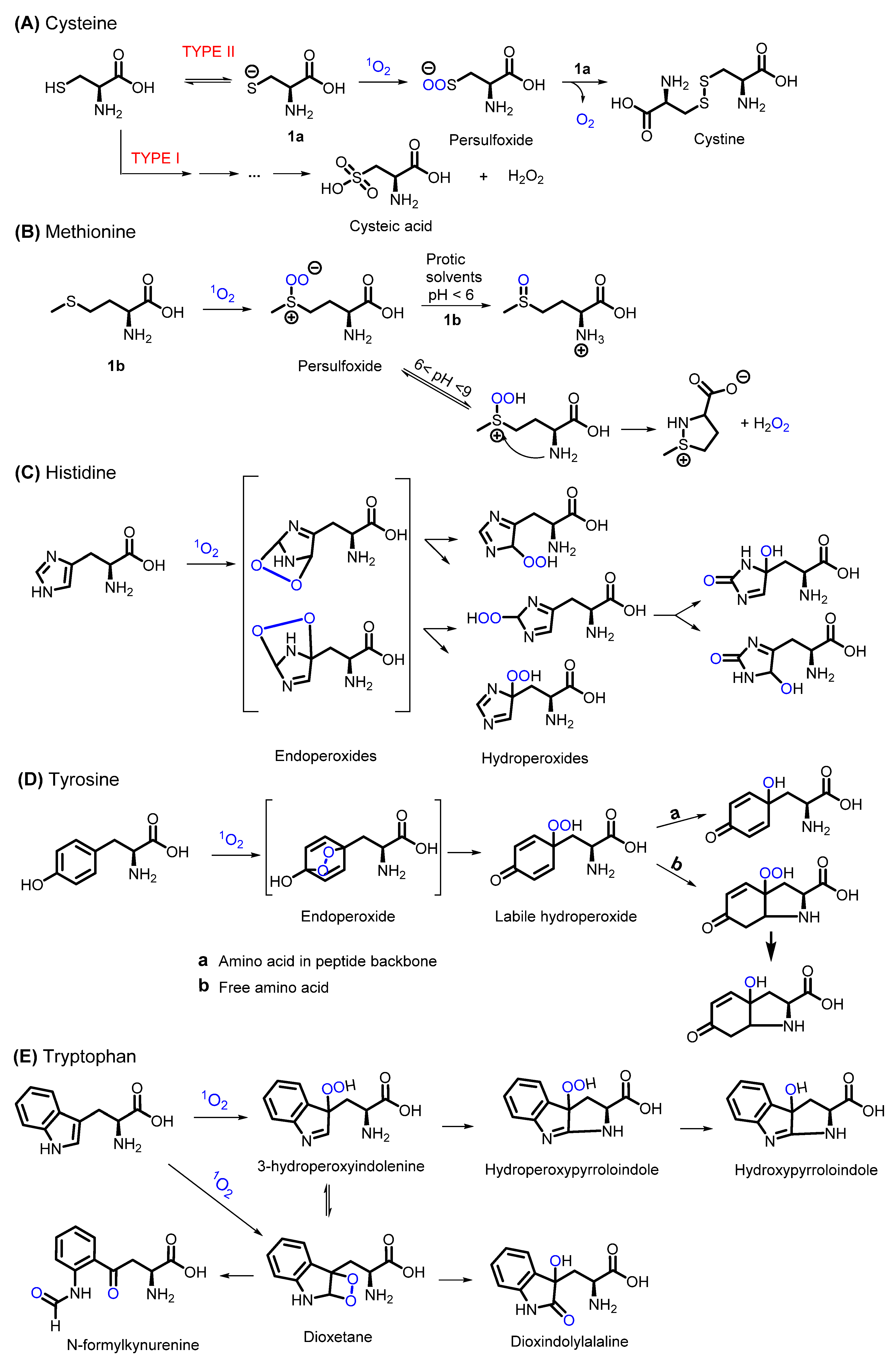
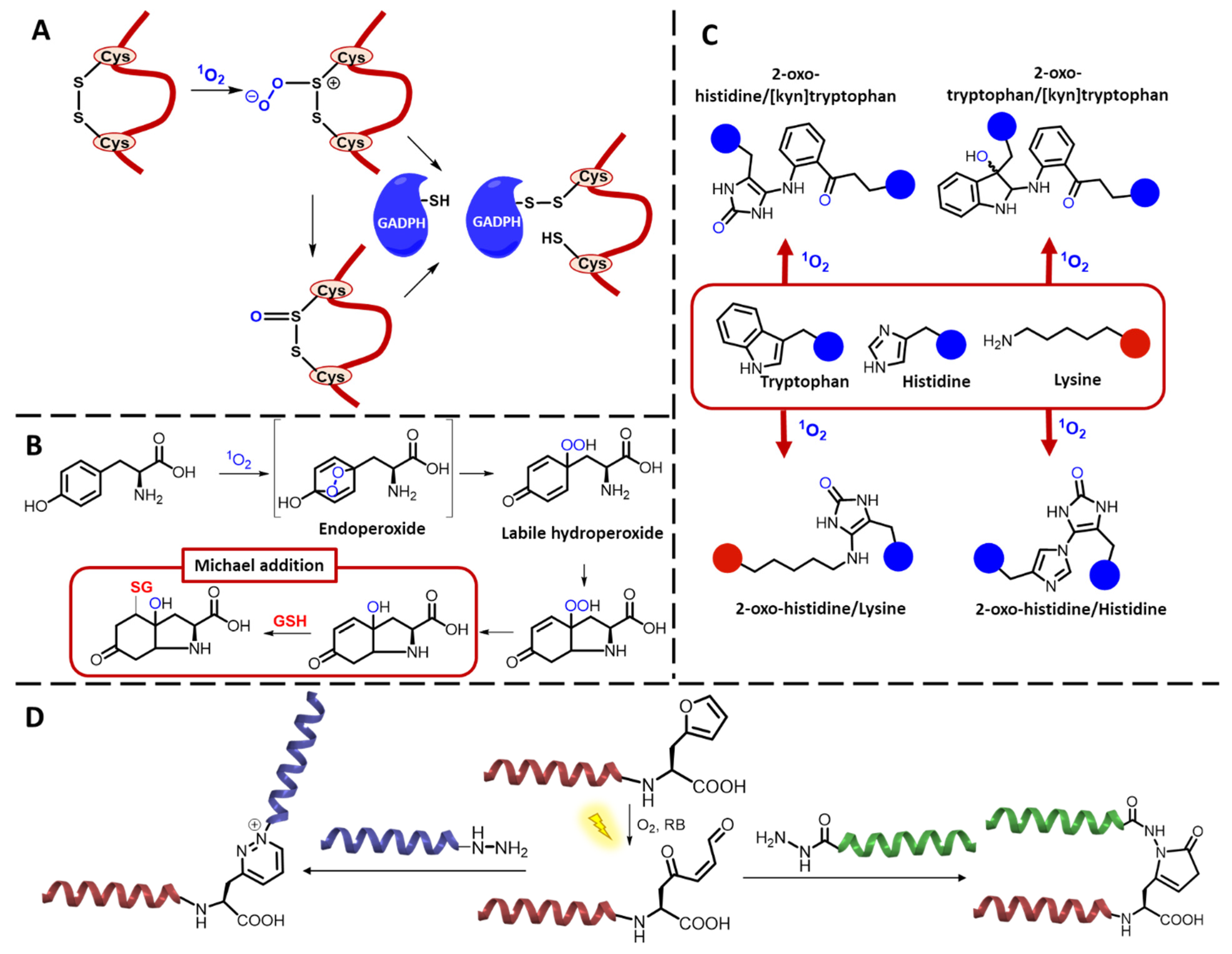
2.1. Peptides and Proteins
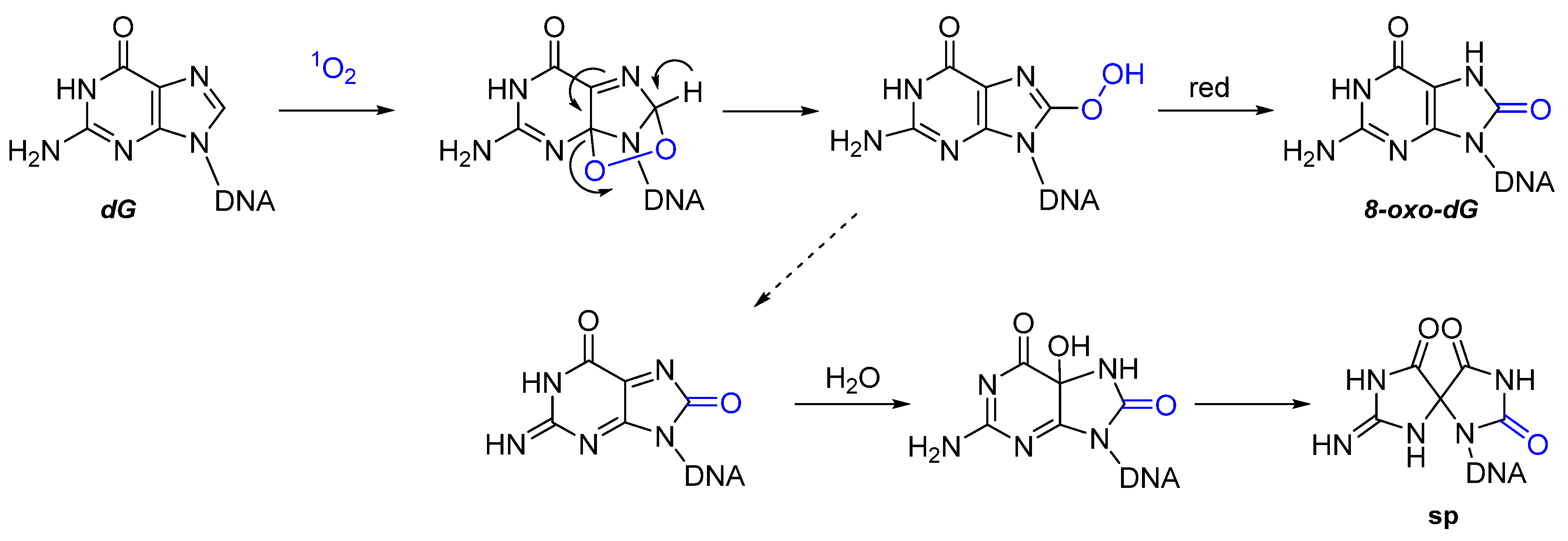
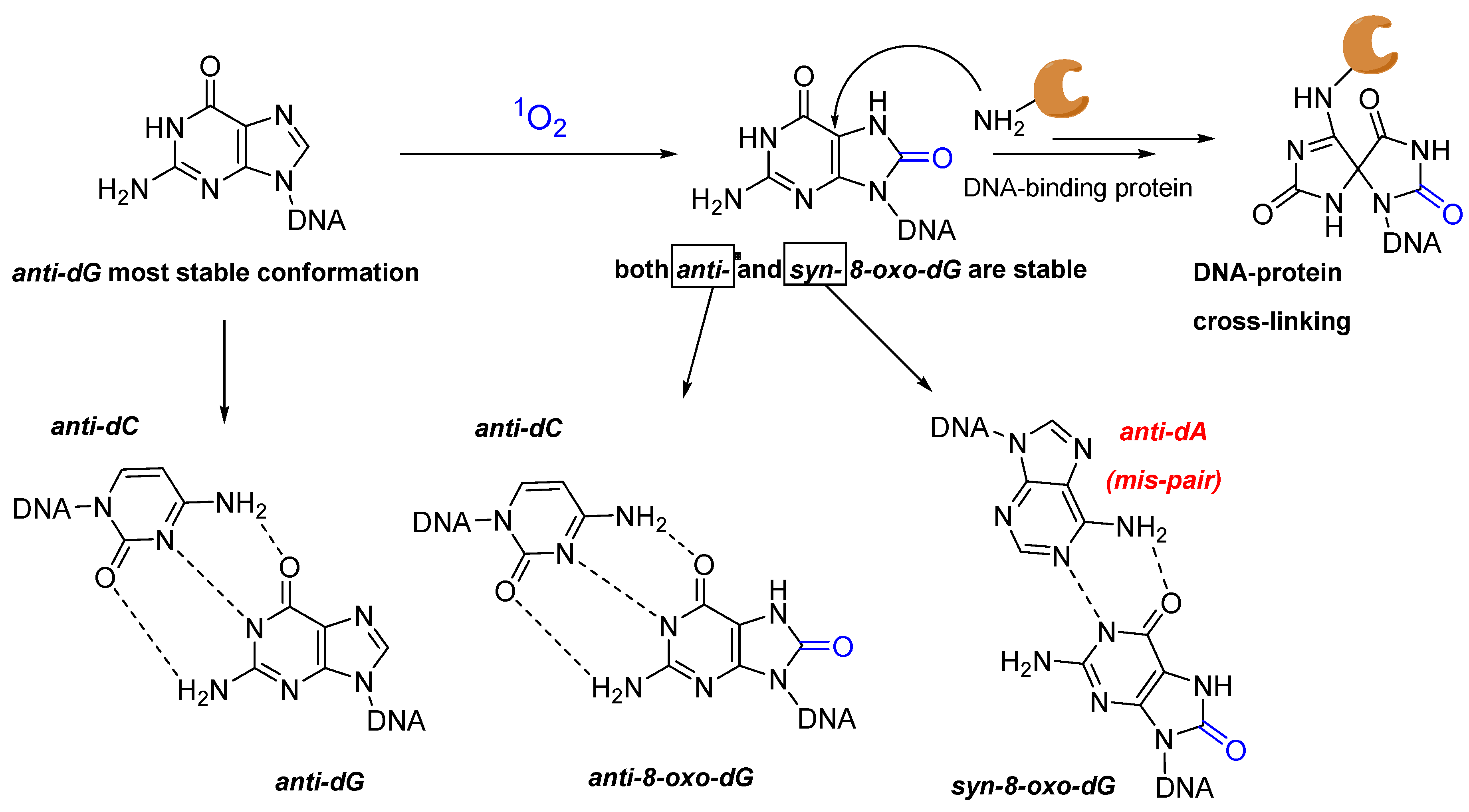
2.2. Nucleic Acids
2.3. Carbohydrates
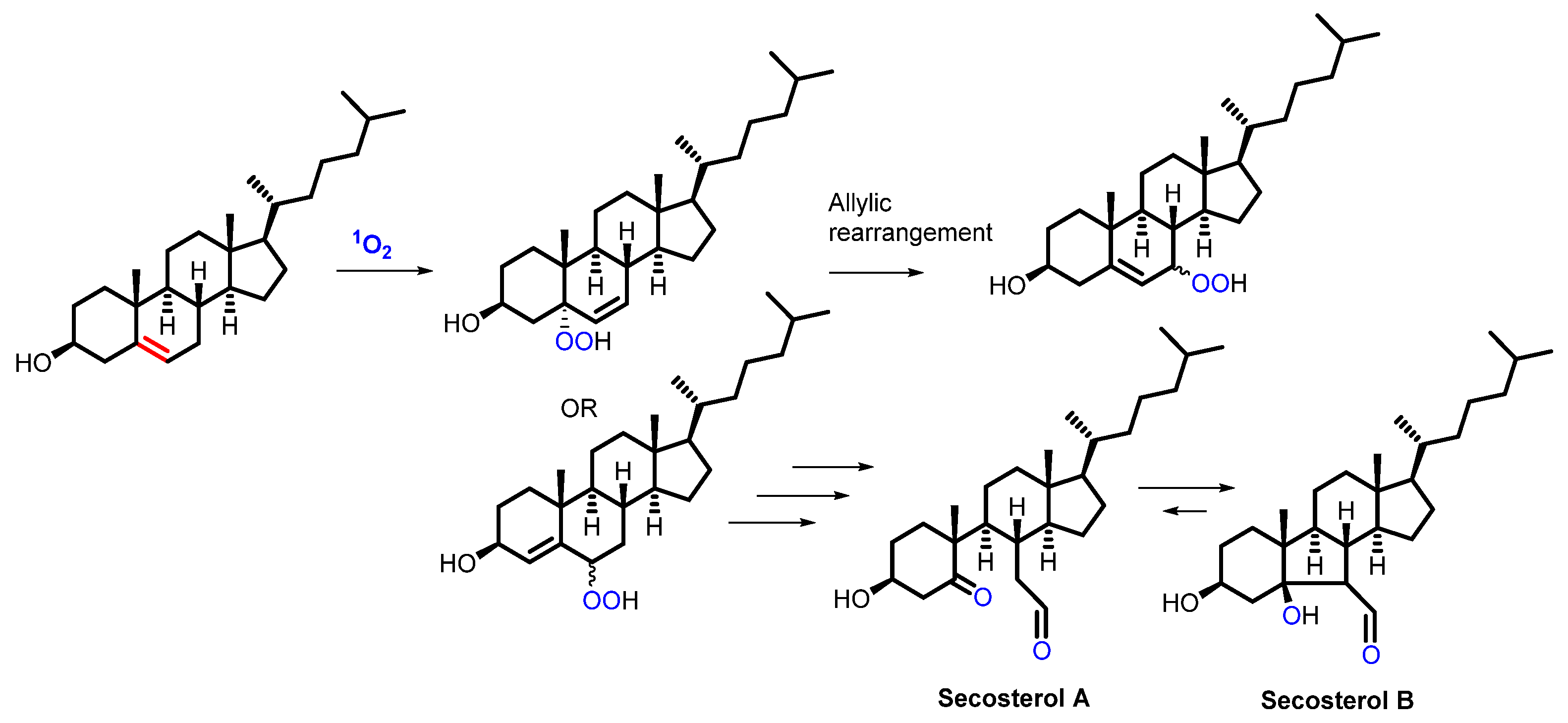
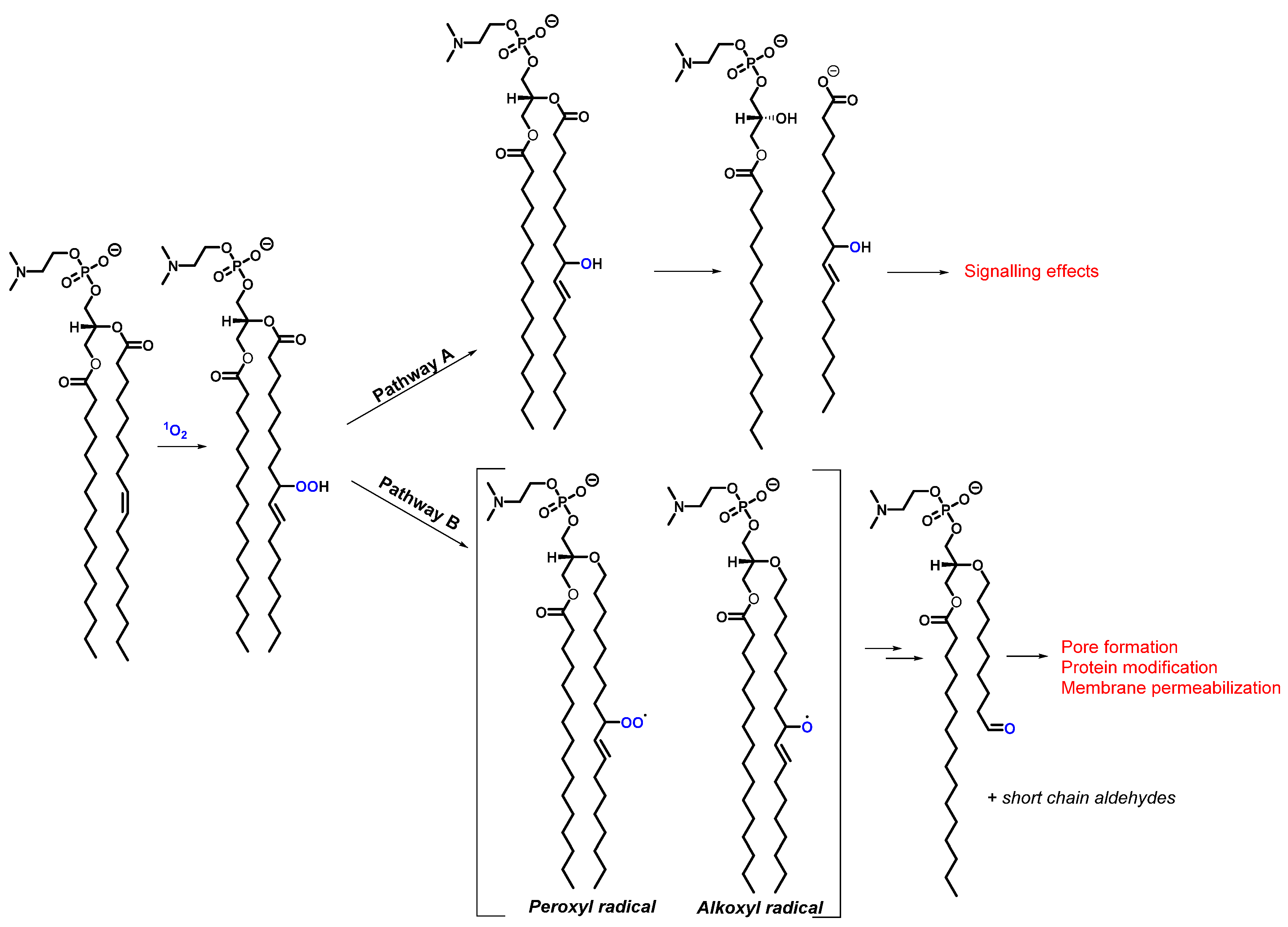
2.4. Lipids
3. 1O2 in Bioorganic Chemistry Applications
3.1. Peptide and Protein Modifications: Labelling, Cross-Linking and Knockdown
3.1.1. 1O2-Based Peptide and Protein Labelling
3.1.2. 1O2-Mediated Peptide and Protein Cross-Linking
Photo-Oxidative Cross-Linking of Peptide/Protein Interactions
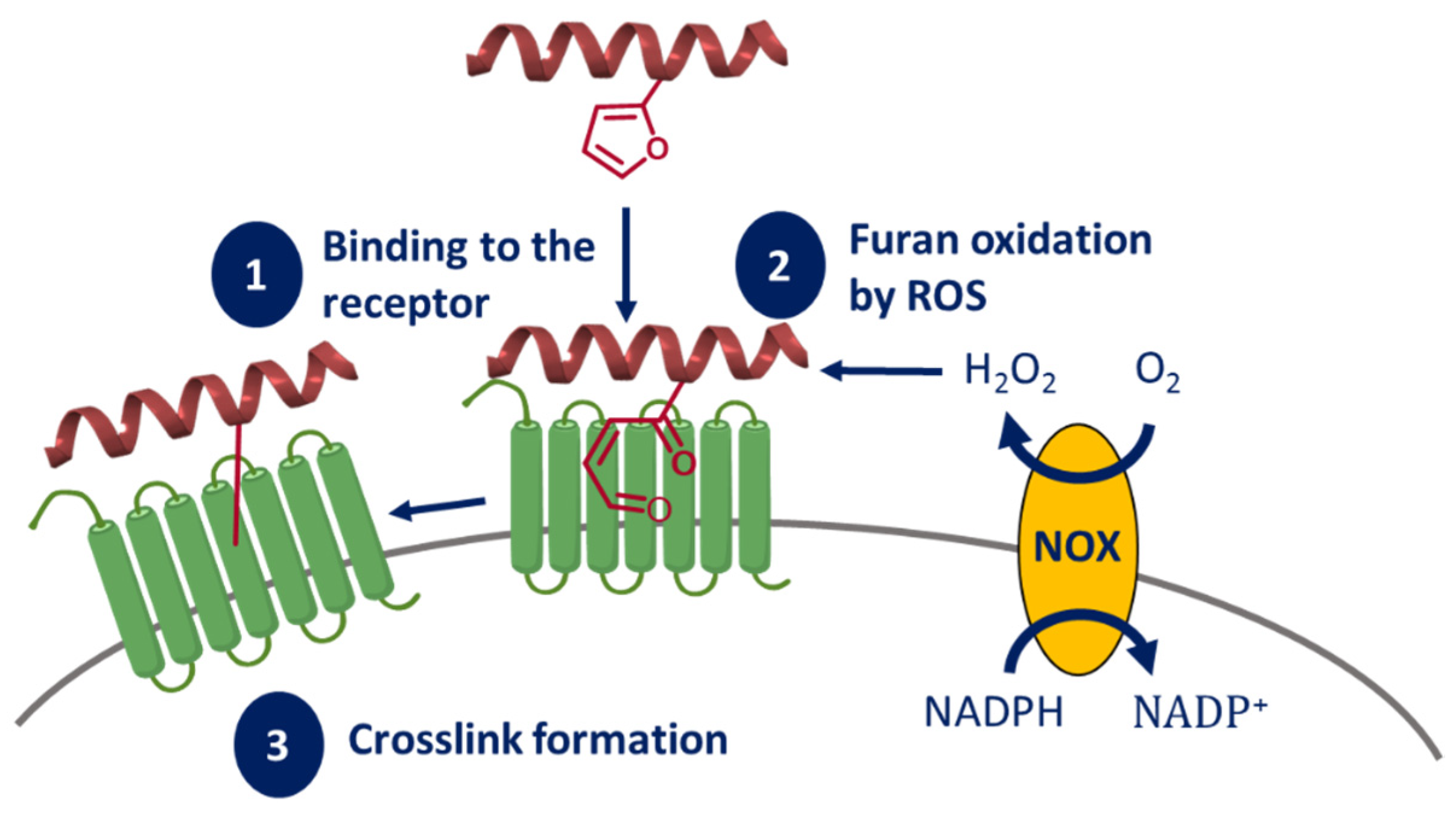
Endogenous ROS-Mediated Ligand-Receptor CL
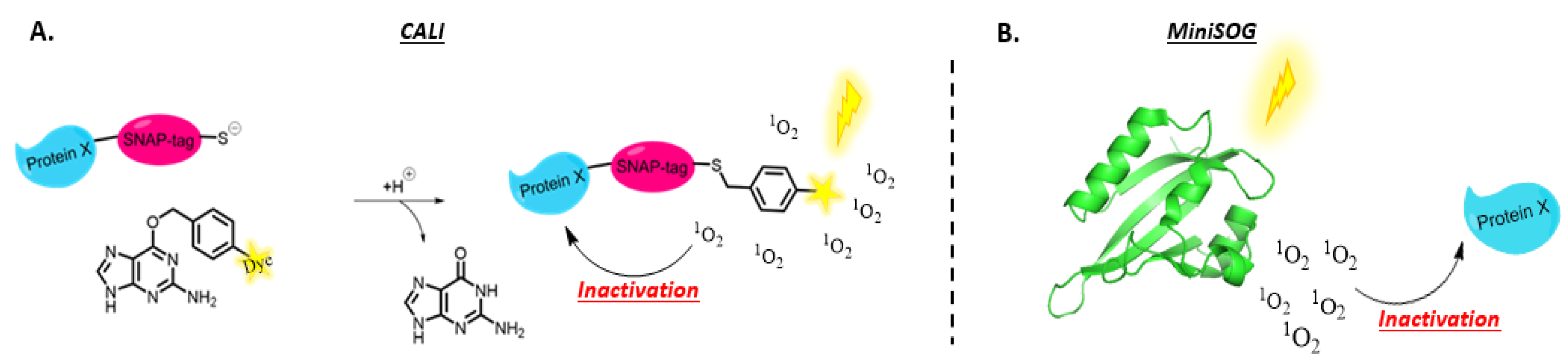
3.1.3. Protein Knockdown
3.2. Oligonucleotide Modifications: DNA and RNA Labelling, Cross-Linking, and Targeting
3.2.1. Oligonucleotide Labelling Methodologies Featuring 1O2
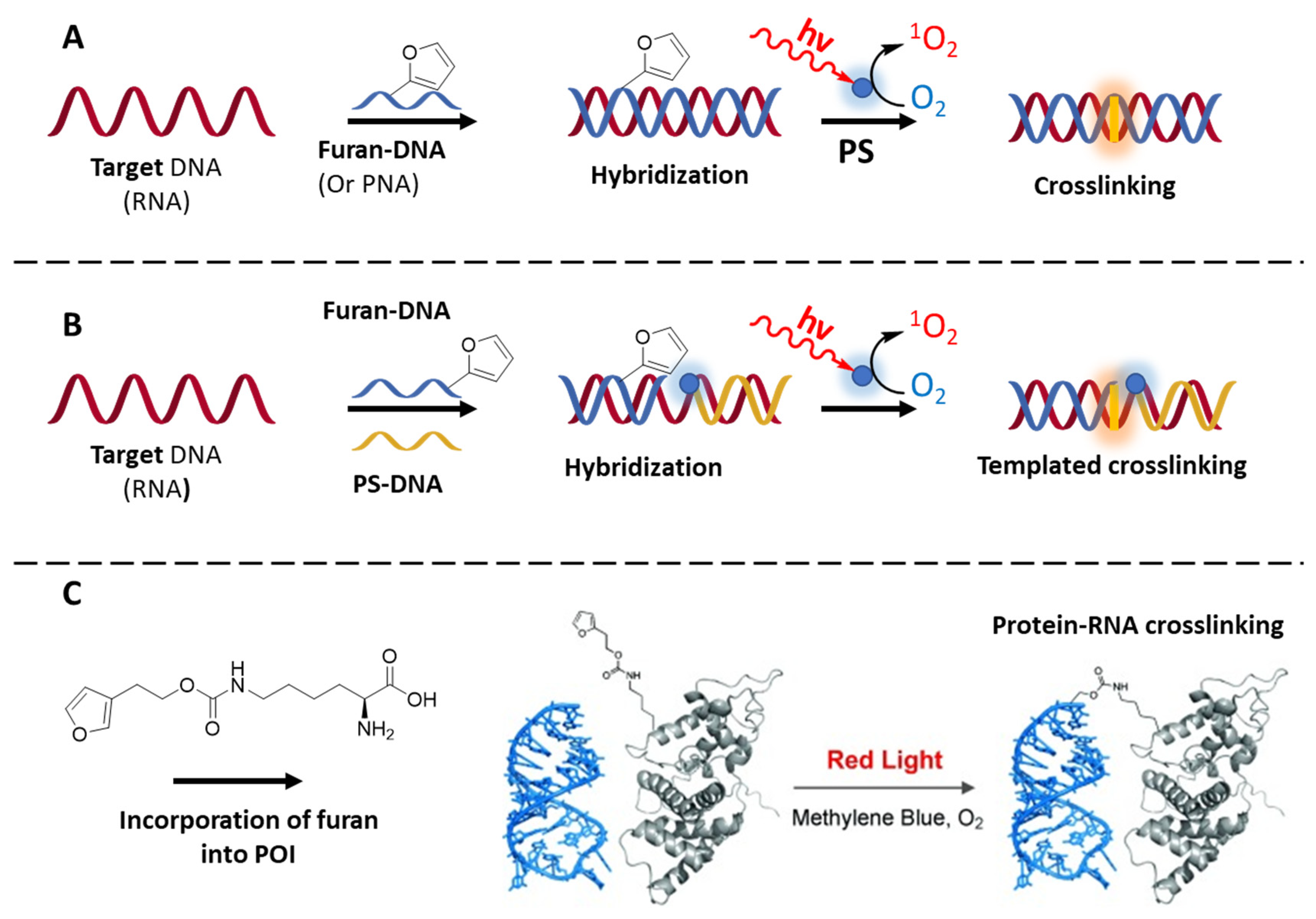
3.2.2. Oligonucleotide Cross-Linking and Targeting Methodologies
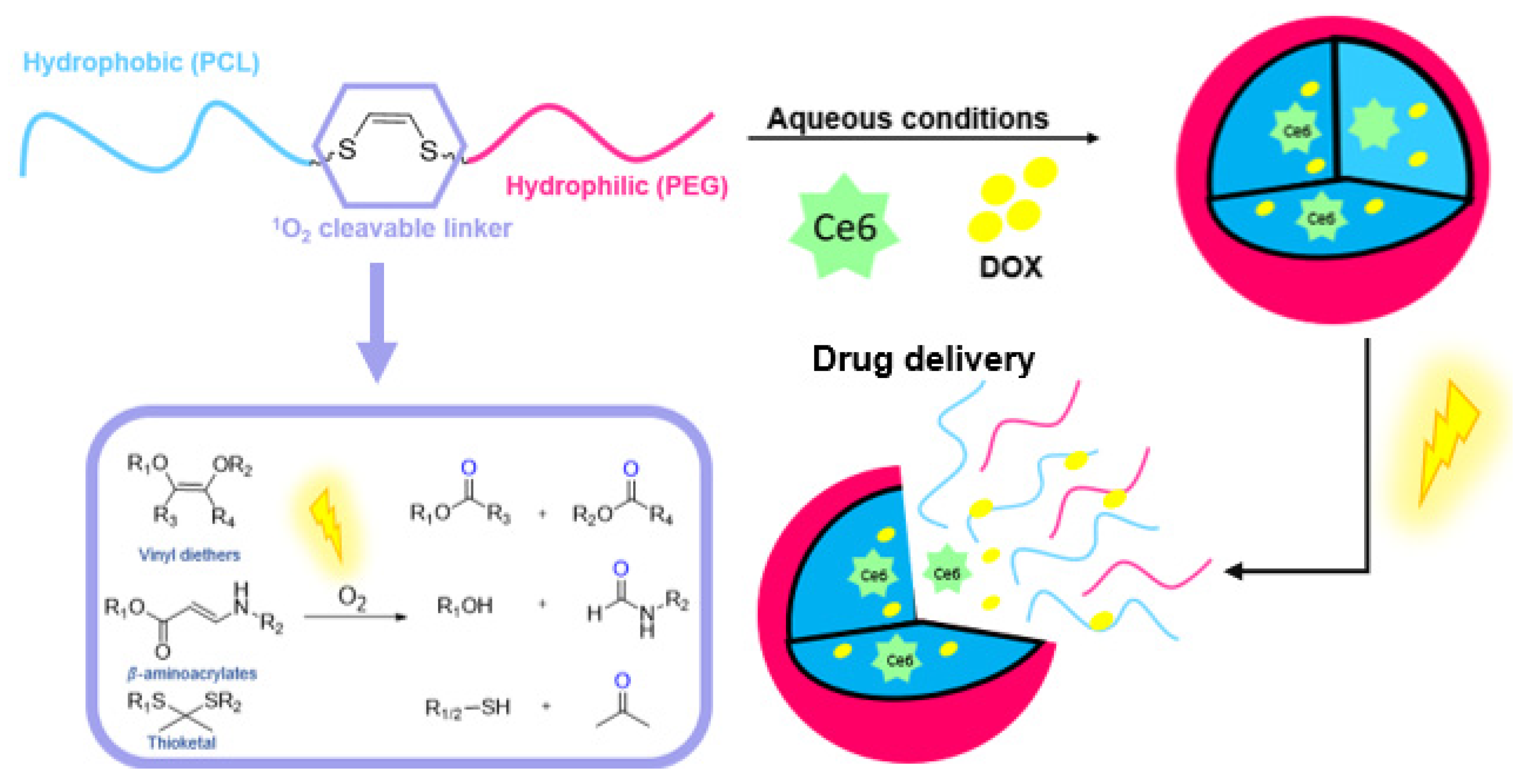
3.3. ROS Cleavable Linkers in Drug Delivery and Prodrug Applications
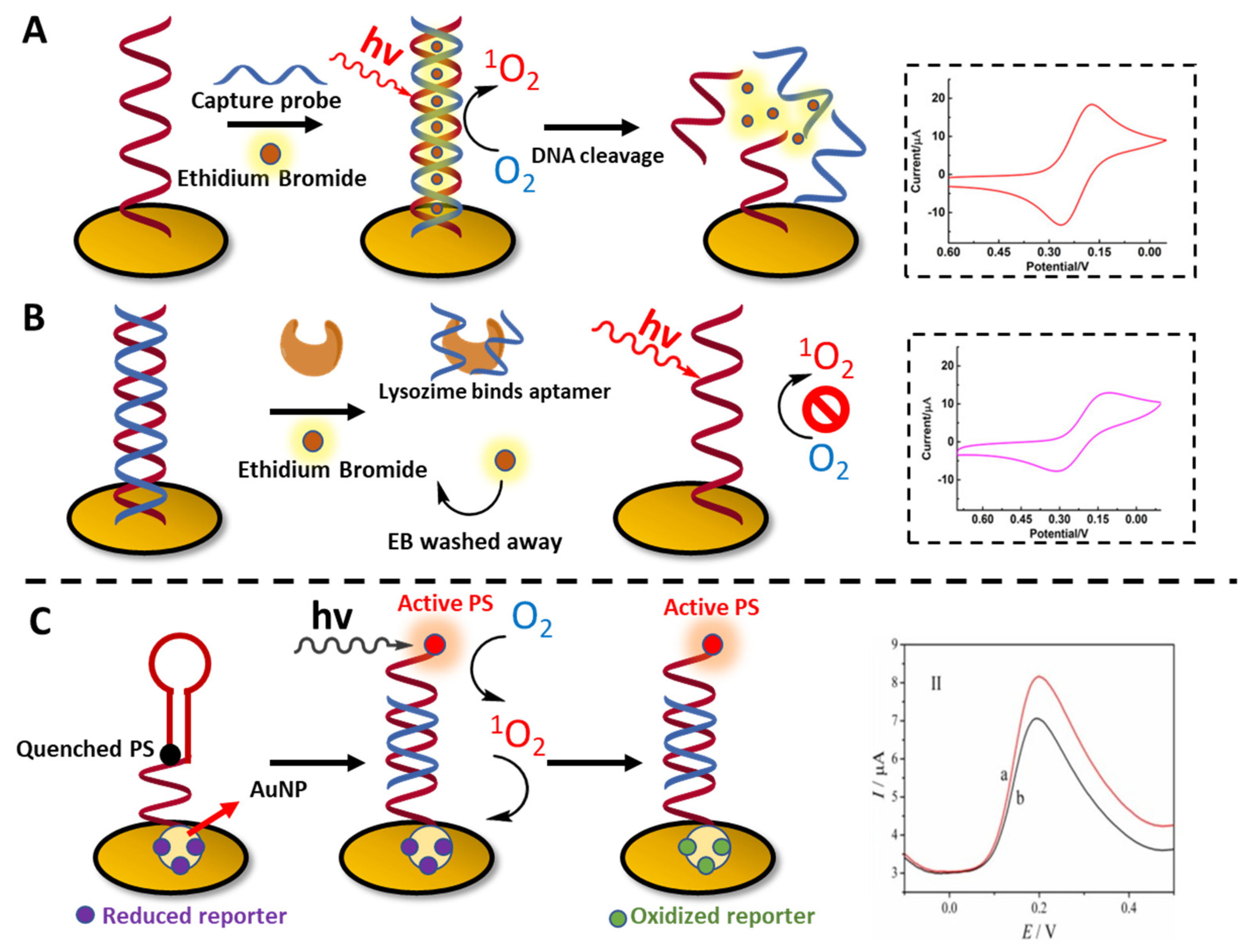
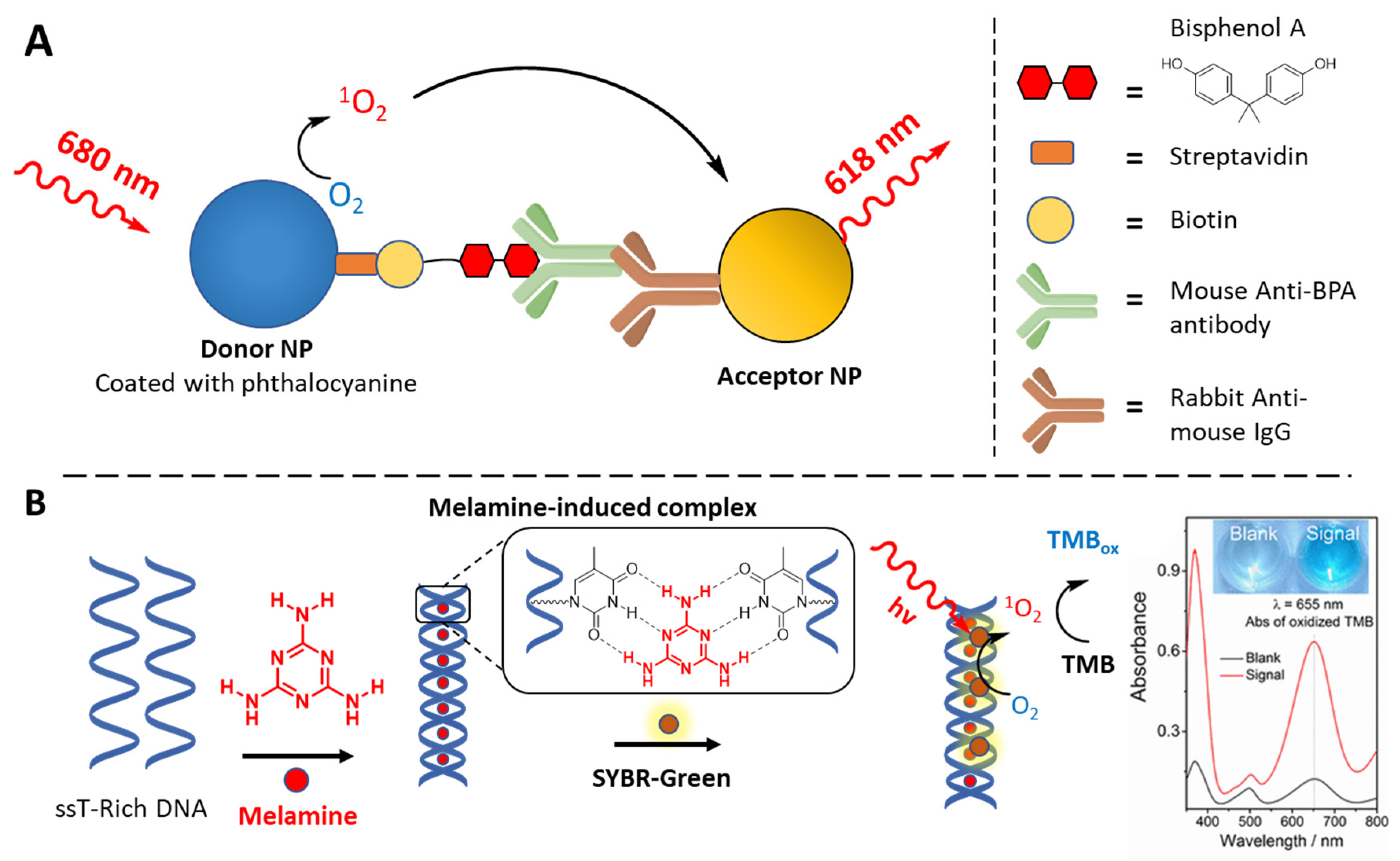
3.4. Biosensing Applications Featuring 1O2 Generation
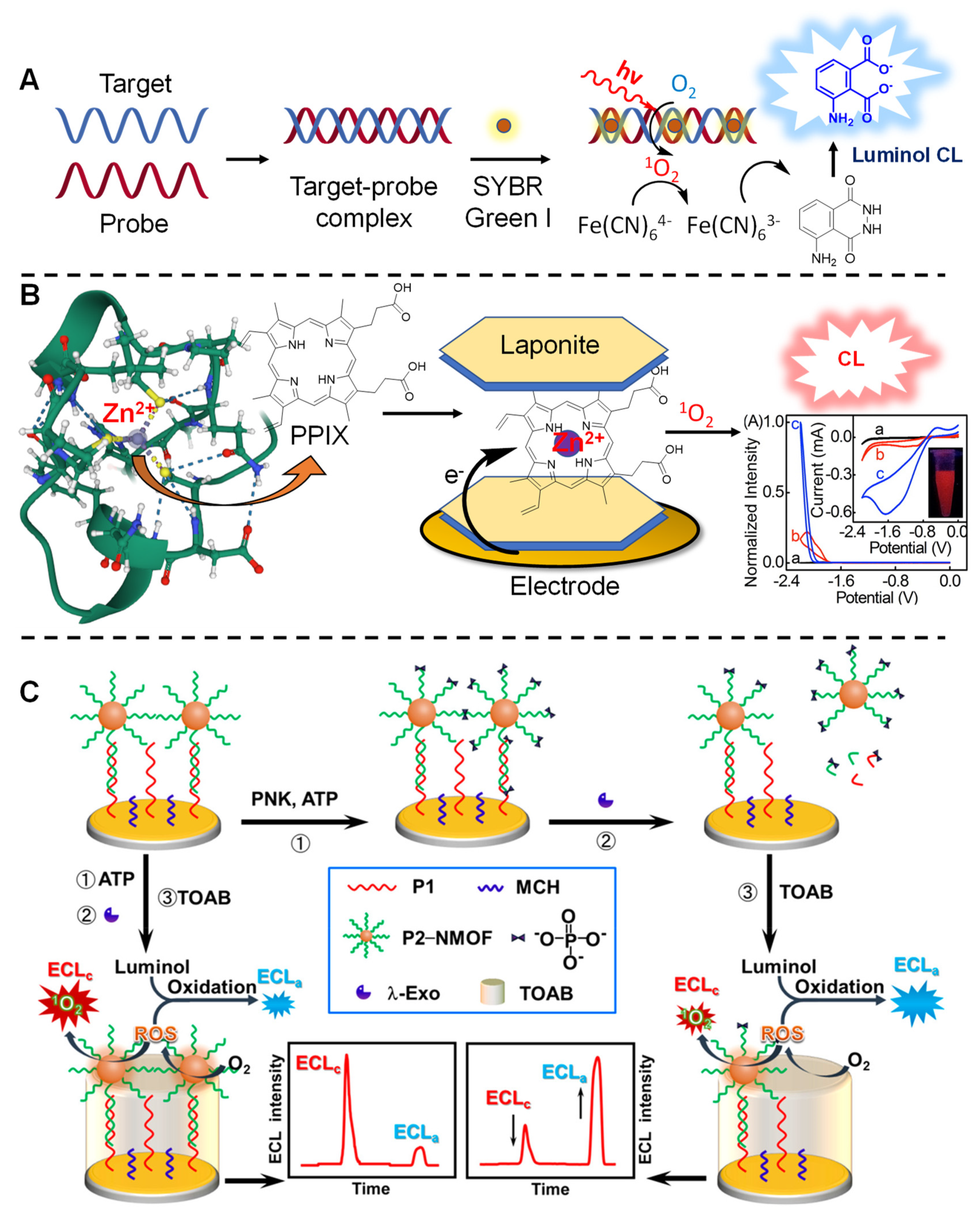
4. How to Tame the Bullet
4.1. Genetically Encoded PSs
4.2. PS Conjugates
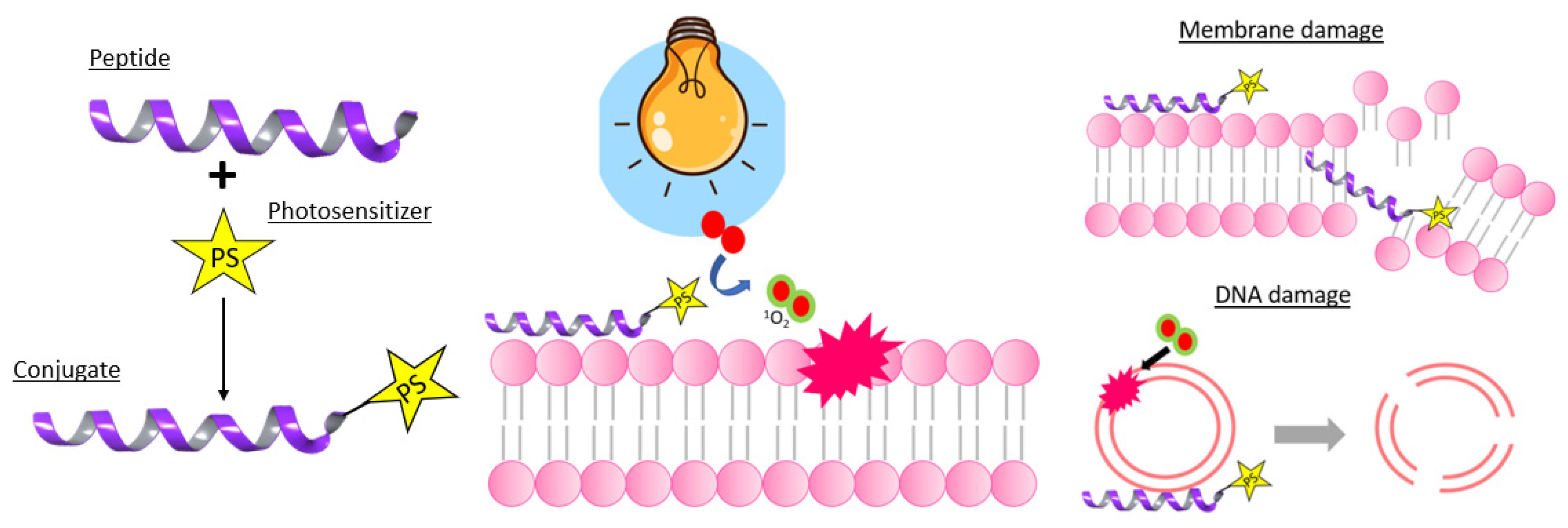
4.2.1. Peptide Conjugates
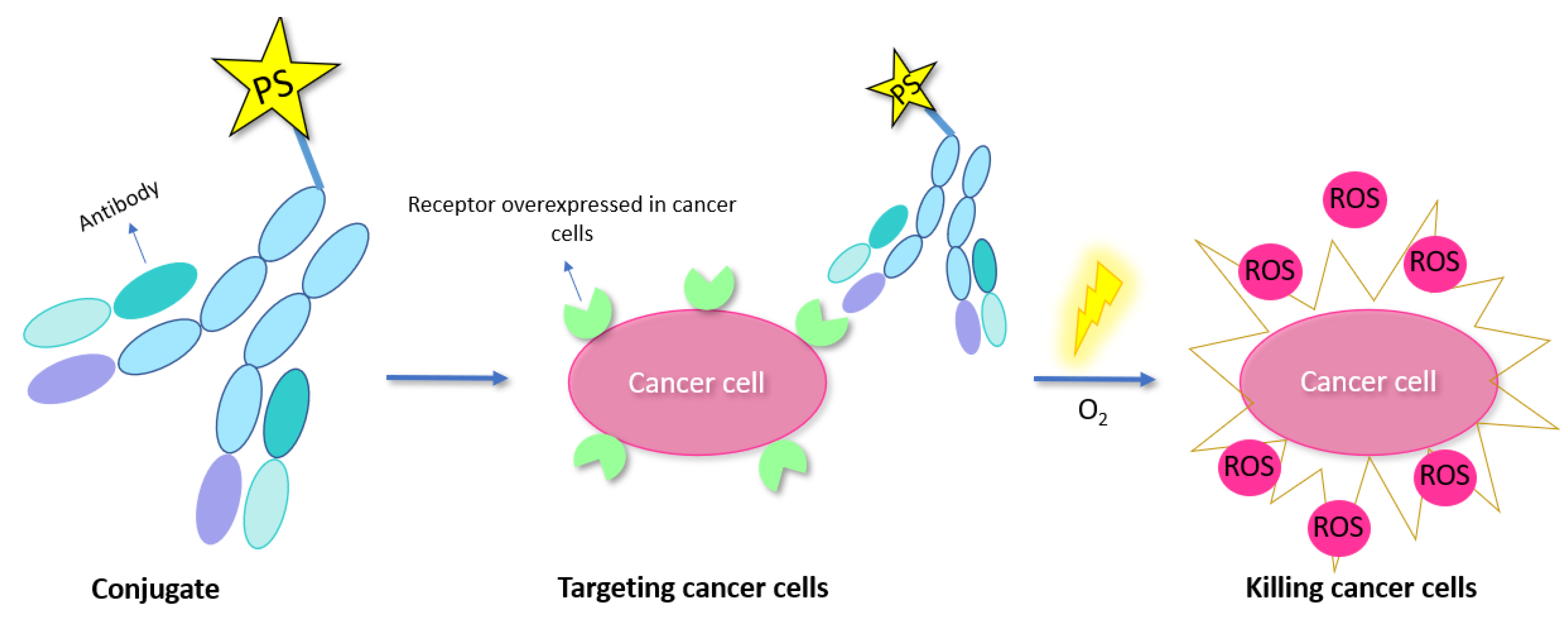
4.2.2. Antibody Conjugates
4.2.3. Oligonucleotide (and Their Analogues) Complexes and Conjugates

5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Fan, T.; Xie, Z.; Zeng, Q.; Xue, P.; Zheng, T.; Chen, Y.; Luo, X.; Zhang, H. Advances in Nanomaterials for Photodynamic Therapy Applications: Status and Challenges. Biomaterials 2020, 237, 119827. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.M.; Ghogare, A.A.; Greer, A.; Zhu, T.C. On the in VIVO Photochemical Rate Parameters for PDT Reactive Oxygen Species Modeling. Phys. Med. Biol. 2017, 62, R1–R48. [Google Scholar] [CrossRef] [PubMed]
- Hone, C.A.; Kappe, C.O. The Use of Molecular Oxygen for Liquid Phase Aerobic Oxidations in Continuous Flow; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; Volume 377, ISBN 0123456789. [Google Scholar]
- Pass, H.I. Photodynamic Therapy in Oncology: Mechanisms and Clinical Use. J. Natl. Cancer Inst. 1993, 85, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Dariva, C.G.; Coelho, J.F.J.; Serra, A.C. Near Infrared Light-Triggered Nanoparticles Using Singlet Oxygen Photocleavage for Drug Delivery Systems. J. Control Release 2019, 294, 337–354. [Google Scholar] [CrossRef]
- Marin, M.L.; Santos-Juanes, L.; Arques, A.; Amat, A.M.; Miranda, M.A. Organic Photocatalysts for the Oxidation of Pollutants and Model Compounds. Chem. Rev. 2012, 112, 1710–1750. [Google Scholar] [CrossRef]
- Shen, Y.; Shuhendler, A.J.; Ye, D.; Xu, J.J.; Chen, H.Y. Two-Photon Excitation Nanoparticles for Photodynamic Therapy. Chem. Soc. Rev. 2016, 45, 6725–6741. [Google Scholar] [CrossRef]
- DeRosa, M.C.; Crutchley, R.J. Photosensitized Singlet Oxygen and Its Applications. Coord. Chem. Rev. 2002, 233–234, 351–371. [Google Scholar] [CrossRef]
- Fudickar, W.; Linker, T. Release of Singlet Oxygen from Organic Peroxides under Mild Conditions. ChemPhotoChem 2018, 2, 548–558. [Google Scholar] [CrossRef]
- Maetzke, A.; Knak Jensen, S.J. Reaction Paths for Production of Singlet Oxygen from Hydrogen Peroxide and Hypochlorite. Chem. Phys. Lett. 2006, 425, 40–43. [Google Scholar] [CrossRef]
- Kanofsky, J.R.; Hoogland, H.; Wever, R.; Weiss, S.J. Singlet Oxygen Production by Human Eosinophils. J. Biol. Chem. 1988, 263, 9692–9696. [Google Scholar] [CrossRef]
- Pibiri, I.; Buscemi, S.; Palumbo Piccionello, A.; Pace, A. Photochemically Produced Singlet Oxygen: Applications and Perspectives. ChemPhotoChem 2018, 2, 535–547. [Google Scholar] [CrossRef]
- Foote, C.S.; Wexler, S. Olefin Oxidations with Excited Singlet Molecular Oxygen. J. Am. Chem. Soc. 1964, 86, 3879–3880. [Google Scholar] [CrossRef]
- Foote, C.S.; Wexler, S. Singlet Oxygen. A Probable Intermediate in Photosensitized Autoxidations. J. Am. Chem. Soc. 1964, 86, 3880–3881. [Google Scholar] [CrossRef]
- Foote, C.S. Mechanisms of Photosensitized Oxidation. Science 1968, 162, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Orfanopoulos, M. Singlet Oxygen: Discovery, Chemistry, C 60 -Sensitization. Photochem. Photobiol. 2021, 97, 1182–1218. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer Science & Business Media: New York, NY, USA, 2006; pp. 1–954. [Google Scholar] [CrossRef]
- Castano, A.P.; Demidova, T.N.; Hamblin, M.R. Mechanisms in Photodynamic Therapy: Part One—Photosensitizers, Photochemistry and Cellular Localization. Photodiagnosis Photodyn. Ther. 2004, 1, 279–293. [Google Scholar] [CrossRef]
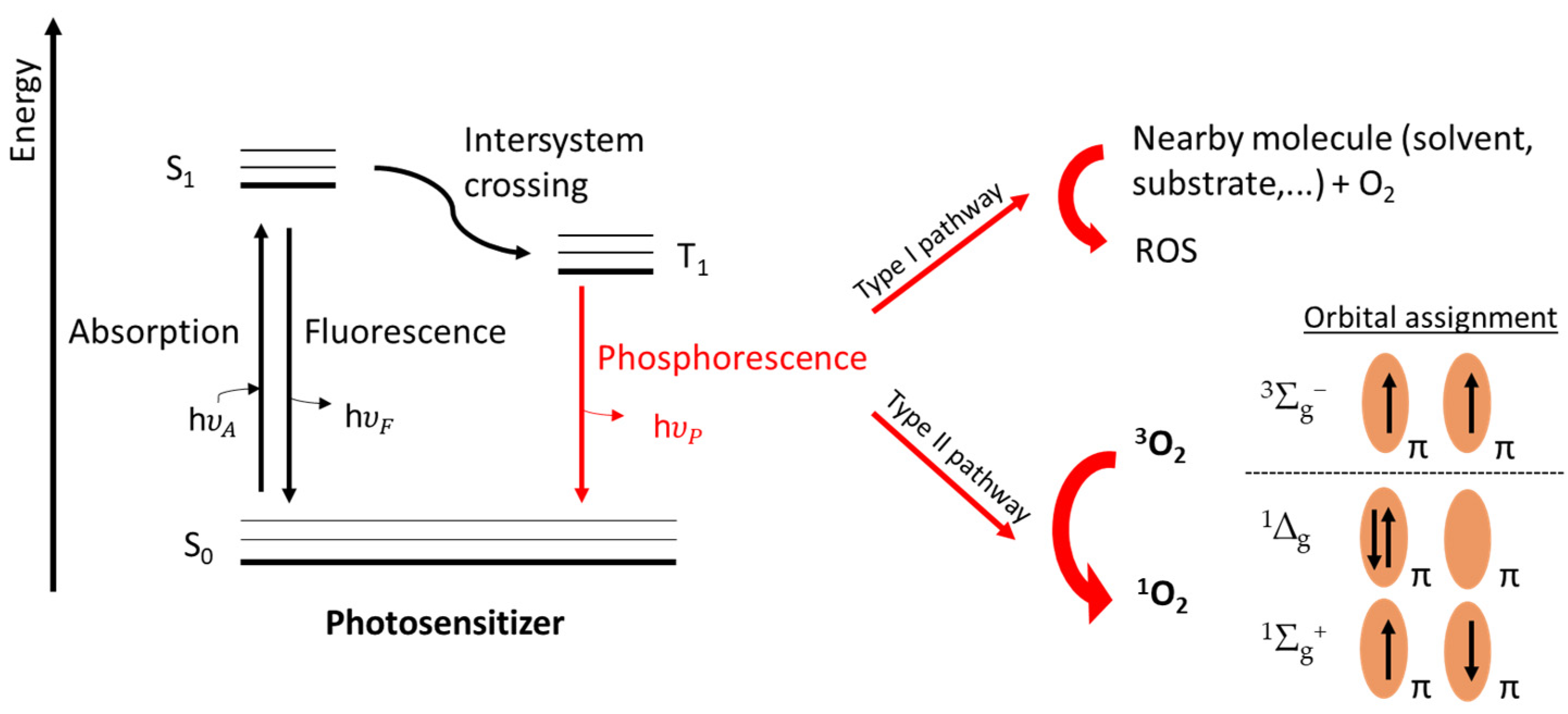
- Baptista, M.S.; Cadet, J.; di Mascio, P.; Ghogare, A.A.; Greer, A.; Hamblin, M.R.; Lorente, C.; Nunez, S.C.; Ribeiro, M.S.; Thomas, A.H.; et al. Type I and Type II Photosensitized Oxidation Reactions: Guidelines and Mechanistic Pathways. Photochem. Photobiol. 2017, 93, 912–919. [Google Scholar] [CrossRef]
- Weldon, D.; Poulsen, T.D.; Mikkelsen, K.V.; Ogilby, P.R. Singlet Sigma: The “Other” Singlet Oxygen in Solution. Photochem. Photobiol. 1999, 70, 369. [Google Scholar] [CrossRef]
- Fekrazad, R.; Nejat, A.H.; Kalhori, K.A.M. Antimicrobial Photodynamic Therapy with Nanoparticles Versus Conventional Photosensitizer in Oral Diseases. In Nanostructures for Antimicrobial Therapy; Nanostructures in Therapeutic Medicine Series; Elsevier: Amsterdam, The Netherlands, 2017; pp. 237–259. [Google Scholar] [CrossRef]
- Oba, T. Photosensitizer Nanoparticles for Photodynamic Therapy. Curr. Bioact. Compd. 2007, 3, 239–251. [Google Scholar] [CrossRef]
- Leach, A.G.; Houk, K.N. Diels–Alder and Ene Reactions of Singlet Oxygen, Nitroso Compounds and Triazolinediones: Transition States and Mechanisms from Contemporary Theory. Chem. Commun. 2002, 2, 1243–1255. [Google Scholar] [CrossRef]
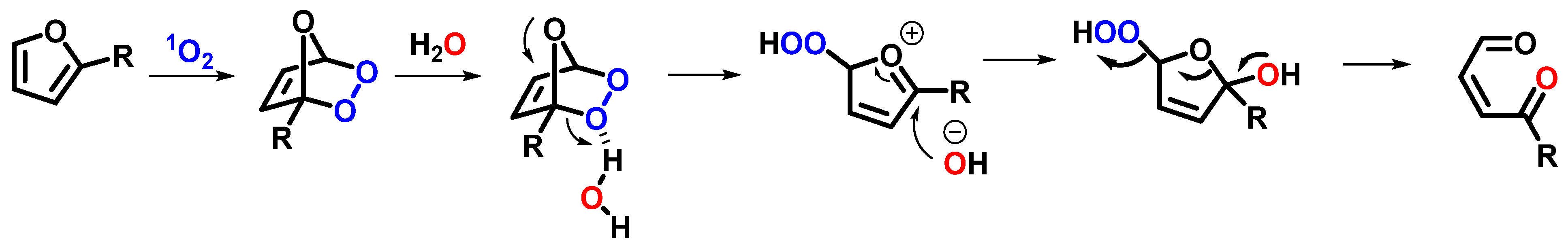
- Montagnon, T.; Kalaitzakis, D.; Triantafyllakis, M.; Stratakis, M.; Vassilikogiannakis, G. Furans and Singlet Oxygen—Why There Is More to Come from This Powerful Partnership. Chem. Commun. 2014, 50, 15480–15498. [Google Scholar] [CrossRef] [PubMed]
- Di Mascio, P.; Martinez, G.R.; Miyamoto, S.; Ronsein, G.E.; Medeiros, M.H.G.; Cadet, J. Singlet Molecular Oxygen Reactions with Nucleic Acids, Lipids, and Proteins. Chem. Rev. 2019, 119, 2043–2086. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Singlet Oxygen-Mediated Damage to Proteins and Its Consequences. Biochem. Biophys. Res. Commun. 2003, 305, 761–770. [Google Scholar] [CrossRef]
- Nonell, S. Singlet Oxygen: Applications in Biosciences and Nanosciences; Nonell, S., Flors, C., Eds.; Royal Society of Chemistry: London, UK, 2016; Volume 1. [Google Scholar]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Protein Function; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Michaeli, A.; Feitelson, J. Reactivity of Singlet Oxygen Toward Amino Acids and Peptides; American Society for Photobiology: Herndon, VA, USA, 1994; Volume 59. [Google Scholar]
- Devasagayam, T.P.; Sundquist, A.R.; Di Mascio, P.; Kaiser, S.; Sies, H. Activity of Thiols as Singlet Molecular Oxygen Quenchers. J. Photochem. Photobiol. B Biol. 1991, 9, 105–116. [Google Scholar] [CrossRef]
- Buettner, G.R.; Hall, R.D. Superoxide, Hydrogen Peroxide and Singlet Oxygen in Hematoporphyrin Derivative-Cysteine, -NADH and -Light Systems. Biochim. Biophys. Acta Gen. Subj. 1987, 923, 501–507. [Google Scholar] [CrossRef]
- Tomita, M.; Irie, M.; Ukita, T. Sensitized Photooxidation of N-Benzoyl Histidine. Tetrahedron Lett. 1968, 9, 4933–4936. [Google Scholar] [CrossRef]
- Tomita, M.; Irie, M.; Ukita, T. Sensitized Photooxidation of Histidine and Its Derivatives. Prod. Mech. Reaction. Biochem. 2002, 8, 5149–5160. [Google Scholar] [CrossRef]
- Kang, P.; Foote, C.S. Synthesis of a 13C,15N Labeled Imidazole and Characterization of the 2,5-Endoperoxide and Its Decomposition. Tetrahedron Lett. 2000, 41, 9623–9626. [Google Scholar] [CrossRef]
- Wright, A.; Bubb, W.A.; Hawkins, C.L.; Davies, M.J. Singlet Oxygen-Mediated Protein Oxidation: Evidence for the Formation of Reactive Side Chain Peroxides on Tyrosine Residues; American Society for Photobiology: Herndon, VA, USA, 2002; Volume 76. [Google Scholar]
- Adam, W.; Ahrweiler, M.; Sauter, M.; Schmiedeskamp, B. Oxidation of Indoles by Singlet Oxygen and Dimethyldioxirane: Isolation of Indole Dioxetanes and Epoxides by Stabilization through Nitrogen Acylation. Tetrahedron Lett. 1993, 34, 5247–5250. [Google Scholar] [CrossRef]
- Mohanty, P.; Matysik, J. Effect of Proline on the Production of Singlet Oxygen. Amino Acids 2001, 21, 195–200. [Google Scholar] [CrossRef]
- Matysik, J.; Bhalu, B.; Mohanty, P. Molecular Mechanisms of Quenching of Reactive Oxygen Species by Proline under Stress in Plants. Curr. Sci. 2002, 82, 525–532. [Google Scholar]
- Schuessler, H.; Schilling, K. Oxygen Effect in the Radiolysis of Proteins. Part 2. Bovine Serum Albumin. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1984, 45, 267–281. [Google Scholar] [CrossRef] [PubMed]
- von Tappeiner, H. Über Die Wirkung Fluoreszierender Substanzen Auf Fermente Und Toxine. Ber. Dtsch. Chem. Ges. 1903, 36, 3035–3038. [Google Scholar] [CrossRef]
- Morgan, P.E.; Dean, R.T.; Davies, M.J. Inhibition of Glyceraldehyde-3-Phosphate Dehydrogenase by Peptide and Protein Peroxides Generated by Singlet Oxygen Attack. Eur. J. Biochem. 2002, 269, 1916–1925. [Google Scholar] [CrossRef]
- Finley, E.L.; Busman, M.; Dillon, J.; Crouch, R.K.; Schey, K.L. Identification of Photooxidation Sites in Bovine α-Crystallin. Photochem. Photobiol. 1997, 66, 635–641. [Google Scholar] [CrossRef]
- Lledías, F.; Rangel, P.; Hansberg, W. Oxidation of Catalase by Singlet Oxygen. J. Biol. Chem. 1998, 273, 10630–10637. [Google Scholar] [CrossRef]
- Lledías, F.; Hansberg, W. Oxidation of Human Catalase by Singlet Oxygen in Myeloid Leukemia Cells. Photochem. Photobiol. 1999, 70, 887–892. [Google Scholar] [CrossRef]
- Gomyo, T.; Sakurai, Y. Studies on Changes of Protein by Dye Sensitized Photooxidation. Agric. Biol. Chem. 2014, 31, 1474–1481. [Google Scholar] [CrossRef]
- Marques, E.F.; Medeiros, M.H.; Di Mascio, P. Singlet Oxygen-Induced Protein Aggregation: Lysozyme Crosslink Formation and NLC-MS/MS Characterization. J. Mass Spectrom. JMS 2019, 54, 894–905. [Google Scholar] [CrossRef]
- Jiang, S.; Carroll, L.; Mariotti, M.; Hägglund, P.; Davies, M.J. Formation of Protein Cross-Links by Singlet Oxygen-Mediated Disulfide Oxidation. Redox Biol. 2021, 41, 101874. [Google Scholar] [CrossRef]
- Jensen, R.L.; Arnbjerg, J.; Ogilby, P.R. Reaction of Singlet Oxygen with Tryptophan in Proteins: A Pronounced Effect of the Local Environment on the Reaction Rate. J. Am. Chem. Soc. 2012, 134, 9820–9826. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 340, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of Mutational Processes in Human Cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free Radicals, Metals and Antioxidants in Oxidative Stress-Induced Cancer. Chem.-Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Ames, B.N.; Shigenaga, M.K.; Hagen, T.M. Oxidants, Antioxidants, and the Degenerative Diseases of Aging. Proc. Natl. Acad. Sci. USA 1993, 90, 7915–7922. [Google Scholar] [CrossRef]
- Sherr, C.J. Cancer Cell Cycles. Science 1996, 274, 1672–1674. [Google Scholar] [CrossRef]
- Lindahl, T.; Nyberg, B. Rate of Depurination of Native Deoxyribonucleic Acid. Biochemistry 1972, 11, 3610–3618. [Google Scholar] [CrossRef]
- Caldecott, K.W. Single-Strand Break Repair and Genetic Disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef]
- Warren, J.J.; Forsberg, L.J.; Beese, L.S. The Structural Basis for the Mutagenicity of O6-Methyl-Guanine Lesions. Proc. Natl. Acad. Sci. USA 2006, 103, 19701–19706. [Google Scholar] [CrossRef]
- David, S.S.; O’Shea, V.L.; Kundu, S. Base-Excision Repair of Oxidative DNA Damage. Nature 2007, 447, 941–950. [Google Scholar] [CrossRef]
- van Loon, B.; Markkanen, E.; Hübscher, U. Oxygen as a Friend and Enemy: How to Combat the Mutational Potential of 8-Oxo-Guanine. DNA Repair 2010, 9, 604–616. [Google Scholar] [CrossRef]
- Martinez, G.R.; Loureiro, A.P.M.; Marques, S.A.; Miyamoto, S.; Yamaguchi, L.F.; Onuki, J.; Almeida, E.A.; Garcia, C.C.M.; Barbosa, L.F.; Medeiros, M.H.G.; et al. Oxidative and Alkylating Damage in DNA. Mutat. Res. Rev. Mutat. Res. 2003, 544, 115–127. [Google Scholar] [CrossRef]
- Yagura, T.; Schuch, A.P.; Garcia, C.C.M.; Rocha, C.R.R.; Moreno, N.C.; Angeli, J.P.F.; Mendes, D.; Severino, D.; Bianchini Sanchez, A.; di Mascio, P.; et al. Direct Participation of DNA in the Formation of Singlet Oxygen and Base Damage under UVA Irradiation. Free Radic. Biol. Med. 2017, 108, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.N.; Simms, C.L.; Keedy, H.E.; Zaher, H.S. Insights into the Base-Pairing Preferences of 8-Oxoguanosine on the Ribosome. Nucleic Acids Res. 2019, 47, 9857–9870. [Google Scholar] [CrossRef] [PubMed]
- Johansen, M.E.; Muller, J.G.; Xu, X.; Burrows, C.J. Oxidatively Induced DNA-Protein Cross-Linking between Single-Stranded Binding Protein and Oligodeoxynucleotides Containing 8-Oxo-7,8-Dihydro-2′- Deoxyguanosine. Biochemistry 2005, 44, 5660–5671. [Google Scholar] [CrossRef]
- Bai, J.; Zhang, Y.; Xi, Z.; Greenberg, M.M.; Zhou, C. Oxidation of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine Leads to Substantial DNA-Histone Cross-Links within Nucleosome Core Particles. Chem. Res. Toxicol. 2018, 31, 1364–1372. [Google Scholar] [CrossRef]
- Barker, S.; Weinfeld, M.; Murray, D. DNA-Protein Crosslinks: Their Induction, Repair, and Biological Consequences. Mutat. Res. Rev. Mutat. Res. 2005, 589, 111–135. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Cadet, J.; Davies, K.J.A.; Medeiros, M.H.; di Mascio, P.; Wagner, J.R. Formation and Repair of Oxidatively Generated Damage in Cellular DNA. Free Radic. Biol. Med. 2017, 107, 13–34. [Google Scholar] [CrossRef]
- Rozelle, A.L.; Cheun, Y.; Vilas, C.K.; Koag, M.C.; Lee, S. DNA Interstrand Cross-Links Induced by the Major Oxidative Adenine Lesion 7,8-Dihydro-8-Oxoadenine. Nat. Commun. 2021, 12, 1897. [Google Scholar] [CrossRef] [PubMed]
- Holesh, J.E.; Aslam, S.; Martin, A. Physiology, Carbohydrates; StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Brandley, B.K.; Schnaar, R.L. Cell-Surface Carbohydrates in Cell Recognition and Response. J. Leukoc. Biol. 1986, 40, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Choe, E.; Min, D.B. Chemistry and Reactions of Reactive Oxygen Species in Foods. J. Food Sci. 2005, 70, R142–R159. [Google Scholar] [CrossRef]
- Hwang, K.O.; Lucia, L.A. Fundamental Insights into the Oxidation of Lignocellulosics Obtained from Singlet Oxygen Photochemistry. J. Photochem. Photobiol. A Chem. 2004, 168, 205–209. [Google Scholar] [CrossRef]
- Griesbeck, A.G.; Miranda, M.A.; Uhlig, J. Sweet Chiral Porphyrins as Singlet Oxygen Sensitizers for Asymmetric Type II Photooxygenation. Photochem. Photobiol. Sci. 2011, 10, 1431–1435. [Google Scholar] [CrossRef]
- Bauch, M.; Fudickar, W.; Linker, T. Stereoselective [4 + 2] Cycloaddition of Singlet Oxygen to Naphthalenes Controlled by Carbohydrates. Molecules 2021, 26, 804. [Google Scholar] [CrossRef]
- Wenk, M.R. The Emerging Field of Lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A Comprehensive Classification System for Lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Scharffetter–Kochanek, K.; Brenneisen, P.; Wenk, J.; Herrmann, G.; Ma, W.; Kuhr, L.; Meewes, C.; Wlaschek, M. Photoaging of the Skin from Phenotype to Mechanisms. Exp. Gerontol. 2000, 35, 307–316. [Google Scholar] [CrossRef]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane Lipids: Where They Are and How They Behave. Nature reviews. Mol. Cell Biol. 2008, 9, 112. [Google Scholar] [CrossRef]
- Yang, S.T.; Kreutzberger, A.J.B.; Lee, J.; Kiessling, V.; Tamm, L.K. The Role of Cholesterol in Membrane Fusion. Chem. Phys. Lipids 2016, 199, 136. [Google Scholar] [CrossRef] [PubMed]
- Kulig, M.J.; Smith, L.L. Sterol Metabolism. XXV. Cholesterol Oxidation by Singlet Molecular Oxygen. J. Org. Chem. 2002, 38, 3639–3642. [Google Scholar] [CrossRef] [PubMed]
- Scheinost, J.C.; Witter, D.P.; Boldt, G.E.; Offer, J.; Wentworth, P. Cholesterol Secosterol Adduction Inhibits the Misfolding of a Mutant Prion Protein Fragment That Induces Neurodegeneration. Angew. Chem. Int. Ed. 2009, 48, 9469–9472. [Google Scholar] [CrossRef] [PubMed]
- Dantas, L.S.; Chaves-Filho, A.B.; Coelho, F.R.; Genaro-Mattos, T.C.; Tallman, K.A.; Porter, N.A.; Augusto, O.; Miyamoto, S. Cholesterol Secosterol Aldehyde Adduction and Aggregation of Cu,Zn-Superoxide Dismutase: Potential Implications in ALS. Redox Biol. 2018, 19, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Girotti, A.W.; Korytowski, W. Cholesterol Hydroperoxide Generation, Translocation, and Reductive Turnover in Biological Systems. Cell Biochem. Biophys. 2017, 75, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Girotti, A.W. Photosensitized Oxidation of Membrane Lipids: Reaction Pathways, Cytotoxic Effects, and Cytoprotective Mechanisms. J. Photochem. Photobiol. B Biol. 2001, 63, 103–113. [Google Scholar] [CrossRef]
- Bacellar, I.O.L.; Oliveira, M.C.; Dantas, L.S.; Costa, E.B.; Junqueira, H.C.; Martins, W.K.; Durantini, A.M.; Cosa, G.; di Mascio, P.; Wainwright, M.; et al. Photosensitized Membrane Permeabilization Requires Contact-Dependent Reactions between Photosensitizer and Lipids. J. Am. Chem. Soc. 2018, 140, 9606–9615. [Google Scholar] [CrossRef]
- Riske, K.A.; Sudbrack, T.P.; Archilha, N.L.; Uchoa, A.F.; Schroder, A.P.; Marques, C.M.; Baptista, M.S.; Itri, R. Giant Vesicles under Oxidative Stress Induced by a Membrane-Anchored Photosensitizer. Biophys. J. 2009, 97, 1362. [Google Scholar] [CrossRef]
- Foret, M.K.; Lincoln, R.; do Carmo, S.; Cuello, A.C.; Cosa, G. Connecting the “Dots”: From Free Radical Lipid Autoxidation to Cell Pathology and Disease. Chem. Rev. 2020, 120, 12757–12787. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Huang, Y.Y.; Hamblin, M.R. Photodynamic Therapy for Localized Infections--State of the Art. Photodiagnosis Photodyn. Ther. 2009, 6, 170–188. [Google Scholar] [CrossRef]
- Klausen, M.; Ucuncu, M.; Bradley, M. Design of Photosensitizing Agents for Targeted Antimicrobial Photodynamic Therapy. Molecules 2020, 25, 5239. [Google Scholar] [CrossRef] [PubMed]
- Castano, A.P.; Mroz, P.; Hamblin, M.R. Photodynamic Therapy and Anti-Tumour Immunity. Nature reviews. Cancer 2006, 6, 535. [Google Scholar] [CrossRef] [PubMed]
- Shadish, J.A.; DeForest, C.A. Site-Selective Protein Modification: From Functionalized Proteins to Functional Biomaterials. Matter 2020, 2, 50–77. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Kim, E.; Koo, H. Biomedical Applications of Copper-Free Click Chemistry: In Vitro, In Vivo, and Ex Vivo. Chem. Sci. 2019, 10, 7835–7851. [Google Scholar] [CrossRef]
- Blizzard, R.J.; Gibson, T.E.; Mehl, R.A. Site-Specific Protein Labeling with Tetrazine Amino Acids. Methods Mol. Biol. 2018, 1728, 201–217. [Google Scholar] [CrossRef]
- Jencks, W.P. Studies on the Mechanism of Oxime and Semicarbazone Formation1. J. Am. Chem. Soc. 2002, 81, 475–481. [Google Scholar] [CrossRef]
- Dorta, D.A.; Deniaud, D.; Mével, M.; Gouin, S.G. Tyrosine Conjugation Methods for Protein Labelling. Chem. A Eur. J. 2020, 26, 14257–14269. [Google Scholar] [CrossRef]
- Joshi, N.S.; Whitaker, L.R.; Francis, M.B. A Three-Component Mannich-Type Reaction for Selective Tyrosine Bioconjugation. J. Am. Chem. Soc. 2004, 126, 15942–15943. [Google Scholar] [CrossRef]
- Antos, J.M.; Francis, M.B. Selective Tryptophan Modification with Rhodium Carbenoids in Aqueous Solution. J. Am. Chem. Soc. 2004, 126, 10256–10257. [Google Scholar] [CrossRef]
- Lundblad, R.L. Chemical Reagents for Protein Modification; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- de Geyter, E.; Antonatou, E.; Kalaitzakis, D.; Smolen, S.; Iyer, A.; Tack, L.; Ongenae, E.; Vassilikogiannakis, G.; Madder, A. 5-Hydroxy-Pyrrolone Based Building Blocks as Maleimide Alternatives for Protein Bioconjugation and Single-Site Multi-Functionalization. Chem. Sci. 2021, 12, 5246–5252. [Google Scholar] [CrossRef] [PubMed]
- Chilamari, M.; Purushottam, L.; Rai, V. Site-Selective Labeling of Native Proteins by a Multicomponent Approach. Chem. A Eur. J. 2017, 23, 3819–3823. [Google Scholar] [CrossRef] [PubMed]
- McKay, C.S.; Finn, M.G. Click Chemistry in Complex Mixtures: Bioorthogonal Bioconjugation. Chem. Biol. 2014, 21, 1075–1101. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhaya, S.; Abu Bakar, F.; Yao, S. Expanding the Chemical Biologists Tool Kit: Chemical Labelling Strategies and Its Applications. Curr. Med. Chem. 2009, 16, 4527–4543. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, B.P.; Zhang, Z.; Hosokawa, A.; Distefano, M.D. Selective Labeling of Proteins by Using Protein Farnesyltransferase. ChemBioChem 2007, 8, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Suárez, M.; Baruah, H.; Martínez-Hernández, L.; Xie, K.T.; Baskin, J.M.; Bertozzi, C.R.; Ting, A.Y. Re-Directing Lipoic Acid Ligase for Cell Surface Protein Labeling with Small-Molecule Probes. Nat. Biotechnol. 2007, 25, 1483. [Google Scholar] [CrossRef] [PubMed]
- Antonatou, E.; Verleysen, Y.; Madder, A. Singlet Oxygen-Mediated One-Pot Chemoselective Peptide-Peptide Ligation. Org. Biomol. Chem. 2017, 15, 8140–8144. [Google Scholar] [CrossRef]
- Zhang, H.; Trout, W.S.; Liu, S.; Andrade, G.A.; Hudson, D.A.; Scinto, S.L.; Dicker, K.T.; Li, Y.; Lazouski, N.; Rosenthal, J.; et al. Rapid Bioorthogonal Chemistry Turn-on through Enzymatic or Long Wavelength Photocatalytic Activation of Tetrazine Ligation. J. Am. Chem. Soc. 2016, 138, 5978–5983. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Zhang, T.; Zou, X.; Wang, H.; Rosenberger, J.E.; Vannam, R.; Trout, W.S.; Grimm, J.B.; Lavis, L.D.; et al. Enabling in Vivo Photocatalytic Activation of Rapid Bioorthogonal Chemistry by Repurposing Silicon-Rhodamine Fluorophores as Cytocompatible Far-Red Photocatalysts. J. Am. Chem. Soc. 2021, 143, 10793–10803. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, F.; Helman, W.P.; Ross, A.B. Quantum Yields for the Photosensitized Formation of the Lowest Electronically Excited Singlet State of Molecular Oxygen in Solution. J. Phys. Chem. Ref. Data 1993, 22, 113–262. [Google Scholar] [CrossRef]
- Miret-Casals, L.; Vannecke, W.; Hoogewijs, K.; Arauz-Garofalo, G.; Gay, M.; Díaz-Lobo, M.; Vilaseca, M.; Ampe, C.; van Troys, M.; Madder, A. Furan Warheads for Covalent Trapping of Weak Protein-Protein Interactions: Cross-Linking of Thymosin Β4 to Actin. Chem. Commun. 2021, 57, 6054–6057. [Google Scholar] [CrossRef] [PubMed]
- Braun, P.; Gingras, A.C. History of Protein-Protein Interactions: From Egg-White to Complex Networks. Proteomics 2012, 12, 1478–1498. [Google Scholar] [CrossRef] [PubMed]
- Schopper, S.; Kahraman, A.; Leuenberger, P.; Feng, Y.; Piazza, I.; Müller, O.; Boersema, P.J.; Picotti, P. Measuring Protein Structural Changes on a Proteome-Wide Scale Using Limited Proteolysis-Coupled Mass Spectrometry. Nat. Protoc. 2017, 12, 2391–2410. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.C.; Abe, K.T.; Raught, B. Getting to Know the Neighborhood: Using Proximity-Dependent Biotinylation to Characterize Protein Complexes and Map Organelles. Curr. Opin. Chem. Biol. 2019, 48, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Matthews, J.M. Protein–Protein Interactions in Human Disease. Curr. Opin. Struct. Biol. 2005, 15, 441–446. [Google Scholar] [CrossRef]
- Sinz, A. Chemical Cross-Linking and Mass Spectrometry to Map Three-Dimensional Protein Structures and Protein–Protein Interactions. Mass Spectrom. Rev. 2006, 25, 663–682. [Google Scholar] [CrossRef]
- Vermeulen, M.; Hubner, N.C.; Mann, M. High Confidence Determination of Specific Protein-Protein Interactions Using Quantitative Mass Spectrometry. Curr. Opin. Biotechnol. 2008, 19, 331–337. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass Spectrometry-Based Proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- Suchanek, M.; Radzikowska, A.; Thiele, C. Photo-Leucine and Photo-Methionine Allow Identification of Protein-Protein Interactions in Living Cells. Nat. Methods 2005, 2, 261–267. [Google Scholar] [CrossRef]
- Hino, N.; Hayashi, A.; Sakamoto, K.; Yokoyama, S. Site-Specific Incorporation of Non-Natural Amino Acids into Proteins in Mammalian Cells with an Expanded Genetic Code. Nat. Protoc. 2007, 1, 2957–2962. [Google Scholar] [CrossRef]
- Chin, J.W.; Santoro, S.W.; Martin, A.B.; King, D.S.; Wang, L.; Schultz, P.G. Addition of P-Azido-l-Phenylalanine to the Genetic Code of Escherichia Coli. J. Am. Chem. Soc. 2002, 124, 9026–9027. [Google Scholar] [CrossRef] [PubMed]
- Reddington, S.; Watson, P.; Rizkallah, P.; Tippmann, E.; Jones, D.D. Genetically Encoding Phenyl Azide Chemistry: New Uses and Ideas for Classical Biochemistry. Biochem. Soc. Trans. 2013, 41, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.M.; Du, X.; Tolić, N.; Wu, S.; Moore, R.J.; Mayer, M.U.; Smith, R.D.; Adkins, J.N. Identification of Cross-Linked Peptides after Click-Based Enrichment Using Sequential Collision-Induced Dissociation and Electron Transfer Dissociation Tandem Mass Spectrometry. Anal. Chem. 2009, 81, 5524–5532. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Y.; Henklein, P.; Li, X.; Hofmann, K.P.; Nakanishi, K.; Ernst, O.P. A Photo-Cross-Linking Strategy to Map Sites of Protein–Protein Interactions. Chem. A Eur. J. 2010, 16, 7389–7394. [Google Scholar] [CrossRef]
- Joiner, C.M.; Breen, M.E.; Clayton, J.; Mapp, A.K. A Bifunctional Amino Acid Enables Both Covalent Chemical Capture and Isolation of in Vivo Protein–Protein Interactions. ChemBioChem 2017, 18, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Stadlmeier, M.; Runtsch, L.S.; Streshnev, F.; Wühr, M.; Carell, T. A Click-Chemistry-Based Enrichable Crosslinker for Structural and Protein Interaction Analysis by Mass Spectrometry. ChemBioChem 2020, 21, 103–107. [Google Scholar] [CrossRef]
- Rey, M.; Dhenin, J.; Kong, Y.; Nouchikian, L.; Filella, I.; Duchateau, M.; Dupré, M.; Pellarin, R.; Duménil, G.; Chamot-Rooke, J. Advanced In Vivo Cross-Linking Mass Spectrometry Platform to Characterize Proteome-Wide Protein Interactions. Anal. Chem. 2021, 93, 4166–4174. [Google Scholar] [CrossRef]
- Hoffmann, J.-E.; Dziuba, D.; Stein, F.; Schultz, C. A Bifunctional Noncanonical Amino Acid: Synthesis, Expression, and Residue-Specific Proteome-Wide Incorporation. Biochemistry 2018, 57, 4747–4752. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Carroll, L.; Rasmussen, L.M.; Davies, M.J. Oxidation of Protein Disulfide Bonds by Singlet Oxygen Gives Rise to Glutathionylated Proteins. Redox Biol. 2021, 38, 101822. [Google Scholar] [CrossRef]
- Nagy, P.; Lechte, T.P.; Das, A.B.; Winterbourn, C.C. Conjugation of Glutathione to Oxidized Tyrosine Residues in Peptides and Proteins. J. Biol. Chem. 2012, 287, 26068–26076. [Google Scholar] [CrossRef]
- Mariotti, M.; Leinisch, F.; Leeming, D.J.; Svensson, B.; Davies, M.J.; Hägglund, P. Mass-Spectrometry-Based Identification of Cross-Links in Proteins Exposed to Photo-Oxidation and Peroxyl Radicals Using 18 O Labeling and Optimized Tandem Mass Spectrometry Fragmentation. J. Proteome Res. 2018, 17, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Kanwar, R. Molecular Pathology of Dityrosine Cross-Links in Proteins: Structural and Functional Analysis of Four Proteins. Mol. Cell. Biochem. 2002, 234, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Lepock, J.R.; Thompson, J.E.; Kruuv, J.; Wallach, D.F.H. Photoinduced Crosslinking of Membrane Proteins by Fluorescein Isothiocyanate. Biochem. Biophys. Res. Commun. 1978, 85, 344–350. [Google Scholar] [CrossRef]
- Goosey, J.D.; Zigler, J.S., Jr.; Kinoshita, J.H. Cross-Linking of Lens Crystallins in a Photodynamic System: A Process Mediated by Singlet Oxygen. Science 1980, 208, 1278–1280. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.R.; Spikes, J.D.; Kopečeková, P.; Kopeček, J. Photodynamic Crosslinking of Proteins. I. Model Studies Using Histidine- and Lysine-Containing N-(2-Hydroxypropyl)Methacrylamide Copolymers. J. Photochem. Photobiol. B Biol. 1996, 34, 203–210. [Google Scholar] [CrossRef]
- Shen, H.R.; Spikes, J.D.; Kopečková, P.; Kopeček, J. Photodynamic Crosslinking of Proteins II. Photocrosslinking of a Model Protein-Ribonuclease A. J. Photochem. Photobiol. B Biol. 1996, 35, 213–219. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, Z.; Cheetham, J.; Ren, D.; Zhou, Z.S. Discovery and Characterization of a Photo-Oxidative Histidine-Histidine Cross-Link in IgG1 Antibody Utilizing 18O-Labeling and Mass Spectrometry. Anal. Chem. 2014, 86, 4940–4948. [Google Scholar] [CrossRef]
- Fuentes-Lemus, E.; López-Alarcón, C. Photo-Induced Protein Oxidation: Mechanisms, Consequences and Medical Applications. Essays Biochem. 2020, 64, 33–44. [Google Scholar] [CrossRef]
- Hoogewijs, K.; Deceuninck, A.; Madder, A. Aromatic Capping Surprisingly Stabilizes Furan Moieties in Peptides against Acidic Degradation. Org. Biomol. Chem. 2012, 10, 3999–4002. [Google Scholar] [CrossRef]
- Decoene, K.W.; Vannecke, W.; Passioura, T.; Suga, H.; Madder, A. Pyrrole-Mediated Peptide Cyclization Identified through Genetically Reprogrammed Peptide Synthesis. Biomedicines 2018, 6, 99. [Google Scholar] [CrossRef]
- Davies, M.J. Reactive Species Formed on Proteins Exposed to Singlet Oxygen. Photochem. Photobiol. Sci. 2004, 3, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, F.; Helman, W.P.; Ross, A.B. Rate Constants for the Decay and Reactions of the Lowest Electronically Excited Singlet State of Molecular Oxygen in Solution. An Expanded and Revised Compilation. J. Phys. Chem. Ref. Data 2009, 24, 663. [Google Scholar] [CrossRef]
- Manicardi, A.; Cadoni, E.; Madder, A. Hydrolysis of 5-Methylfuran-2-Yl to 2,5-Dioxopentanyl Allows for Stable Bio-Orthogonal Proximity-Induced Ligation. Commun. Chem. 2021, 4, 146. [Google Scholar] [CrossRef]
- Rabinowitz, Y.S. Keratoconus. Surv. Ophthalmol. 1998, 42, 297–319. [Google Scholar] [CrossRef]
- Santhiago, M.R.; Randleman, J.B. The Biology of Corneal Cross-Linking Derived from Ultraviolet Light and Riboflavin. Exp. Eye Eye Res. 2021, 202, 108355. [Google Scholar] [CrossRef]
- Zhang, Y.; Conrad, A.H.; Conrad, G.W. Effects of Ultraviolet-A and Riboflavin on the Interaction of Collagen and Proteoglycans during Corneal Cross-Linking. J. Biol. Chem. 2011, 286, 13011–13022. [Google Scholar] [CrossRef]
- McCall, A.S.; Kraft, S.; Edelhauser, H.F.; Kidder, G.W.; Lundquist, R.R.; Bradshaw, H.E.; Dedeic, Z.; Dionne, M.J.C.; Clement, E.M.; Conrad, G.W. Mechanisms of Corneal Tissue Cross-Linking in Response to Treatment with Topical Riboflavin and Long-Wavelength Ultraviolet Radiation (UVA). Investig. Ophthalmol. Vis. Sci. 2010, 51, 129–138. [Google Scholar] [CrossRef]
- Wollensak, G.; Spoerl, E.; Wilsch, M.; Seiler, T. Keratocyte Apoptosis after Corneal Collagen Cross-Linking Using Riboflavin/UVA Treatment. Cornea 2004, 23, 43–49. [Google Scholar] [CrossRef]
- Wollensak, G.; Spoerl, E.; Reber, F.; Seiler, T. Keratocyte Cytotoxicity of Riboflavin/UVA-Treatment in Vitro. Eye 2004, 18, 718–722. [Google Scholar] [CrossRef]
- Jordan, C.; Patel, D.V.; Abeysekera, N.; McGhee, C.N. In Vivo Confocal Microscopy Analyses of Corneal Microstructural Changes in a Prospective Study of Collagen Cross-Linking in Keratoconus. Ophthalmology 2014, 121, 469–474. [Google Scholar] [CrossRef]
- Nathan, C.; Cunningham-Bussel, A. Beyond Oxidative Stress: An Immunologist’s Guide to Reactive Oxygen Species. Nat. Rev. Immunol. 2013, 13, 349. [Google Scholar] [CrossRef] [PubMed]
- Galli, F.; Piroddi, M.; Annetti, C.; Aisa, C.; Floridi, E.; Floridi, A. Oxidative Stress and Reactive Oxygen Species. Contrib. Nephrol. 2005, 149, 240–260. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.-Y.; Storz, P. Reactive Oxygen Species in Cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Vannecke, W.; Ampe, C.; van Troys, M.; Beltramo, M.; Madder, A. Cross-Linking Furan-Modified Kisspeptin-10 to the KISS Receptor. ACS Chem. Biol. 2017, 12, 2191–2200. [Google Scholar] [CrossRef]
- Jacobson, K.; Rajfur, Z.; Vitriol, E.; Hahn, K. Chromophore-Assisted Laser Inactivation in Cell Biology. Trends Cell Biol. 2008, 18, 443–450. [Google Scholar] [CrossRef]
- Sano, Y.; Watanabe, W.; Matsunaga, S. Chromophore-Assisted Laser Inactivation—Towards a Spatiotemporal-Functional Analysis of Proteins, and the Ablation of Chromatin, Organelle and Cell Function. J. Cell Sci. 2014, 127, 1621–1629. [Google Scholar] [CrossRef]
- Beck, S.; Sakurai, T.; Eustace, B.K.; Beste, G.; Schier, R.; Rudert, F.; Jay, D.G. Fluorophore-Assisted Light Inactivation: A High-Throughput Tool for Direct Target Validation of Proteins. Proteomics 2002, 2, 247–255. [Google Scholar] [CrossRef]
- Jay, D.G. Selective Destruction of Protein Function by Chromophore-Assisted Laser Inactivation. Proc. Natl. Acad. Sci. USA 1988, 85, 5454–5458. [Google Scholar] [CrossRef]
- Jean, B.; Schmolz, M.W.; Schöllhorn, V.G. Selective Laser-Induced Inactivation of Proteins (SLIP) by Labelling with Chromophores. Med. Biol. Eng. Comput. 1992, 30, CE17–CE20. [Google Scholar] [CrossRef]
- Yogo, T.; Urano, Y.; Mizushima, A.; Sunahara, H.; Inoue, T.; Hirose, K.; Iino, M.; Kikuchi, K.; Nagano, T. Selective Photo Inactivation of Protein Function through Environment-Sensitive Switching of Singlet Oxygen Generation by Photosensitizer. Proc. Natl. Acad. Sci. USA 2008, 105, 28–32. [Google Scholar] [CrossRef]
- Takemoto, K.; Matsuda, T.; McDougall, M.; Klaubert, D.H.; Hasegawa, A.; Los, G.V.; Wood, K.V.; Miyawaki, A.; Nagai, T. Chromophore-Assisted Light Inactivation of HaloTag Fusion Proteins Labeled with Eosin in Living Cells. ACS Chem. Biol. 2011, 6, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Marek, K.W.; Davis, G.W. Neurotechnique Transgenically Encoded Protein Photoinactivation (FlAsH-FALI): Acute Inactivation of Synaptotagmin I the Consequence. Neuron 2002, 36, 805–813. [Google Scholar] [CrossRef]
- Tour, O.; Meijer, R.M.; Zacharias, D.A.; Adams, S.R.; Tsien, R.Y. Genetically Targeted Chromophore-Assisted Light Inactivation. Nat. Biotechnol. 2003, 21, 1505–1508. [Google Scholar] [CrossRef] [PubMed]
- Keppler, A.; Ellenberg, J. Chromophore-Assisted Laser Inactivation of α- and γ-Tubulin SNAP-Tag Fusion Proteins inside Living Cells. ACS Chem. Biol. 2009, 4, 127–138. [Google Scholar] [CrossRef]
- Rajfur, Z.; Roy, P.; Otey, C.; Romer, L.; Jacobson, K. Dissecting the Link between Stress Fibres and Focal Adhesions by CALI Wit EGFP Fusion Proteins. Nat. Cell Biol. 2002, 4, 286–293. [Google Scholar] [CrossRef]
- Wojtovich, A.P.; Wei, A.Y.; Sherman, T.A.; Foster, T.H.; Nehrke, K. Chromophore-Assisted Light Inactivation of Mitochondrial Electron Transport Chain Complex II in Caenorhabditis Elegans. Sci. Rep. 2016, 6, 29695. [Google Scholar] [CrossRef]
- Kassis, A.I. Therapeutic Radionuclides: Biophysical and Radiobiologic Principles. Semin. Nucl. Med. 2008, 38, 358–366. [Google Scholar] [CrossRef]
- Yao, S.; Belfield, K.D. Two-Photon Fluorescent Probes for Bioimaging. Eur. J. Org. Chem. 2012, 2012, 3199–3217. [Google Scholar] [CrossRef]
- Wilchek, M.; Bayer, E.A. The Avidin-Biotin Complex in Bioanalytical Applications. J. Clin. Chem. Clin. Biochem. 1989, 27, 889–890. [Google Scholar] [CrossRef]
- Liang, J.; Jia, H.; Li, L.; Li, X.; Li, Y. β-Difluoroalkylamine as a Motif for Singlet Oxygen-Mediated Proximity Labeling in Living Cells. Org. Lett. 2021, 23, 4640–4644. [Google Scholar] [CrossRef]
- de Beeck, M.O.; Madder, A. Unprecedented C-Selective Interstrand Cross-Linking through in Situ Oxidation of Furan-Modified Oligodeoxynucleotides. J. Am. Chem. Soc. 2011, 133, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Stevens, K.; Madder, A. Furan-Modified Oligonucleotides for Fast, High-Yielding and Site-Selective DNA Inter-Strand Cross-Linking with Non-Modified Complements. Nucleic Acids Res. 2009, 37, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- de Beeck, M.O.; Madder, A. Sequence Specific DNA Cross-Linking Triggered by Visible Light. J. Am. Chem. Soc. 2012, 134, 10737–10740. [Google Scholar] [CrossRef] [PubMed]
- Carrette, L.L.G.; Gyssels, E.; Loncke, J.; Madder, A. A Mildly Inducible and Selective Cross-Link Methodology for RNA Duplexes. Org. Biomol. Chem. 2014, 12, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Llamas, E.M.; Tome, J.P.C.; Rodrigues, J.M.M.; Torres, T.; Madder, A. Porphyrin-Based Photosensitizers and Their DNA Conjugates for Singlet Oxygen Induced Nucleic Acid Interstrand Crosslinking. Org. Biomol. Chem. 2017, 15, 5402–5409. [Google Scholar] [CrossRef]
- De Laet, N.; Llamas, E.M.; Madder, A. Templated DNA Cross-Linking: Towards a Non-Invasive Singlet-Oxygen-Based Triggering Method. ChemPhotoChem 2018, 2, 575–579. [Google Scholar] [CrossRef]
- Cadoni, E.; Manicardi, A.; Fossépré, M.; Heirwegh, K.; Surin, M.; Madder, A. Teaching Photosensitizers a New Trick: Red Light-Triggered G-Quadruplex Alkylation by Ligand Co-Localization. Chem. Commun. 2021, 57, 1010–1013. [Google Scholar] [CrossRef]
- Véliz Montes, C.; Memczak, H.; Gyssels, E.; Torres, T.; Madder, A.; Schneider, R.J. Photoinduced Cross-Linking of Short Furan-Modified DNA on Surfaces. Langmuir 2017, 33, 1197–1201. [Google Scholar] [CrossRef]
- Muangkaew, P.; Vilaivan, T. Pyrrolidinyl Peptide Nucleic Acid Probes Capable of Crosslinking with DNA: Effects of Terminal and Internal Modifications on Crosslink Efficiency. ChemBioChem 2021, 22, 241–252. [Google Scholar] [CrossRef]
- Manicardi, A.; Cadoni, E.; Madder, A. Visible-Light Triggered Templated Ligation on Surface Using Furan-Modified PNAs. Chem. Sci. 2020, 11, 11729–11739. [Google Scholar] [CrossRef]
- Schmidt, M.J.; Summerer, D. Red-Light-Controlled Protein-RNA Crosslinking with a Genetically Encoded Furan. Angew. Chem.-Int. Ed. 2013, 52, 4690–4693. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Cui, C.; Cheng, X.; Zhao, M.; Mao, Q.; Zhang, Y.; Wang, A.; Fang, J.; Zhao, Y.; Shi, H. Red Light-Initiated Cross-Linking of NIR Probes to Cytoplasmic RNA: An Innovative Strategy for Prolonged Imaging and Unexpected Tumor Suppression. J. Am. Chem. Soc. 2020, 142, 21502–21512. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Baldino, C.M. D,l- and Meso-Isochrysohermidin: Total Synthesis and Interstrand DNA Cross-Linking. J. Am. Chem. Soc. 1993, 115, 11418–11425. [Google Scholar] [CrossRef]
- Hong, I.S.; Greenberg, M.M. Efficient DNA Interstrand Cross-Link Formation from a Nucleotide Radical. J. Am. Chem. Soc. 2005, 127, 3692–3693. [Google Scholar] [CrossRef]
- Hong, I.S.; Greenberg, M.M. DNA Interstrand Cross-Link Formation Initiated by Reaction between Singlet Oxygen and a Modified Nucleotide. J. Am. Chem. Soc. 2005, 127, 10510–10511. [Google Scholar] [CrossRef]
- Shemesh, Y.; Yavin, E. PNA-Rose Bengal Conjugates as Efficient DNA Photomodulators. Bioconjugate Chem. 2015, 26, 1916–1922. [Google Scholar] [CrossRef]
- Solivio, M.J.; Nemera, D.B.; Sallans, L.; Merino, E.J. Biologically Relevant Oxidants Cause Bound Proteins to Readily Oxidatively Cross-Link at Guanine. Chem. Res. Toxicol. 2012, 25, 326–336. [Google Scholar] [CrossRef]
- Lamb, B.M.; Barbas, C.F. Selective Arylthiolane Deprotection by Singlet Oxygen: A Promising Tool for Sensors and Prodrugs. Chem. Commun. 2015, 51, 3196–3199. [Google Scholar] [CrossRef]
- Frimer, A.A. The Reaction of Singlet Oxygen with Olefins: The Question of Mechanism. Chem. Rev. 1979, 79, 359–387. [Google Scholar] [CrossRef]
- Murthy, R.S.; Bio, M.; You, Y. Low Energy Light-Triggered Oxidative Cleavage of Olefins. Tetrahedron Lett. 2009, 50, 1041–1044. [Google Scholar] [CrossRef]
- Dinache, A.; Smarandache, A.; Simon, A.; Nastasa, V.; Tozar, T.; Pascu, A.; Enescu, M.; Khatyr, A.; Sima, F.; Pascu, M.-L.; et al. Photosensitized Cleavage of Some Olefins as Potential Linkers to Be Used in Drug Delivery. Appl. Surf. Sci. 2017, 417, 136–142. [Google Scholar] [CrossRef]
- Filgueiras, C.A.L.; Celso, C.; Coelho, G.H.; Johnson, B.F.G. The Synthesis and Properties of Cis-[Rac-1,2-Cis-Bis (Phenylsulphinyl)Ethene]-Dichloro-Platinum(II). Inorg. Nucl. Chem. Lett. 1981, 17, 283–285. [Google Scholar] [CrossRef]
- Bio, M.; Nkepang, G.; You, Y. Click and Photo-Unclick Chemistry of Aminoacrylate for Visible Light-Triggered Drug Release. Chem. Commun. 2012, 48, 6517–6519. [Google Scholar] [CrossRef] [PubMed]
- Nkepang, G.; Bio, M.; Rajaputra, P.; Awuah, S.G.; You, Y. Folate Receptor-Mediated Enhanced and Specific Delivery of Far-Red Light-Activatable Prodrugs of Combretastatin A-4 to FR-Positive Tumor. Bioconjugate Chem. 2014, 25, 2175–2188. [Google Scholar] [CrossRef] [PubMed]
- Thapa, P.; Li, M.; Bio, M.; Rajaputra, P.; Nkepang, G.; Sun, Y.; Woo, S.; You, Y. Far-Red Light-Activatable Prodrug of Paclitaxel for the Combined Effects of Photodynamic Therapy and Site-Specific Paclitaxel Chemotherapy. J. Med. Chem. 2016, 59, 3204–3214. [Google Scholar] [CrossRef] [PubMed]
- Bio, M.; Rajaputra, P.; Lim, I.; Thapa, P.; Tienabeso, B.; Hurst, R.E.; You, Y. Efficient Activation of a Visible Light-Activatable CA4 Prodrug through Intermolecular Photo-Unclick Chemistry in Mitochondria. Chem. Commun. 2017, 53, 1884–1887. [Google Scholar] [CrossRef]
- Rajaputra, P.; Bio, M.; Nkepang, G.; Thapa, P.; Woo, S.; You, Y. Anticancer Drug Released from near IR-Activated Prodrug Overcomes Spatiotemporal Limits of Singlet Oxygen. Bioorg. Med. Chem. 2016, 24, 1540–1549. [Google Scholar] [CrossRef]
- Saravanakumar, G.; Lee, J.; Kim, J.; Kim, W.J. Visible Light-Induced Singlet Oxygen-Mediated Intracellular Disassembly of Polymeric Micelles Co-Loaded with a Photosensitizer and an Anticancer Drug for Enhanced Photodynamic Therapy. Chem. Commun. 2015, 51, 9995–9998. [Google Scholar] [CrossRef]
- Chen, L.; Li, G.; Wang, X.; Li, J.; Zhang, Y. Spherical Nucleic Acids for Near-Infrared Light-Responsive Self-Delivery of Small-Interfering RNA and Antisense Oligonucleotide. ACS Nano 2021, 15, 11929–11939. [Google Scholar] [CrossRef]
- Tabero, A.; Planas, O.; Gallavardin, T.; Nieves, I.; Nonell, S.; Villanueva, A. Smart Dual-Functionalized Gold Nanoclusters for Spatio-Temporally Controlled Delivery of Combined Chemo-and Photodynamic Therapy. Nanomaterials 2020, 10, 2474. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, C.; Li, J.; Cui, D.; Jiang, Y.; Pu, K. Activatable Polymer Nanoenzymes for Photodynamic Immunometabolic Cancer Therapy. Adv. Mater. 2021, 33, 2007247. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.C.H.; Ng, D.K.P.; Fong, W.P.; Lo, P.C. Glutathione- And Light-Controlled Generation of Singlet Oxygen for Triggering Drug Release in Mesoporous Silica Nanoparticles. J. Mater. Chem. B 2020, 8, 4460–4468. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Sun, C.; Yu, C. Highly-Controllable Drug Release from Core Cross-Linked Singlet Oxygen-Responsive Nanoparticles for Cancer Therapy. RSC Adv. 2020, 10, 19997–20008. [Google Scholar] [CrossRef]
- Martínez-Carmona, M.; Lozano, D.; Baeza, A.; Colilla, M.; Vallet-Regí, M. A Novel Visible Light Responsive Nanosystem for Cancer Treatment. Nanoscale 2017, 9, 15967–15973. [Google Scholar] [CrossRef] [PubMed]
- Sassolas, A.; Leca-Bouvier, B.D.; Blum, L.J. DNA Biosensors and Microarrays. Chem. Rev. 2008, 108, 109–139. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical Biosensors: Recommended Definitions and Classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Strianese, M.; Staiano, M.; Ruggiero, G.; Labella, T.; Pellecchia, C.; D’Auria, S. Fluorescence-Based Biosensors. Methods Mol. Biol. 2012, 875, 193–216. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold Nanoparticle-Based Colorimetric Biosensors. Nanoscale 2018, 10, 18–33. [Google Scholar] [CrossRef]
- Han, T.; Cao, Y.; Chen, H.-Y.; Zhu, J.-J. Versatile Porous Nanomaterials for Electrochemiluminescence Biosensing: Recent Advances and Future Perspective. J. Electroanal. Chem. 2021, 902, 115821. [Google Scholar] [CrossRef]
- Qin, X.; Xu, S.; Deng, L.; Huang, R.; Zhang, X. Photocatalytic Electrosensor for Label-Free and Ultrasensitive Detection of BRCA1 Gene. Biosens. Bioelectron. 2016, 85, 957–963. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, Q.; Tang, G.; Liu, S.; Xu, S.; Zhang, X. A Facile Electrochemical Aptasensor for Lysozyme Detection Based on Target-Induced Turn-off of Photosensitization. Biosens. Bioelectron. 2019, 126, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Arken, G.; Li, G.; Guan, M.; Tian, S. A Novel Photo-Induced Electrochemical Biosensing Method Based on Fluorescent Labeled Molecular Beacon. Electroanalysis 2017, 29, 1310–1315. [Google Scholar] [CrossRef]
- Shanmugam, S.T.; Trashin, S.; de Wael, K. Singlet Oxygen-Based Photoelectrochemical Detection of DNA. Biosens. Bioelectron. 2022, 195, 113652. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Mansour, H.; Hu, H.; Wang, G.A.; Watson, C.J.F.; Yousef, M.; Matamoros, G.; Sanchez, A.L.; Macneil, A.J.; Wu, P.; et al. Colorimetric Polymerase Chain Reaction Enabled by a Fast Light-Activated Substrate Chromogenic Detection Platform. Anal. Chem. 2020, 92, 6456–6461. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Wang, Y.; Deng, L.; Li, L.; Zhang, X.; Wu, P. Oxidative Capacity Storage of Transient Singlet Oxygen from Photosensitization with a Redox Mediator for Improved Chemiluminescent Sensing. Anal. Chem. 2019, 91, 9407–9412. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yang, T.; Liu, M.; Hu, Y.; Wang, J. An Aptamer-Based Biosensor for Sensitive Thrombin Detection with Phthalocyanine@SiO2 Mesoporous Nanoparticles. Biosens. Bioelectron. 2014, 53, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Zhang, T.; Ji, X.; Wan, Y.; Xin, P.; Shan, D.; Zhang, X. Detection of Zinc Finger Protein (EGR1) Based on Electrogenerated Chemiluminescence from Singlet Oxygen Produced in a Nanoclay-Supported Porphyrin Environment. Anal. Chem. 2015, 87, 9155–9162. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, P.; Zhang, Z.; Zhai, X.; Liang, J.; Chen, Q.; Li, K.; Lin, G.; Liu, T.; Wu, Y. Ultrasensitive Sensor Using Quantum Dots-Doped Polystyrene Nanospheres for Clinical Diagnostics of Low-Volume Serum Samples. Anal. Chem. 2019, 91, 5777–5785. [Google Scholar] [CrossRef]
- Gefter, M.L.; Becker, A.; Hurwitz, J. The Enzymatic Repair of DNA. I. Formation of Circular Lambda-DNA. Proc. Natl. Acad. Sci. USA 1967, 58, 240–247. [Google Scholar] [CrossRef]
- Zhang, G.; Chai, H.; Tian, M.; Zhu, S.; Qu, L.; Zhang, X. Zirconium-Metalloporphyrin Frameworks-Luminol Competitive Electrochemiluminescence for Ratiometric Detection of Polynucleotide Kinase Activity. Anal. Chem. 2020, 92, 7354–7362. [Google Scholar] [CrossRef]
- Zhang, G.; Li, M.; Yu, K.; Chai, H.; Xu, S.; Xu, T.; Qu, L.; Zhang, X. Two-Dimensional Metalloporphyrinic Framework Nanosheet-Based Dual-Mechanism-Driven Ratiometric Electrochemiluminescent Biosensing of Protein Kinase Activity. ACS Appl. Bio Mater. 2021, 4, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Zhao, L.; Geng, F.; Wang, D.; Guo, L.H. Donor/Acceptor Nanoparticle Pair-Based Singlet Oxygen Channeling Homogenous Chemiluminescence Immunoassay for Quantitative Determination of Bisphenol A. Anal. Bioanal. Chem. 2016, 408, 8795–8804. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, J.; Ding, Y.; Zhang, X.; Xu, K.; Hou, X.; Wu, P. Modulation of the Singlet Oxygen Generation from the Double Strand DNA-SYBR Green i Complex Mediated by T-Melamine-T Mismatch for Visual Detection of Melamine. Anal. Chem. 2017, 89, 5101–5106. [Google Scholar] [CrossRef] [PubMed]
- Ling, P.; Sun, X.; Chen, N.; Cheng, S.; Gao, X.; Gao, F. Electrochemical Biosensor Based on Singlet Oxygen Generated by Molecular Photosensitizers. Anal. Chim. Acta 2021, 1183, 338970. [Google Scholar] [CrossRef]
- Mironova, K.E.; Proshkina, G.M.; Ryabova, A.V.; Stremovskiy, O.A.; Lukyanov, S.A.; Petrov, R.V.; Deyev, S.M. Genetically Encoded Immunophotosensitizer 4D5scFv-MiniSOG Is a Highly Selective Agent for Targeted Photokilling of Tumor Cells in Vitro. Theranostics 2013, 3, 831–840. [Google Scholar] [CrossRef]
- Bulina, M.E.; Chudakov, D.M.; Britanova, O.V.; Yanushevich, Y.G.; Staroverov, D.B.; Chepurnykh, T.V.; Merzlyak, E.M.; Shkrob, M.A.; Lukyanov, S.; Lukyanov, K.A. A Genetically Encoded Photosensitizer. Nat. Biotechnol. 2006, 24, 95–99. [Google Scholar] [CrossRef]
- Bulina, M.E.; Lukyanov, K.A.; Britanova, O.V.; Onichtchouk, D.; Lukyanov, S.; Chudakov, D.M. Chromophore-Assisted Light Inactivation (CALI) Using the Phototoxic FLuorescent Protein KillerRed. 7. Nat. Protoc. 2006, 1, 947–953. [Google Scholar] [CrossRef]
- Pletnev, S.; Gurskaya, N.G.; Pletneva, N.V.; Lukyanov, K.A.; Chudakov, D.M.; Martynov, V.I.; Popov, V.O.; Kovalchuk, M.V.; Wlodawer, A.; Dauter, Z.; et al. Structural Basis for Phototoxicity of the Genetically Encoded Photosensitizer KillerRed. J. Biol. Chem. 2009, 284, 32028–32039. [Google Scholar] [CrossRef]
- Shu, X.; Lev-Ram, V.; Deerinck, T.J.; Qi, Y.; Ramko, E.B.; Davidson, M.W.; Jin, Y.; Ellisman, M.H.; Tsien, R.Y. A Genetically Encoded Tag for Correlated Light and Electron Microscopy of Intact Cells, Tissues, and Organisms. PLoS Biol. 2011, 9, e1001041. [Google Scholar] [CrossRef]
- Ryumina, A.P.; Serebrovskaya, E.O.; Shirmanova, M.V.; Snopova, L.B.; Kuznetsova, M.M.; Turchin, I.V.; Ignatova, N.I.; Klementieva, N.V.; Fradkov, A.F.; Shakhov, B.E.; et al. Flavoprotein MiniSOG as a Genetically Encoded Photosensitizer for Cancer Cells. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 5059–5067. [Google Scholar] [CrossRef]
- Lin, J.Y.; Sann, S.B.; Zhou, K.; Nabavi, S.; Proulx, C.D.; Malinow, R.; Jin, Y.; Tsien, R.Y. Optogenetic Inhibition of Synaptic Release with Chromophore-Assisted Light Inactivation (CALI). Neuron 2013, 79, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Alonso-de Castro, S.; Cortajarena, A.L.; López-Gallego, F.; Salassa, L. Bioorthogonal Catalytic Activation of Platinum and Ruthenium Anticancer Complexes by FAD and Flavoproteins. Angew. Chem. 2018, 130, 3197–3201. [Google Scholar] [CrossRef]
- Kuzichkina, E.O.; Shilova, O.N.; Deyev, S.M.; Petrov, R.V. The Application of Recombinant Phototoxins 4D5scFv-MiniSOG and DARPin-MiniSOG to Study the HER2 Receptor Internalization. Dokl. Biochem. Biophys. 2018, 482, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Gurruchaga-Pereda, J.; Martínez-Martínez, V.; Rezabal, E.; Lopez, X.; Garino, C.; Mancin, F.; Cortajarena, A.L.; Salassa, L. Flavin Bioorthogonal Photocatalysis toward Platinum Substrates. ACS Catal. 2020, 10, 187–196. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, W.; Zeng, K.; Ao, Y.; Wang, M.; Yu, Z.; Qi, F.; Yu, W.; Mao, H.; Tao, L.; et al. Upconversion Nanoparticles–Based Multiplex Protein Activation to Neuron Ablation for Locomotion Regulation. Small 2020, 16, 1906797. [Google Scholar] [CrossRef]
- Diaz, D.; Vidal, X.; Sunna, A.; Care, A. Bioengineering a Light-Responsive Encapsulin Nanoreactor: A Potential Tool for in Vitro Photodynamic Therapy. ACS Appl. Mater. Interfaces 2021, 13, 7977–7986. [Google Scholar] [CrossRef]
- van de Steen, A.; Khalife, R.; Colant, N.; Mustafa Khan, H.; Deveikis, M.; Charalambous, S.; Robinson, C.M.; Dabas, R.; Esteban Serna, S.; Catana, D.A.; et al. Bioengineering Bacterial Encapsulin Nanocompartments as Targeted Drug Delivery System. Synth. Syst. Biotechnol. 2021, 6, 231–241. [Google Scholar] [CrossRef]
- Yang, W.; Yoon, Y.; Lee, Y.; Oh, H.; Choi, J.; Shin, S.; Lee, S.; Lee, H.; Lee, Y.; Seo, J. Photosensitizer-Peptoid Conjugates for Photoinactivation of Gram-Negative Bacteria: Structure-Activity Relationship and Mechanistic Studies. Org. Biomol. Chem. 2021, 19, 6546–6557. [Google Scholar] [CrossRef]
- Soukos, N.S.; Ximenez-Fyvie, L.A.; Hamblin, M.R.; Socransky, S.S.; Hasan, T. Targeted Antimicrobial Photochemotherapy. Antimicrob. Agents Chemother. 1998, 42, 2595–2601. [Google Scholar] [CrossRef]
- Liu, F.; Soh Yan Ni, A.; Lim, Y.; Mohanram, H.; Bhattacharjya, S.; Xing, B. Lipopolysaccharide Neutralizing Peptide–Porphyrin Conjugates for Effective Photoinactivation and Intracellular Imaging of Gram-Negative Bacteria Strains. Bioconjugate Chem. 2012, 23, 1639–1647. [Google Scholar] [CrossRef]
- Dosselli, R.; Ruiz-González, R.; Moret, F.; Agnolon, V.; Compagnin, C.; Mognato, M.; Sella, V.; Agut, M.; Nonell, S.; Gobbo, M.; et al. Synthesis, Spectroscopic, and Photophysical Characterization and Photosensitizing Activity toward Prokaryotic and Eukaryotic Cells of Porphyrin-Magainin and -Buforin Conjugates. J. Med. Chem. 2014, 57, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.A.; Muthukrishnan, N.; Pellois, J.P. Photoinactivation of Gram Positive and Gram Negative Bacteria with the Antimicrobial Peptide (KLAKLAK)2 Conjugated to the Hydrophilic Photosensitizer Eosin y. Bioconjugate Chem. 2013, 24, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.A.; Ellis, E.A.; Kim, H.; Muthukrishnan, N.; Snavely, T.; Pellois, J.P. Photoinduced Membrane Damage of E. Coli and S. Aureus by the Photosensitizer-Antimicrobial Peptide Conjugate Eosin-(KLAKLAK)2. PLoS ONE 2014, 9, e91220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.N.; Wu, W.; Zhang, C.; Wang, Q.Y.; Zhuang, Z.N.; Cheng, H.; Zhang, X.Z. A Versatile Bacterial Membrane-Binding Chimeric Peptide with Enhanced Photodynamic Antimicrobial Activity. J. Mater. Chem. B 2019, 7, 1087–1095. [Google Scholar] [CrossRef]
- Lei, X.; Qiu, L.; Lan, M.; Du, X.; Zhou, S.; Cui, P.; Zheng, R.; Jiang, P.; Wang, J.; Xia, J. Antibacterial Photodynamic Peptides for Staphylococcal Skin Infection. Biomater. Sci. 2020, 8, 6695–6702. [Google Scholar] [CrossRef]
- le Guern, F.; Sol, V.; Ouk, C.; Arnoux, P.; Frochot, C.; Ouk, T.S. Enhanced Photobactericidal and Targeting Properties of a Cationic Porphyrin Following the Attachment of Polymyxin B. Bioconjugate Chem. 2017, 28, 2493–2506. [Google Scholar] [CrossRef]
- le Guern, F.; Ouk, T.S.; Grenier, K.; Joly, N.; Lequart, V.; Sol, V. Enhancement of Photobactericidal Activity of Chlorin-E6-Cellulose Nanocrystals by Covalent Attachment of Polymyxin B. J. Mater. Chem. B 2017, 5, 6953–6962. [Google Scholar] [CrossRef]
- Chu, J.C.H.; Yang, C.; Fong, W.-P.; Wong, C.T.T.; Ng, D.K.P. Facile One-Pot Synthesis of Cyclic Peptide-Conjugated Photosensitisers for Targeted Photodynamic Therapy. Chem. Commun. 2020, 56, 11941–11944. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, J.; Jing, Q.; Chen, Z.; Ullah, A.; Jiang, L.; Zheng, K.; Yuan, C.; Huang, M. Development of a Potent Antimicrobial Peptide With Photodynamic Activity. Front. Microbiol. 2021, 12, 1013. [Google Scholar] [CrossRef]
- Pierce, S.; Jennings, M.P.; Juliano, S.A.; Angeles-Boza, A.M. Peptide-Ruthenium Conjugate as an Efficient Photosensitizer for the Inactivation of Multidrug-Resistant Bacteria. Inorg. Chem. 2020, 59, 14866–14870. [Google Scholar] [CrossRef]
- Diogo, P.; Faustino, M.F.A.; Neves, G.M.P.M.S.; Palma, P.J.; Baptista, I.P.; Gonçalves, T.; Santos, J.M. An Insight into Advanced Approaches for Photosensitizer Optimization in Endodontics—A Critical Review. J. Funct. Biomater. 2019, 10, 44. [Google Scholar] [CrossRef]
- de Freitas, L.M.; Lorenzón, E.N.; Santos-Filho, N.A.; Zago, L.H.D.P.; Uliana, M.P.; de Oliveira, K.T.; Cilli, E.M.; Fontana, C.R. Antimicrobial Photodynamic Therapy Enhanced by the Peptide Aurein. Sci. Rep. 2018, 8, 4212. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, W.; Hu, P.; Wang, D.; Lin, F.; Xue, J.; Chen, Z.; Iqbal, Z.; Huang, M. Dual Antimicrobial Actions on Modified Fabric Leads to Inactivation of Drug-Resistant Bacteria. Dye. Pigment. 2017, 140, 236–243. [Google Scholar] [CrossRef]
- Dosselli, R.; Gobbo, M.; Bolognini, E.; Campestrini, S.; Reddi, E. Porphyrin-Apidaecin Conjugate as a New Broad Spectrum Antibacterial Agent. ACS Med. Chem. Lett. 2010, 1, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Ucuncu, M.; Mills, B.; Duncan, S.; Staderini, M.; Dhaliwal, K.; Bradley, M. Polymyxin-Based Photosensitizer for the Potent and Selective Killing of Gram-Negative Bacteria. Chem. Commun. 2020, 56, 3757–3760. [Google Scholar] [CrossRef]
- le Guern, F.; Ouk, T.S.; Ouk, C.; Vanderesse, R.; Champavier, Y.; Pinault, E.; Sol, V. Lysine Analogue of Polymyxin B as a Significant Opportunity for Photodynamic Antimicrobial Chemotherapy. ACS Med. Chem. Lett. 2018, 9, 11–16. [Google Scholar] [CrossRef]
- Berg, K.; Selbo, P.K.; Prasmickaite, L.; Tjelle, T.E.; Sandvig, K.; Moan, J.; Gaudernack, G.; Fodstad, O.; Kjølsrud, S.; Anholt, H.; et al. Photochemical Internalization: A Novel Technology for Delivery of Macromolecules into Cytosol. Cancer Res. 1999, 59, 1180–1183. [Google Scholar]
- Watanabe, K.; Fujiwara, H.; Kitamatsu, M.; Ohtsuki, T. Photoinduced Apoptosis Using a Peptide Carrying a Photosensitizer. Bioorg. Med. Chem. Lett. 2016, 26, 3115–3118. [Google Scholar] [CrossRef]
- Jerjes, W.; Theodossiou, T.A.; Hirschberg, H.; Høgset, A.; Weyergang, A.; Selbo, P.K.; Hamdoon, Z.; Hopper, C.; Berg, K. Photochemical Internalization for Intracellular Drug Delivery. From Basic Mechanisms to Clinical Research. J. Clin. Med. 2020, 9, 528. [Google Scholar] [CrossRef]
- Muthukrishnan, N.; Johnson, G.A.; Erazo-Oliveras, A.; Pellois, J.-P. Synergy Between Cell-Penetrating Peptides and Singlet Oxygen Generators Leads to Efficient Photolysis of Membranes. Photochem. Photobiol. 2013, 89, 625–630. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yaghini, E.; Dondi, R.; Tewari, K.M.; Loizidou, M.; Eggleston, I.M.; MacRobert, A.J. Endolysosomal Targeting of a Clinical Chlorin Photosensitiser for Light-Triggered Delivery of Nano-Sized Medicines. Sci. Rep. 2017, 7, 6059. [Google Scholar] [CrossRef] [PubMed]
- Soe, T.H.; Watanabe, K.; Ohtsuki, T. Photoinduced Endosomal Escape Mechanism: A View from Photochemical Internalization Mediated by CPP-Photosensitizer Conjugates. Molecules 2021, 26, 36. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, Y.; Kadono, M.; Okazaki, S.; Nishimura, A.; Kitamatsu, M.; Watanabe, K.; Ohtsuki, T. Endosomal Escape of Peptide-Photosensitizer Conjugates Is Affected by Amino Acid Sequences near the Photosensitizer. Bioconjugate Chem. 2020, 31, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Tirand, L.; Frochot, C.; Vanderesse, R.; Thomas, N.; Trinquet, E.; Pinel, S.; Viriot, M.L.; Guillemin, F.; Barberi-Heyob, M. A Peptide Competing with VEGF165 Binding on Neuropilin-1 Mediates Targeting of a Chlorin-Type Photosensitizer and Potentiates Its Photodynamic Activity in Human Endothelial Cells. J. Control Release 2006, 111, 153–164. [Google Scholar] [CrossRef]
- Tirand, L.; Thomas, N.; Dodeller, M.; Dumas, D.; Frochot, C.; Maunit, B.; Guillemin, F.; Barberi-Heyob, M. Metabolic Profile of a Peptide-Conjugated Chlorin-Type Photosensitizer Targeting Neuropilin-1: An in Vivo and in Vitro Study. Drug Metab. Dispos. 2007, 35, 806–813. [Google Scholar] [CrossRef]
- Thomas, N.; Tirand, L.; Chatelut, E.; Plenat, F.; Frochot, C.; Dodeller, M.; Guillemin, F.; Barberi-Heyob, M. Tissue Distribution and Pharmacokinetics of an ATWLPPR-Conjugated Chlorin-Type Photosensitizer Targeting Neuropilin-1 in Glioma-Bearing Nude Mice. Photochem. Photobiol. Sci. 2008, 7, 433–441. [Google Scholar] [CrossRef]
- Kamarulzaman, E.; Gazzali, A.; Acherar, S.; Frochot, C.; Barberi-Heyob, M.; Boura, C.; Chaimbault, P.; Sibille, E.; Wahab, H.; Vanderesse, R. New Peptide-Conjugated Chlorin-Type Photosensitizer Targeting Neuropilin-1 for Anti-Vascular Targeted Photodynamic Therapy. Int. J. Mol. Sci. 2015, 16, 24059–24080. [Google Scholar] [CrossRef]
- Fontenot, K.R.; Ongarora, B.G.; LeBlanc, L.E.; Zhou, Z.; Jois, S.D.; Vicente, M.G.H. Targeting of the Epidermal Growth Factor Receptor with Mesoporphyrin IX-Peptide Conjugates. J. Porphyr. Phthalocyanines 2016, 20, 352–366. [Google Scholar] [CrossRef]
- Yu, L.; Wang, Q.; Wong, R.C.-H.; Zhao, S.; Ng, D.K.P.; Lo, P.-C. Synthesis and Biological Evaluation of Phthalocyanine-Peptide Conjugate for EGFR-Targeted Photodynamic Therapy and Bioimaging. Dye. Pigment. 2019, 163, 197–203. [Google Scholar] [CrossRef]
- Pethő, L.; Murányi, J.; Pénzes, K.; Gurbi, B.; Brauswetter, D.; Halmos, G.; Csík, G.; Mező, G. Suitability of GnRH Receptors for Targeted Photodynamic Therapy in Head and Neck Cancers. Int. J. Mol. Sci. 2019, 20, 5027. [Google Scholar] [CrossRef]
- Ying, M.; Shen, Q.; Zhan, C.; Wei, X.; Gao, J.; Xie, C.; Yao, B.; Lu, W. A Stabilized Peptide Ligand for Multifunctional Glioma Targeted Drug Delivery. J. Control Release 2016, 243, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, L.; Zhang, Y.; Zhou, S.; Cai, H.-H.; Li, T.; Jin, H.; Cai, J.; Zhou, H.; Pi, J. GE11 Peptide Conjugated Liposomes for EGFR-Targeted and Chemophotothermal Combined Anticancer Therapy. Bioinorg. Chem. Appl. 2021, 2021, 5534870. [Google Scholar] [CrossRef] [PubMed]
- Dhaini, B.; Kenzhebayeva, B.; Ben-Mihoub, A.; Gries, M.; Acherar, S.; Baros, F.; Thomas, N.; Daouk, J.; Schohn, H.; Hamieh, T.; et al. Peptide-Conjugated Nanoparticles for Targeted Photodynamic Therapy. Nanophotonics 2021, 10, 3089–3134. [Google Scholar] [CrossRef]
- Verwilst, P.; David, C.C.; Leen, V.; Hofkens, J.; de Witte, P.A.M.; de Borggraeve, W.M. Synthesis and in Vitro Evaluation of a PDT Active BODIPY-NLS Conjugate. Bioorg. Med. Chem. Lett. 2013, 23, 3204–3207. [Google Scholar] [CrossRef]
- Chen, D.; Brahimi, F.; Angell, Y.; Li, Y.C.; Moscowicz, J.; Saragovi, H.U.; Burgess, K. Bivalent Peptidomimetic Ligands of TrkC Are Biased Agonists and Selectively Induce Neuritogenesis or Potentiate Neurotrophin-3 Trophic Signals. ACS Chem. Biol. 2009, 4, 769–781. [Google Scholar] [CrossRef]
- Kue, C.S.; Kamkaew, A.; Voon, S.H.; Kiew, L.V.; Chung, L.Y.; Burgess, K.; Lee, H.B. Tropomyosin Receptor Kinase C Targeted Delivery of a Peptidomimetic Ligand-Photosensitizer Conjugate Induces Antitumor Immune Responses Following Photodynamic Therapy. Sci. Rep. 2016, 6, 37209. [Google Scholar] [CrossRef]
- Ng, S.Y.; Kamkaew, A.; Fu, N.; Kue, C.S.; Chung, L.Y.; Kiew, L.V.; Wittayakun, J.; Burgess, K.; Lee, H.B. Active Targeted Ligand-Aza-BODIPY Conjugate for near-Infrared Photodynamic Therapy in Melanoma. Int. J. Pharm. 2020, 579, 119189. [Google Scholar] [CrossRef]
- Mew, D.; Wat, C.K.; Towers, G.H.; Levy, J.G. Photoimmunotherapy: Treatment of Animal Tumors with Tumor-Specific Monoclonal Antibody-Hematoporphyrin Conjugates. 6. J. Immunol. 1983, 130, 1473–1477. [Google Scholar] [PubMed]
- Mew, D.; Lum, V.; Wat, C.-K.; Towers, G.H.N.; Sun, C.-H.C.; Walter, R.J.; Wright, W.; Berns, M.W.; Levy, J.G. Ability of Specific Monoclonal Antibodies and Conventional Antisera Conjugated to Hematoporphyrin to Label and Kill Selected Cell Lines Subsequent to Light Activation. Cancer Res. 1985, 45, 8. [Google Scholar]
- Hasan, T.; Lin, A.; Yarmush, D.; Oseroff, A.; Yarmush, M. Monoclonal Antibody-Chromophore Conjugates as Selective Phototoxins. J. Control Release 1989, 10, 107–117. [Google Scholar] [CrossRef]
- Jiang, F.N.; Jiang, S.; Liu, D.; Richter, A.; Levy, J.G. Development of Technology for Linking Photosensitizers to a Model Monoclonal Antibody. J. Immunol. Methods 1990, 134, 139–149. [Google Scholar] [CrossRef]
- Poiroux, G.; Barre, A.; Rouge, P.; Benoist, H. Targeting Glycosylation Aberrations to Improve the Efficiency of Cancer Phototherapy. Curr. Cancer Drug Targets 2019, 19, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.R.G.; Fernandes, R.; Sarmento, B.; Pereira, P.M.R.; Tome, J.P.C. Photoimmunoconjugates: Novel Synthetic Strategies to Target and Treat Cancer by Photodynamic Therapy. Org. Biomol. Chem. 2019, 17, 2579–2593. [Google Scholar] [CrossRef] [PubMed]
- Sandland, J.; Boyle, R.W. Photosensitizer Antibody-Drug Conjugates: Past, Present, and Future. Bioconjugate Chem. 2019, 30, 975–993. [Google Scholar] [CrossRef]
- Wang, X.; Luo, D.; Basilion, J.P. Photodynamic Therapy: Targeting Cancer Biomarkers for the Treatment of Cancers. Cancers 2021, 13, 2992. [Google Scholar] [CrossRef]
- Monaco, H.; Yokomizo, S.; Choi, H.S.; Kashiwagi, S. Quickly Evolving Near-infrared Photoimmunotherapy Provides Multifaceted Approach to Modern Cancer Treatment. VIEW 2021, 20200110. [Google Scholar] [CrossRef]
- Pereira, P.M.R.; Korsak, B.; Sarmento, B.; Schneider, R.J.; Fernandes, R.; Tomé, J.P.C. Antibodies Armed with Photosensitizers: From Chemical Synthesis to Photobiological Applications. Org. Biomol. Chem. 2015, 13, 2518–2529. [Google Scholar] [CrossRef]
- Mehraban, N.; Freeman, H.S. Developments in PDT Sensitizers for Increased Selectivity and Singlet Oxygen Production. Materials 2015, 8, 4421–4456. [Google Scholar] [CrossRef]
- Watanabe, R.; Hanaoka, H.; Sato, K.; Nagaya, T.; Harada, T.; Mitsunaga, M.; Kim, I.; Paik, C.H.; Wu, A.M.; Choyke, P.L.; et al. Photoimmunotherapy Targeting Prostate-Specific Membrane Antigen: Are Antibody Fragments as Effective as Antibodies? J. Nucl. Med. 2015, 56, 140–144. [Google Scholar] [CrossRef]
- Wei, D.; Tao, Z.; Shi, Q.; Wang, L.; Liu, L.; She, T.; Yi, Q.; Wen, X.; Liu, L.; Li, S.; et al. Selective Photokilling of Colorectal Tumors by Near-Infrared Photoimmunotherapy with a GPA33-Targeted Single-Chain Antibody Variable Fragment Conjugate. Mol. Pharm. 2020, 17, 2508–2517. [Google Scholar] [CrossRef]
- Zhen, Z.; Tang, W.; Wang, M.; Zhou, S.; Wang, H.; Wu, Z.; Hao, Z.; Li, Z.; Liu, L.; Xie, J. Protein Nanocage Mediated Fibroblast-Activation Protein Targeted Photoimmunotherapy to Enhance Cytotoxic T Cell Infiltration and Tumor Control. Nano Lett. 2017, 17, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Heukers, R.; van Bergen en Henegouwen, P.M.P.; Oliveira, S. Nanobody-Photosensitizer Conjugates for Targeted Photodynamic Therapy. Nanomed.-Nanotechnol. Biol. Med. 2014, 10, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- van Driel, P.B.A.A.; Boonstra, M.C.; Slooter, M.D.; Heukers, R.; Stammes, M.A.; Snoeks, T.J.A.; de Bruijn, H.S.; van Diest, P.J.; Vahrmeijer, A.L.; van Bergen en Henegouwen, P.M.P.; et al. EGFR Targeted Nanobody-Photosensitizer Conjugates for Photodynamic Therapy in a Pre-Clinical Model of Head and Neck Cancer. J. Control Release 2016, 229, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Spelier, S.; Hernandez, I.B.; de Bree, R.; Willems, S.M.; Clevers, H.; Oliveira, S. Patient-Derived Head and Neck Cancer Organoids Recapitulate EGFR Expression Levels of Respective Tissues and Are Responsive to EGFR-Targeted Photodynamic Therapy. J. Clin. Med. 2019, 8, 1880. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, H.S.; Mashayekhi, V.; Schreurs, T.J.L.; van Driel, P.B.A.A.; Strijkers, G.J.; van Diest, P.J.; Lowik, C.W.G.M.; Seynhaeve, A.L.B.; ten Hagen, T.L.M.; Prompers, J.J.; et al. Acute Cellular and Vascular Responses to Photodynamic Therapy Using EGFR-Targeted Nanobody-Photosensitizer Conjugates Studied with Intravital Optical Imaging and Magnetic Resonance Imaging. Theranostics 2020, 10, 2436–2452. [Google Scholar] [CrossRef] [PubMed]
- Beltrán Hernández, I.; Grinwis, G.C.M.; di Maggio, A.; van Bergen en Henegouwen, P.M.P.; Hennink, W.E.; Teske, E.; Hesselink, J.W.; van Nimwegen, S.A.; Mol, J.A.; Oliveira, S. Nanobody-Targeted Photodynamic Therapy for the Treatment of Feline Oral Carcinoma: A Step towards Translation to the Veterinary Clinic. Nanophotonics 2021, 10, 3075–3087. [Google Scholar] [CrossRef]
- Xiong, T.; Peng, Q.; Chen, Y.; Li, M.; Du, J.; Fan, J.; Jia, L.; Peng, X. A Novel Nanobody-Photosensitizer Conjugate for Hypoxia Resistant Photoimmunotherapy. Adv. Funct. Mater. 2021, 31, 2103629. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, S.; Na, H.K.; Min, D.H. MicroRNA-Responsive Drug Release System for Selective Fluorescence Imaging and Photodynamic Therapy In Vivo. Adv. Healthc. Mater. 2016, 5, 2386–2395. [Google Scholar] [CrossRef]
- Kruspe, S.; Meyer, C.; Hahn, U. Chlorin E6 Conjugated Interleukin-6 Receptor Aptamers Selectively Kill Target Cells upon Irradiation. Mol. Ther.-Nucleic Acids 2014, 2, e143. [Google Scholar] [CrossRef]
- Shi, J.; Wang, D.; Ma, Y.; Liu, J.; Li, Y.; Reza, R.; Zhang, Z.; Liu, J.; Zhang, K.; Shi, J.; et al. Photoactivated Self-Disassembly of Multifunctional DNA Nanoflower Enables Amplified Autophagy Suppression for Low-Dose Photodynamic Therapy. Small 2021, 17, 2104722. [Google Scholar] [CrossRef]
- Chang, R.; Nikoloudakis, E.; Zou, Q.; Mitraki, A.; Coutsolelos, A.G.; Yan, X. Supramolecular Nanodrugs Constructed by Self-Assembly of Peptide Nucleic Acid-Photosensitizer Conjugates for Photodynamic Therapy. ACS Appl. Bio Mater. 2020, 3, 2–9. [Google Scholar] [CrossRef] [PubMed]
- de Laet, N.; Madder, A. Synthesis and Evaluation of Methylene Blue Oligonucleotide Conjugates for DNA Interstrand Cross-Linking. J. Photochem. Photobiol. A Chem. 2016, 318, 64–70. [Google Scholar] [CrossRef][Green Version]
- Liu, T.W.; Akens, M.K.; Chen, J.; Wise-Milestone, L.; Wilson, B.C.; Zheng, G. Imaging of Specific Activation of Photodynamic Molecular Beacons in Breast Cancer Vertebral Metastases. Bioconjugate Chem. 2011, 22, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lu, C.; Chen, Q.; Xing, D. DNA Duplex-Based Photodynamic Molecular Beacon for Targeted Killing of Retinoblastoma Cell. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6011–6019. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zheng, G.; Chen, J.; Stefflova, K.; Jarvi, M.; Li, H.; Wilson, B.C. Photodynamic Molecular Beacon as an Activatable Photosensitizer Based on Protease-Controlled Singlet Oxygen Quenching and Activation. Proc. Natl. Acad. Sci. USA 2007, 104, 8989–8994. [Google Scholar] [CrossRef]
- Lo, P.C.; Chen, J.; Stefflova, K.; Warren, M.S.; Navab, R.; Bandarchi, B.; Mullins, S.; Tsao, M.; Cheng, J.D.; Zheng, G. Photodynamic Molecular Beacon Triggered by Fibroblast Activation Protein on Cancer-Associated Fibroblasts for Diagnosis and Treatment of Epithelial Cancers. J. Med. Chem. 2009, 52, 358–368. [Google Scholar] [CrossRef]
- Wu, D.; Song, G.; Li, Z.; Zhang, T.; Wei, W.; Chen, M.; He, X.; Ma, N. A Two-Dimensional Molecular Beacon for MRNA-Activated Intelligent Cancer Theranostics. Chem. Sci. 2015, 6, 3839–3844. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Chen, W.; Zhang, Y.; Zhang, X.; Liu, Y.; Ju, H. A Near-Infrared Photo-Switched MicroRNA Amplifier for Precise Photodynamic Therapy of Early-Stage Cancers. Angew. Chem. Int. Ed. 2020, 59, 21454–21459. [Google Scholar] [CrossRef]
- Liu, J.; Ding, G.; Chen, S.; Xue, C.; Chen, M.; Wu, X.; Yuan, Q.; Zheng, J.; Yang, R. Multifunctional Programmable DNA Nanotrain for Activatable Hypoxia Imaging and Mitochondrion-Targeted Enhanced Photodynamic Therapy. ACS Appl. Mater. Interfaces 2021, 13, 9681–9690. [Google Scholar] [CrossRef]
- Fudickar, W.; Bauch, M.; Ihmels, H.; Linker, T. DNA-Triggered Enhancement of Singlet Oxygen Production by Pyridinium Alkynylanthracenes. Chem. A Eur. J. 2021, 27, 13591–13604. [Google Scholar] [CrossRef]
- Fudickar, W.; Roder, P.; Listek, M.; Hanack, K.; Linker, T. Pyridinium Alkynylanthracenes as Sensitizers for Photodynamic Therapy. Photochem. Photobiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Xodo, L.E.; Cogoi, S.; Rapozzi, V. Photosensitizers Binding to Nucleic Acids as Anticancer Agents. Future Med. Chem. 2010, 8, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Ferino, A.; Nicoletto, G.; D’este, F.; Zorzet, S.; Lago, S.; Richter, S.N.; Tikhomirov, A.; Shchekotikhin, A.; Xodo, L.E. Photodynamic Therapy for Ras-Driven Cancers: Targeting G-Quadruplex RNA Structures with Bifunctional Alkyl-Modified Porphyrins. J. Med. Chem. 2020, 63, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Caterino, M.; D’Aria, F.; Kustov, A.V.; Belykh, D.V.; Khudyaeva, I.S.; Starseva, O.M.; Berezin, D.B.; Pylina, Y.I.; Usacheva, T.; Amato, J.; et al. Selective Binding of a Bioactive Porphyrin-Based Photosensitizer to the G-Quadruplex from the KRAS Oncogene Promoter. Int. J. Biol. Macromol. 2020, 145, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, K.; Urano, R.; Kinoshita, N.; Kuwamoto, S.; Torii, T.; Hashimoto, Y.; Taniguchi, S.; Tsuruta, M.; Miyoshi, D. Photosensitizers Based on G-Quadruplex Ligand for Cancer Photodynamic Therapy. Genes 2020, 11, 1340. [Google Scholar] [CrossRef] [PubMed]
- Cadoni, E.; de Paepe, L.; Manicardi, A.; Madder, A. Beyond Small Molecules: Targeting G-Quadruplex Structures with Oligonucleotides and Their Analogues. Nucleic Acids Res. 2021, 49, 6638–6659. [Google Scholar] [CrossRef]

| Classes | Examples |
|---|---|
| Organic dyes and aromatic hydrocarbons | Rose Bengal (RB), methylene blue (MB), quinones |
| Tetrapyrroles | Porphyrins, Phtalocyanines |
| Transition metal complexes | Metal complexes of Ruthenium (Ru(II) tris-bipyridine), Pt- and Pd(II) mixed ligands |
| Semiconductors | TiO2, ZnO |
| Inorganic nanoparticles | (CdSe, CdSe/ZnS) quantum dots, porous silica nanocrystals, gold, silver, platinum and nichel nanoconstructs (nanoparticles, nanoclusters, nanorods) |
| Antimicrobial Peptides | PS | References |
|---|---|---|
| KLA(KLAKLAK)2 | Eosin Y | [241,242,243] |
| YVLWKRKRKFCFI-NH2 | protophophyrin IX | [244] |
| (GKRWWKWWRR)2KGGK | Chlorin e6 | [244] |
| Polymyxin B | Chlorin e6, Porphyrins | [245,246] |
| GGGKKKKKRWRWRW | Phthalocyanine | [247,248] |
| Cyclic Bactenicin | ||
| Buforin II | [Ru(bpy)3]2+ | [249] |
| Poly-L-Lysine | Chlorine e6, BOHTMPn, GlamTMPn | [238,250] |
| Aurein 1.2 | Methylene blue, Chlorin e6, Curcumin | [251] |
| epsilon-poly-Lysine | CPZ | [252] |
| Peptoid | TMPyp | [237] |
| Apidaecin | Porphyrins | [240,245,246,248,253,254,255] |
| WRF | ||
| RWRW | ||
| FRWWRR | ||
| Polymyxin Magainin Buforin |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aerssens, D.; Cadoni, E.; Tack, L.; Madder, A. A Photosensitized Singlet Oxygen (1O2) Toolbox for Bio-Organic Applications: Tailoring 1O2 Generation for DNA and Protein Labelling, Targeting and Biosensing. Molecules 2022, 27, 778. https://doi.org/10.3390/molecules27030778
Aerssens D, Cadoni E, Tack L, Madder A. A Photosensitized Singlet Oxygen (1O2) Toolbox for Bio-Organic Applications: Tailoring 1O2 Generation for DNA and Protein Labelling, Targeting and Biosensing. Molecules. 2022; 27(3):778. https://doi.org/10.3390/molecules27030778
Chicago/Turabian StyleAerssens, Dorien, Enrico Cadoni, Laure Tack, and Annemieke Madder. 2022. "A Photosensitized Singlet Oxygen (1O2) Toolbox for Bio-Organic Applications: Tailoring 1O2 Generation for DNA and Protein Labelling, Targeting and Biosensing" Molecules 27, no. 3: 778. https://doi.org/10.3390/molecules27030778
APA StyleAerssens, D., Cadoni, E., Tack, L., & Madder, A. (2022). A Photosensitized Singlet Oxygen (1O2) Toolbox for Bio-Organic Applications: Tailoring 1O2 Generation for DNA and Protein Labelling, Targeting and Biosensing. Molecules, 27(3), 778. https://doi.org/10.3390/molecules27030778






