General Method to Increase Carboxylic Acid Content on Nanodiamonds
Abstract
:1. Introduction
2. Results and Discussion
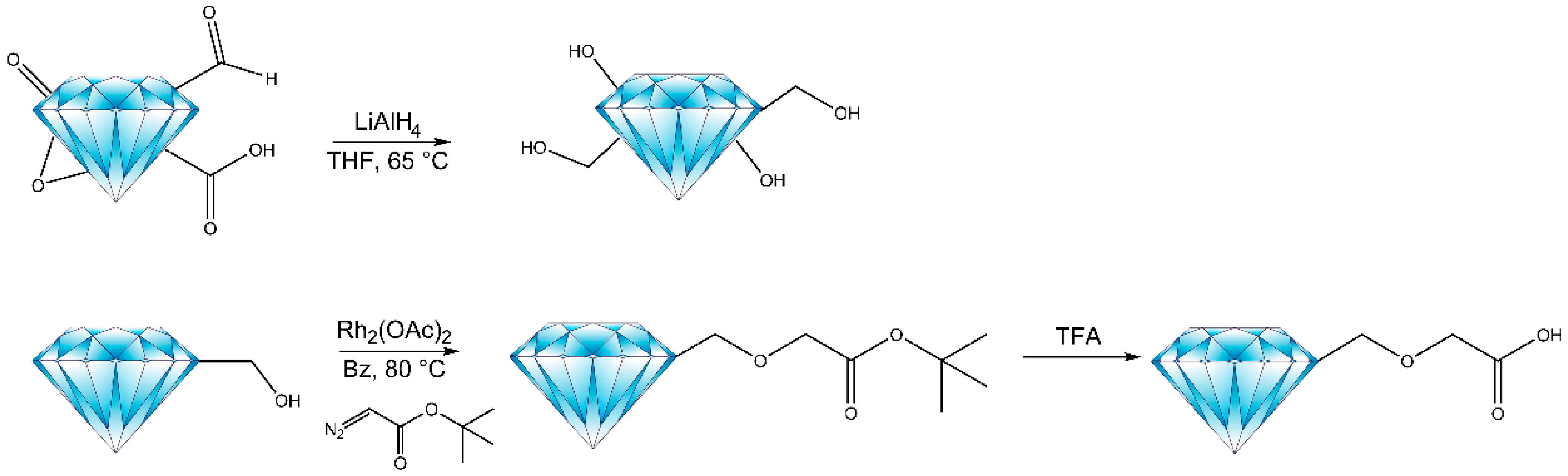
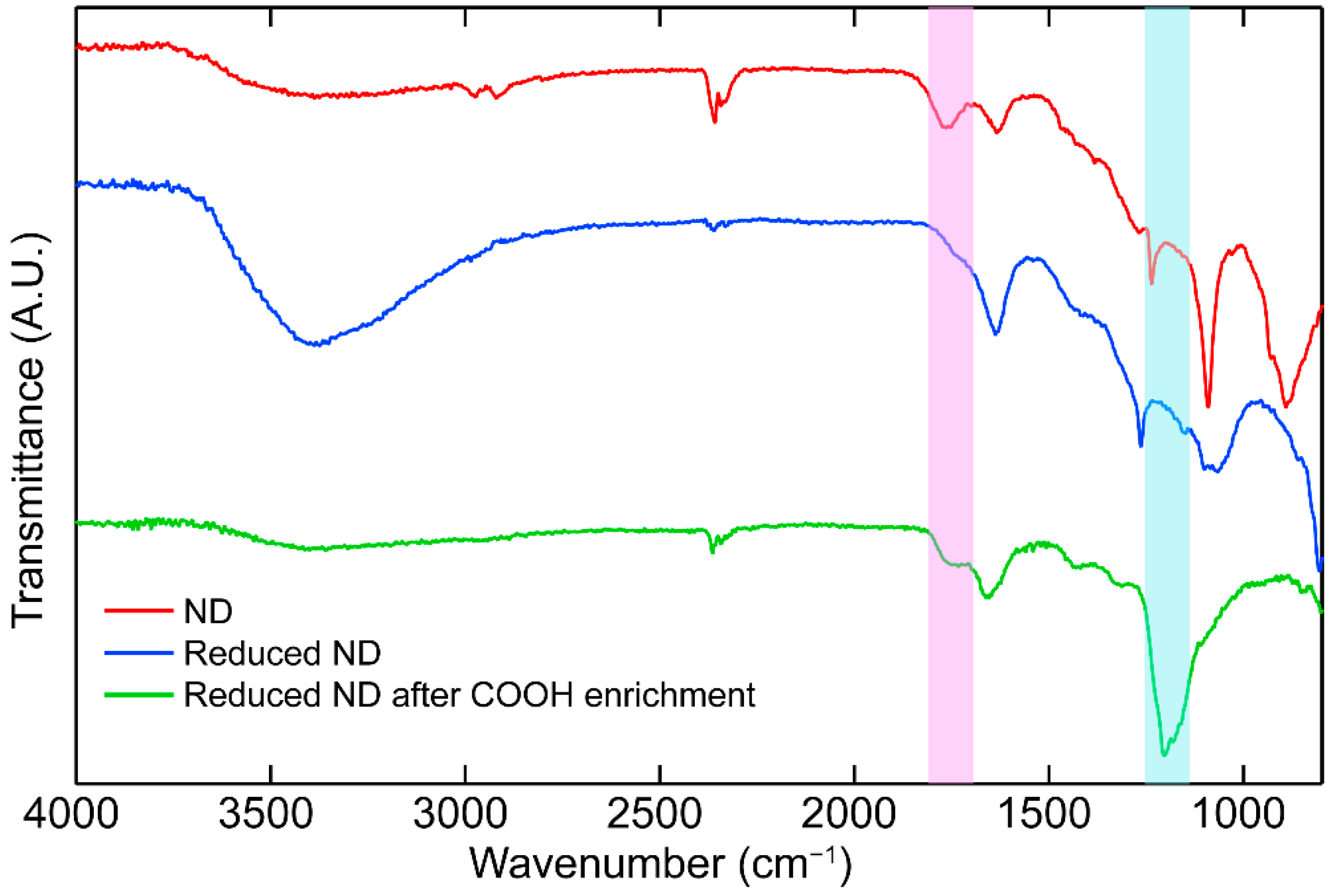
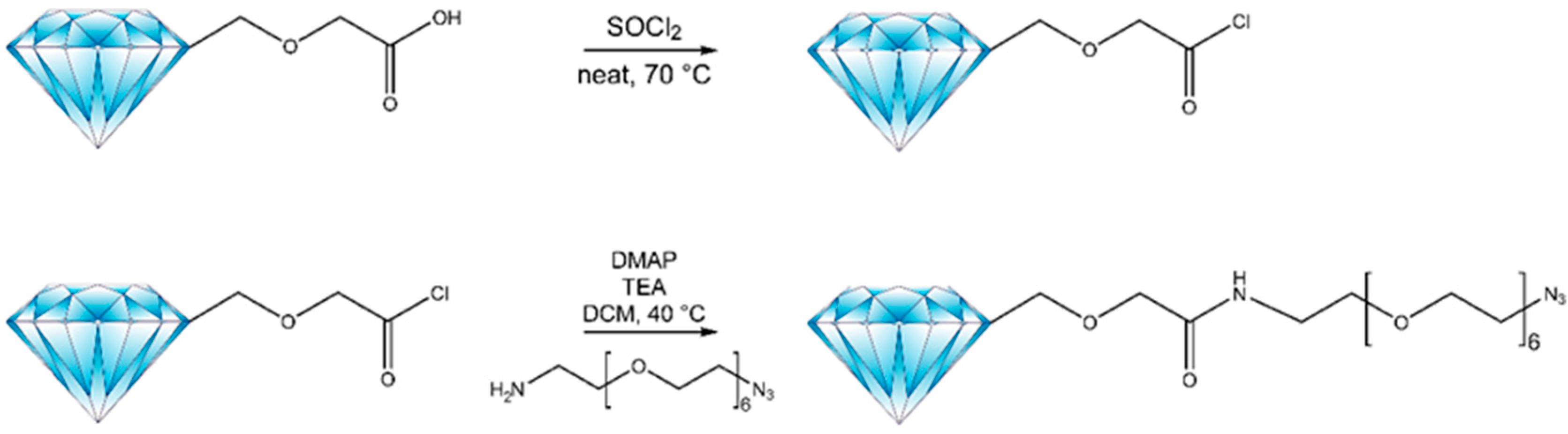
2.1. Synthetic Method and Characterization
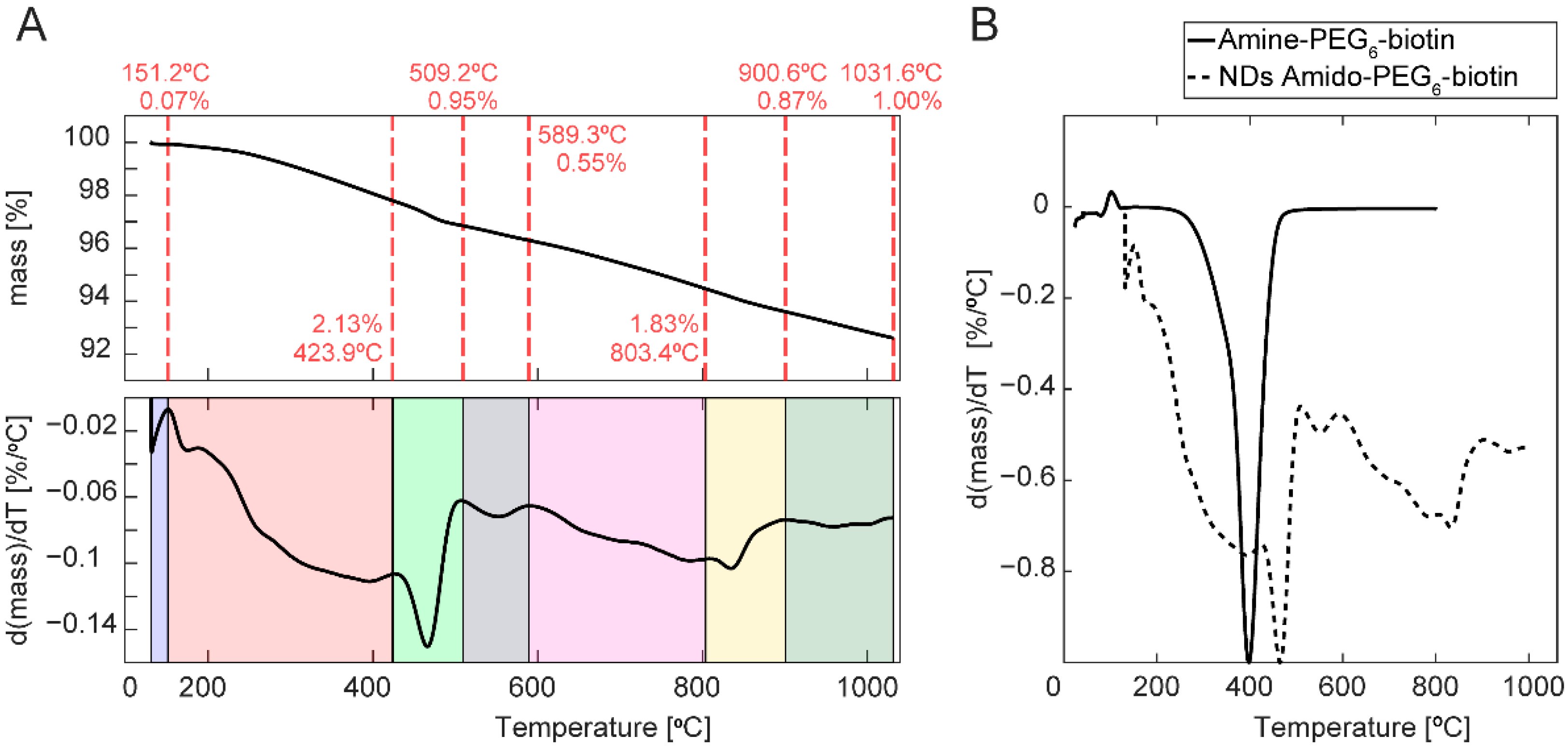
2.2. Thermogravimetric Analysis (TGA) of FND Surface Functionalization
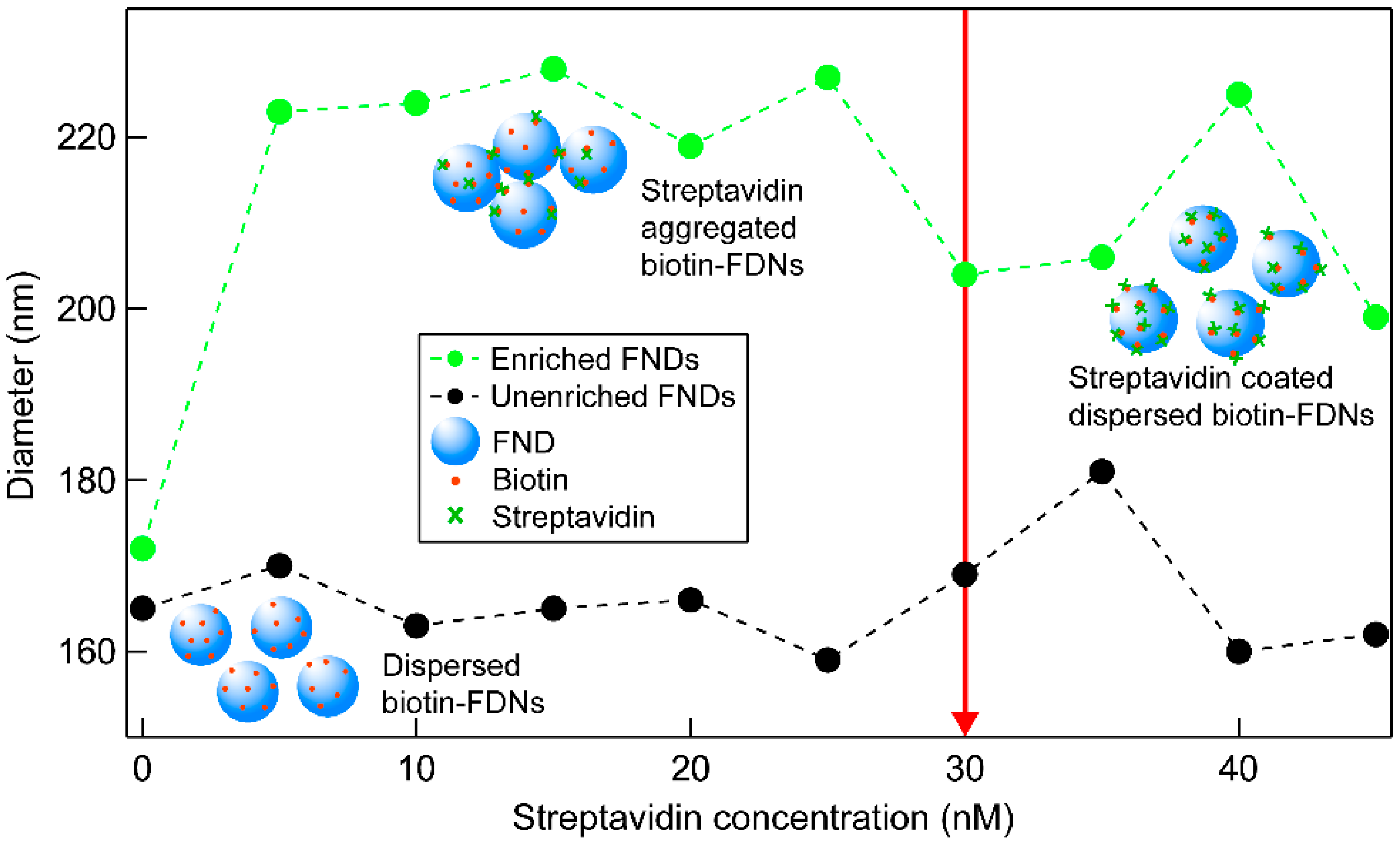
2.3. Streptavidin Biotin–FND Coupling
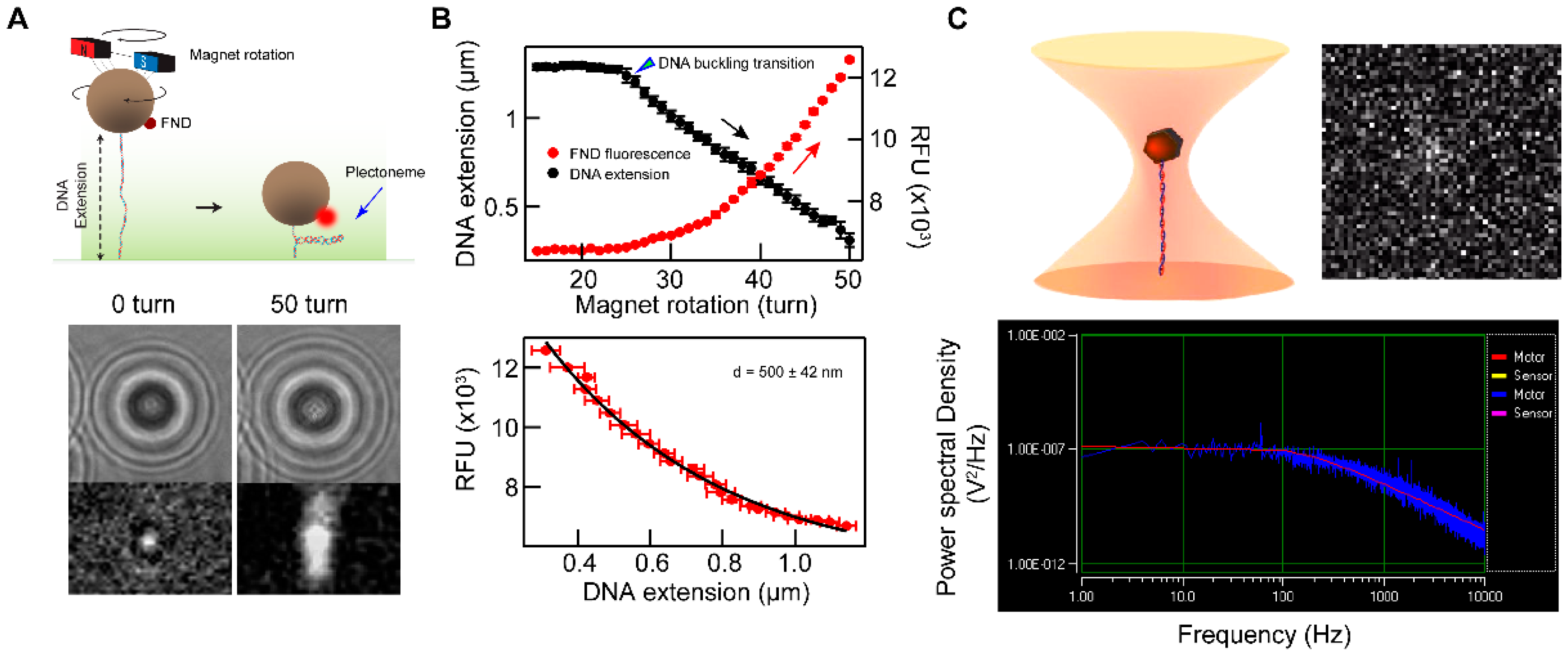
2.4. Functionalized FNDs as Single-Molecule Probes
3. Materials and Methods
3.1. Materials
3.2. General Method for Reduction of FNDs
3.3. General Method for Carboxylic Acid Enrichment on Reduced FNDs
3.4. Coupling Carboxylic Acid Enriched FNDs with Tags
3.5. Optical Trap
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Danilenko, V.V. On the history of the discovery of nanodiamond synthesis. Phys. Solid State 2004, 46, 595–599. [Google Scholar] [CrossRef]
- Ivanov, M.G.; Pavlyshko, S.V.; Ivanov, D.M.; Petrov, I.; Shenderova, O. Synergistic Compositions of Colloidal Nanodiamond as Lubricant-additive. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom. 2010, 28, 869–877. [Google Scholar] [CrossRef]
- Mochalin, V.N.; Shenderova, O.; Ho, D.; Gogotsi, Y. The properties and applications of nanodiamonds. Nat. Nanotechnol. 2012, 7, 11–23. [Google Scholar] [CrossRef]
- Shenderova, O.A.; Shames, A.I.; Nunn, N.A.; Torelli, M.D.; Vlasov, I.; Zaitsev, A. Review Article: Synthesis, properties, and applications of fluorescent diamond particles. J. Vac. Sci. Technol. B 2019, 37, 030802. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.-S.; Neuman, K.C. Surface Modification of Fluorescent Nanodiamonds for Biological Applications. Nanomaterials 2021, 11, 153. [Google Scholar] [CrossRef]
- Wrachtrup, J.; Jelezko, F. Processing quantum information in diamond. J. Phys. Condens. Matter 2006, 18, S807–S824. [Google Scholar] [CrossRef]
- Gupta, A.; Hacquebard, L.; Childress, L. Efficient signal processing for time-resolved fluorescence detection of nitrogen-vacancy spins in diamond. J. Opt. Soc. Am. B 2016, 33, B28. [Google Scholar] [CrossRef] [Green Version]
- Aharonovich, I.; Neu, E. Diamond Nanophotonics. Adv. Opt. Mater. 2014, 2, 911–928. [Google Scholar] [CrossRef]
- Aharonovich, I.; Greentree, A.D.; Prawer, S. Diamond photonics. Nat. Photonics 2011, 5, 397–405. [Google Scholar] [CrossRef]
- Kayci, M.; Mor, F.M.; Radenovic, A. Fluorescent Nanodiamonds in Biological and Biomedical Imaging. In Super-Resolution Imaging in Biomedicine; Series in Cellular and Clinical Imaging; Taylor & Francis: Boca Raton, FL, USA, 2016; Volume 263, pp. 215–232. ISBN 9781482244342. [Google Scholar]
- Claveau, S.; Bertrand, J.R.; Treussart, F. Fluorescent nanodiamond applications for cellular process sensing and cell tracking. Micromachines 2018, 9, 247. [Google Scholar] [CrossRef] [Green Version]
- Mochalin, V.N.; Pentecost, A.; Li, X.-M.; Neitzel, I.; Nelson, M.; Wei, C.; He, T.; Guo, F.; Gogotsi, Y. Adsorption of Drugs on Nanodiamond: Toward Development of a Drug Delivery Platform. Mol. Pharm. 2013, 10, 3728–3735. [Google Scholar] [CrossRef] [PubMed]
- Neugart, F.; Zappe, A.; Jelezko, F.; Tietz, C.; Boudou, J.P.; Krueger, A.; Wrachtrup, J. Dynamics of diamond nanoparticles in solution and cells. Nano Lett. 2007, 7, 3588–3591. [Google Scholar] [CrossRef] [PubMed]
- Hemelaar, S.R.; Saspaanithy, B.; L’Hommelet, S.R.M.; Martinez, F.P.P.; van der Laan, K.J.; Schirhagl, R. The Response of HeLa Cells to Fluorescent NanoDiamond Uptake. Sensors 2018, 18, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigaeva, A.; Morita, A.; Hemelaar, S.R.; Schirhagl, R. Nanodiamond uptake in colon cancer cells: The influence of direction and trypsin-EDTA treatment. Nanoscale 2019, 11, 17357–17367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.-R.; Lee, H.-Y.; Chen, K.; Chang, C.-C.; Tsai, D.-S.; Fu, C.-C.; Lim, T.-S.; Tzeng, Y.-K.; Fang, C.-Y.; Han, C.-C.; et al. Mass production and dynamic imaging of fluorescent nanodiamonds. Nat. Nanotechnol. 2008, 3, 284–288. [Google Scholar] [CrossRef]
- Boudou, J.-P.; Curmi, P.A.; Jelezko, F.; Wrachtrup, J.; Aubert, P.; Sennour, M.; Balasubramanian, G.; Reuter, R.; Thorel, A.; Gaffet, E. High yield fabrication of fluorescent nanodiamonds. Nanotechnology 2009, 20, 235602. [Google Scholar] [CrossRef]
- Yu, S.-J.; Kang, M.-W.; Chang, H.-C.; Chen, K.-M.; Yu, Y.-C. Bright Fluorescent Nanodiamonds: No Photobleaching and Low Cytotoxicity. J. Am. Chem. Soc. 2005, 127, 17604–17605. [Google Scholar] [CrossRef]
- Schrand, A.M.; Dai, L.; Schlager, J.J.; Hussain, S.M.; Osawa, E. Differential biocompatibility of carbon nanotubes and nanodiamonds. Diam. Relat. Mater. 2007, 16, 2118–2123. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Li, X.; Dolan, P.R.; Gu, M. Nonlinear absorption properties of the charge states of nitrogen-vacancy centers in nanodiamonds. Opt. Lett. 2013, 38, 1358. [Google Scholar] [CrossRef]
- Mochalin, V.N.; Gogotsi, Y. Wet Chemistry Route to Hydrophobic Blue Fluorescent Nanodiamond. J. Am. Chem. Soc. 2009, 131, 4594–4595. [Google Scholar] [CrossRef]
- Williams, O.A.; Hees, J.; Dieker, C.; Jäger, W.; Kirste, L.; Nebel, C.E. Size-Dependent Reactivity of Diamond Nanoparticles. ACS Nano 2010, 4, 4824–4830. [Google Scholar] [CrossRef] [PubMed]
- Heyer, S.; Janssen, W.; Turner, S.; Lu, Y.-G.; Yeap, W.S.; Verbeeck, J.; Haenen, K.; Krueger, A. Toward Deep Blue Nano Hope Diamonds: Heavily Boron-Doped Diamond Nanoparticles. ACS Nano 2014, 8, 5757–5764. [Google Scholar] [CrossRef]
- Wu, X.; Bruschi, M.; Waag, T.; Schweeberg, S.; Tian, Y.; Meinhardt, T.; Stigler, R.; Larsson, K.; Funk, M.; Steinmüller-Nethl, D.; et al. Functionalization of bone implants with nanodiamond particles and angiopoietin-1 to improve vascularization and bone regeneration. J. Mater. Chem. B 2017, 5, 6629–6636. [Google Scholar] [CrossRef] [PubMed]
- Happel, P.; Waag, T.; Schimke, M.; Schweeberg, S.; Muzha, A.; Fortak, K.; Heesch, D.; Klask, L.; Pilscheur, M.; Hoppe, F.; et al. Intrinsically 32 P-Labeled Diamond Nanoparticles for In Vivo Imaging and Quantification of Their Biodistribution in Chicken Embryos. Adv. Funct. Mater. 2018, 28, 1802873. [Google Scholar] [CrossRef]
- Ackermann, J.; Krueger, A. Highly sensitive and reproducible quantification of oxygenated surface groups on carbon nanomaterials. Carbon N. Y. 2020, 163, 56–62. [Google Scholar] [CrossRef]
- Shenderova, O.; Koscheev, A.; Zaripov, N.; Petrov, I.; Skryabin, Y.; Detkov, P.; Turner, S.; Tendeloo, G. Van Surface Chemistry and Properties of Ozone-Purified Detonation Nanodiamonds. J. Phys. Chem. C 2011, 115, 9827–9837. [Google Scholar] [CrossRef]
- Ackermann, J.; Krueger, A. Efficient surface functionalization of detonation nanodiamond using ozone under ambient conditions. Nanoscale 2019, 11, 8012–8019. [Google Scholar] [CrossRef]
- Krüger, A.; Liang, Y.; Jarre, G.; Stegk, J. Surface functionalisation of detonation diamond suitable for biological applications. J. Mater. Chem. 2006, 16, 2322–2328. [Google Scholar] [CrossRef]
- Bumb, A.; Sarkar, S.K.; Billington, N.; Brechbiel, M.W.; Neuman, K.C. Silica Encapsulation of Fluorescent Nanodiamonds for Colloidal Stability and Facile Surface Functionalization. J. Am. Chem. Soc. 2013, 135, 7815–7818. [Google Scholar] [CrossRef] [Green Version]
- Torelli, M.D.; Rickard, A.G.; Backer, M.V.; Filonov, D.S.; Nunn, N.A.; Kinev, A.V.; Backer, J.M.; Palmer, G.M.; Shenderova, O.A. Targeting Fluorescent Nanodiamonds to Vascular Endothelial Growth Factor Receptors in Tumor. Bioconjug. Chem. 2019, 30, 604–613. [Google Scholar] [CrossRef]
- Jariwala, D.H.; Patel, D.; Wairkar, S. Surface functionalization of nanodiamonds for biomedical applications. Mater. Sci. Eng. C 2020, 113, 110996. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Alemany, L.B.; Margrave, J.L.; Khabashesku, V.N. Sidewall Carboxylic Acid Functionalization of Single-Walled Carbon Nanotubes. J. Am. Chem. Soc. 2003, 125, 15174–15182. [Google Scholar] [CrossRef] [PubMed]
- Shenderova, O.; Panich, A.M.; Moseenkov, S.; Hens, S.C.; Kuznetsov, V.; Vieth, H.M. Hydroxylated Detonation Nanodiamond: FTIR, XPS, and NMR Studies. J. Phys. Chem. C 2011, 115, 19005–19011. [Google Scholar] [CrossRef]
- Zou, Q.; Li, Y.G.; Zou, L.H.; Wang, M.Z. Characterization of structures and surface states of the nanodiamond synthesized by detonation. Mater. Charact. 2009, 60, 1257–1262. [Google Scholar] [CrossRef]
- Paci, J.T.; Man, H.B.; Saha, B.; Ho, D.; Schatz, G.C. Understanding the surfaces of nanodiamonds. J. Phys. Chem. C 2013, 117, 17256–17267. [Google Scholar] [CrossRef]
- Dreyer, D.R.; Park, S.; Bielawski, C.W.; Ruoff, R.S. The chemistry of graphene oxide. Chem. Soc. Rev. 2010, 39, 228–240. [Google Scholar] [CrossRef]
- Mykhaylyk, O.O.; Solonin, Y.M.; Batchelder, D.N.; Brydson, R. Transformation of nanodiamond into carbon onions: A comparative study by high-resolution transmission electron microscopy, electron energy-loss spectroscopy, X-ray diffraction, small-angle x-ray scattering, and ultraviolet Raman spectroscopy. J. Appl. Phys. 2005, 97, 074302. [Google Scholar] [CrossRef]
- Turner, S.; Lebedev, O.I.; Shenderova, O.; Vlasov, I.I.; Verbeeck, J.; Van Tendeloo, G. Determination of size, morphology, and nitrogen impurity location in treated detonation nanodiamond by transmission electron microscopy. Adv. Funct. Mater. 2009, 19, 2116–2124. [Google Scholar] [CrossRef]
- Barnard, A.S.; Sternberg, M. Crystallinity and surface electrostatics of diamond nanocrystals. J. Mater. Chem. 2007, 17, 4811–4819. [Google Scholar] [CrossRef]
- Aller, E.; Brown, D.S.; Cox, G.G.; Miller, D.J.; Moody, C.J. Diastereoselectivity in the O-H Insertion Reactions of Rhodium Carbenoids Derived from Phenyldiazoacetates of Chiral Alcohols. Preparation of .alpha.-Hydroxy and .alpha.-Alkoxy Esters. J. Org. Chem. 1995, 60, 4449–4460. [Google Scholar] [CrossRef]
- Cox, G.G.; Haigh, D.; Hindley, R.M.; Miller, D.J.; Moody, C.J. Competing OH insertion and β-elimination in rhodium carbenoid reactions; synthesis of 2-alkoxy-3-arylpropanoates. Tetrahedron Lett. 1994, 35, 3139–3142. [Google Scholar] [CrossRef]
- Lieber, E.; Rao, C.N.R.; Chao, T.S.; Hoffman, C.W.W. Infrared Spectra of Organic Azides. Anal. Chem. 1957, 29, 916–918. [Google Scholar] [CrossRef]
- Miyazawa, T.; Blout, E.R. The Infrared Spectra of Polypeptides in Various Conformations: Amide I and II Bands. J. Am. Chem. Soc. 1961, 83, 712–719. [Google Scholar] [CrossRef]
- Niu, Y.H.; Munro, A.M.; Cheng, Y.-J.; Tian, Y.Q.; Liu, M.S.; Zhao, J.L.; Bardecker, J.A.; Jen-La Plante, I.; Ginger, D.S.; Jen, A.K.-Y. Improved Performance from Multilayer Quantum Dot Light-Emitting Diodes via Thermal Annealing of the Quantum Dot Layer. Adv. Mater. 2007, 19, 3371–3376. [Google Scholar] [CrossRef]
- Neelgund, G.M.; Oki, A. Photocatalytic activity of CdS and Ag2S quantum dots deposited on poly(amidoamine) functionalized carbon nanotubes. Appl. Catal. B Environ. 2011, 110, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Dong, A.; Ye, X.; Chen, J.; Kang, Y.; Gordon, T.; Kikkawa, J.M.; Murray, C.B. A Generalized Ligand-Exchange Strategy Enabling Sequential Surface Functionalization of Colloidal Nanocrystals. J. Am. Chem. Soc. 2011, 133, 998–1006. [Google Scholar] [CrossRef]
- Yu, W.W.; Chang, E.; Falkner, J.C.; Zhang, J.; Al-Somali, A.M.; Sayes, C.M.; Johns, J.; Drezek, R.; Colvin, V.L. Forming Biocompatible and Nonaggregated Nanocrystals in Water Using Amphiphilic Polymers. J. Am. Chem. Soc. 2007, 129, 2871–2879. [Google Scholar] [CrossRef]
- Pang, L.S.K.; Saxby, J.D.; Chatfield, S.P. Thermogravimetric analysis of carbon nanotubes and nanoparticles. J. Phys. Chem. 1993, 97, 6941–6942. [Google Scholar] [CrossRef]
- Mansfield, E.; Kar, A.; Hooker, S.A. Applications of TGA in quality control of SWCNTs. Anal. Bioanal. Chem. 2010, 396, 1071–1077. [Google Scholar] [CrossRef]
- Bom, D.; Andrews, R.; Jacques, D.; Anthony, J.; Chen, B.; Meier, M.S.; Selegue, J.P. Thermogravimetric Analysis of the Oxidation of Multiwalled Carbon Nanotubes: Evidence for the Role of Defect Sites in Carbon Nanotube Chemistry. Nano Lett. 2002, 2, 615–619. [Google Scholar] [CrossRef]
- Jiao, Y.; Liu, S.; Sun, Y.; Yue, W.; Zhang, H. Bioinspired Surface Functionalization of Nanodiamonds for Enhanced Lubrication. Langmuir 2018, 34, 12436–12444. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, H.S.N.; Liyanage, S.H.; Rathnayake, K.; Patel, U.; Yan, M. Analytical Methods for Characterization of Nanomaterial Surfaces. Anal. Chem. 2021, 93, 1889–1911. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wang, Y.-K.; Zhao, J.; Lu, H.; Stenzel, M.H.; Xiao, P. PEG Grafted-Nanodiamonds for the Delivery of Gemcitabine. Macromol. Rapid Commun. 2016, 37, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yao, P.; Zhu, Q.; Wang, L.; Zhu, Y. The polystyrene-divinylbenzene stationary phase hybridized with oxidized nanodiamonds for liquid chromatography. Talanta 2018, 185, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, A.; Blomstergren, A.; Nord, O.; Lukacs, M.; Lundeberg, J.; Uhlén, M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis 2005, 26, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-S.; Cho, K.-J.; Seol, Y.; Takagi, Y.; Dittmore, A.; Roche, P.A.; Neuman, K.C. Polydopamine Encapsulation of Fluorescent Nanodiamonds for Biomedical Applications. Adv. Funct. Mater. 2018, 28, 1801252. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 1981, 89, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Seol, Y.; Neuman, K.C. Combined magnetic tweezers and micro-mirror total internal reflection fluorescence microscope for single-molecule manipulation and visualization. In Single Molecule Analysis; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; pp. 297–316. [Google Scholar]
- Neuman, K.C.; Block, S.M. Optical trapping. Rev. Sci. Instrum. 2004, 75, 2787–2809. [Google Scholar] [CrossRef]
- Jannasch, A.; Abdosamadi, M.K.; Ramaiya, A.; De, S.; Ferro, V.; Sonnberger, A.; Schäffer, E. Custom-made microspheres for optical tweezers. In Optical Tweezers; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; pp. 137–155. [Google Scholar]
- Hui, Y.Y.; Zhang, B.; Chang, Y.-C.; Chang, C.-C.; Chang, H.-C.; Hsu, J.-H.; Chang, K.; Chang, F.-H. Two-photon fluorescence correlation spectroscopy of lipid-encapsulated fluorescent nanodiamonds in living cells. Opt. Express 2010, 18, 5896–5905. [Google Scholar] [CrossRef]
- Wee, T.-L.; Tzeng, Y.-K.; Han, C.-C.; Chang, H.-C.; Fann, W.; Hsu, J.-H.; Chen, K.-M.; Yu, Y.-C. Two-photon Excited Fluorescence of Nitrogen-Vacancy Centers in Proton-Irradiated Type Ib Diamond. J. Phys. Chem. A 2007, 111, 9379–9386. [Google Scholar] [CrossRef]
- Florin, E.-L.; Pralle, A.; Hörber, J.K.H.; Stelzer, E.H.K. Photonic Force Microscope Based on Optical Tweezers and Two-Photon Excitation for Biological Applications. J. Struct. Biol. 1997, 119, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.P.; McElroy, C.A.; Amero, C.D. Solution NMR of Large Molecules and Assemblies. Biochemistry 2007, 46, 331–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presti, C.; Thankamony, A.S.L.; Alauzun, J.G.; Mutin, P.H.; Carnevale, D.; Lion, C.; Vezin, H.; Laurencin, D.; Lafon, O. NMR and EPR Characterization of Functionalized Nanodiamonds. J. Phys. Chem. C 2015, 119, 12408–12422. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shenoy, G.; Ettedgui, J.; Mushti, C.; Hong, J.; Lane, K.; Blackman, B.; Jung, H.-S.; Takagi, Y.; Seol, Y.; Brechbiel, M.; et al. General Method to Increase Carboxylic Acid Content on Nanodiamonds. Molecules 2022, 27, 736. https://doi.org/10.3390/molecules27030736
Shenoy G, Ettedgui J, Mushti C, Hong J, Lane K, Blackman B, Jung H-S, Takagi Y, Seol Y, Brechbiel M, et al. General Method to Increase Carboxylic Acid Content on Nanodiamonds. Molecules. 2022; 27(3):736. https://doi.org/10.3390/molecules27030736
Chicago/Turabian StyleShenoy, Ganesh, Jessica Ettedgui, Chandrasekhar Mushti, Jennifer Hong, Kelly Lane, Burchelle Blackman, Hak-Sung Jung, Yasuharu Takagi, Yeonee Seol, Martin Brechbiel, and et al. 2022. "General Method to Increase Carboxylic Acid Content on Nanodiamonds" Molecules 27, no. 3: 736. https://doi.org/10.3390/molecules27030736
APA StyleShenoy, G., Ettedgui, J., Mushti, C., Hong, J., Lane, K., Blackman, B., Jung, H.-S., Takagi, Y., Seol, Y., Brechbiel, M., Swenson, R. E., & Neuman, K. C. (2022). General Method to Increase Carboxylic Acid Content on Nanodiamonds. Molecules, 27(3), 736. https://doi.org/10.3390/molecules27030736





