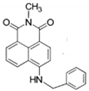
Cholesteric Chiral Molecular Tweezer for Rapid Detection of F− in Food Samples
Abstract
:1. Introduction
2. Results
2.1. Selective Recognition of Anions by Host Molecular Tweezer 7d
2.2. Influences of Coexisting Ions and Response Time Test
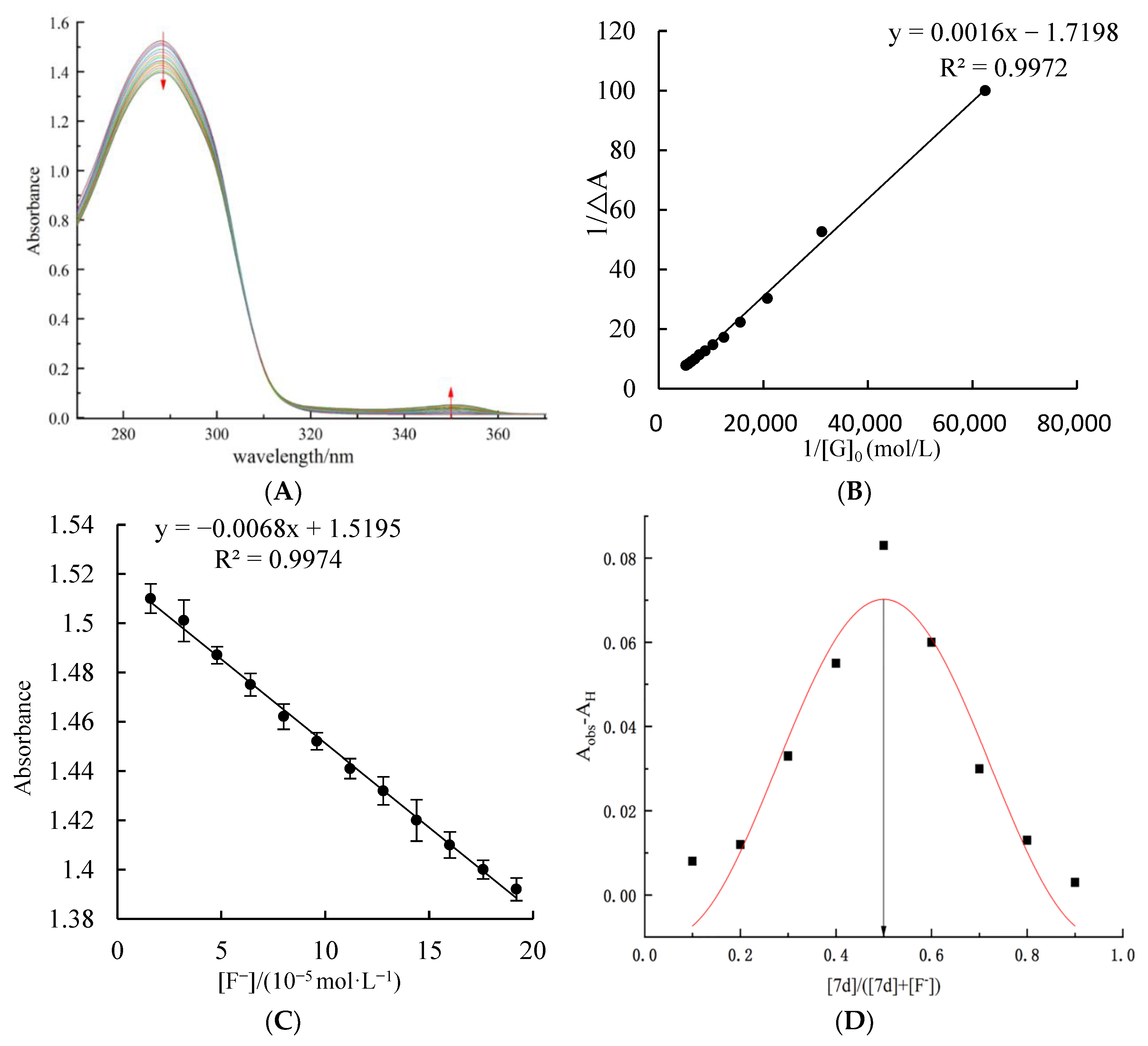
2.3. UV Titration Test at Different Concentrations of F− Ions by Host Molecular Tweezer 7d
2.4. Microstructure Observation and Energy Spectrum Imaging Analysis of the Complex
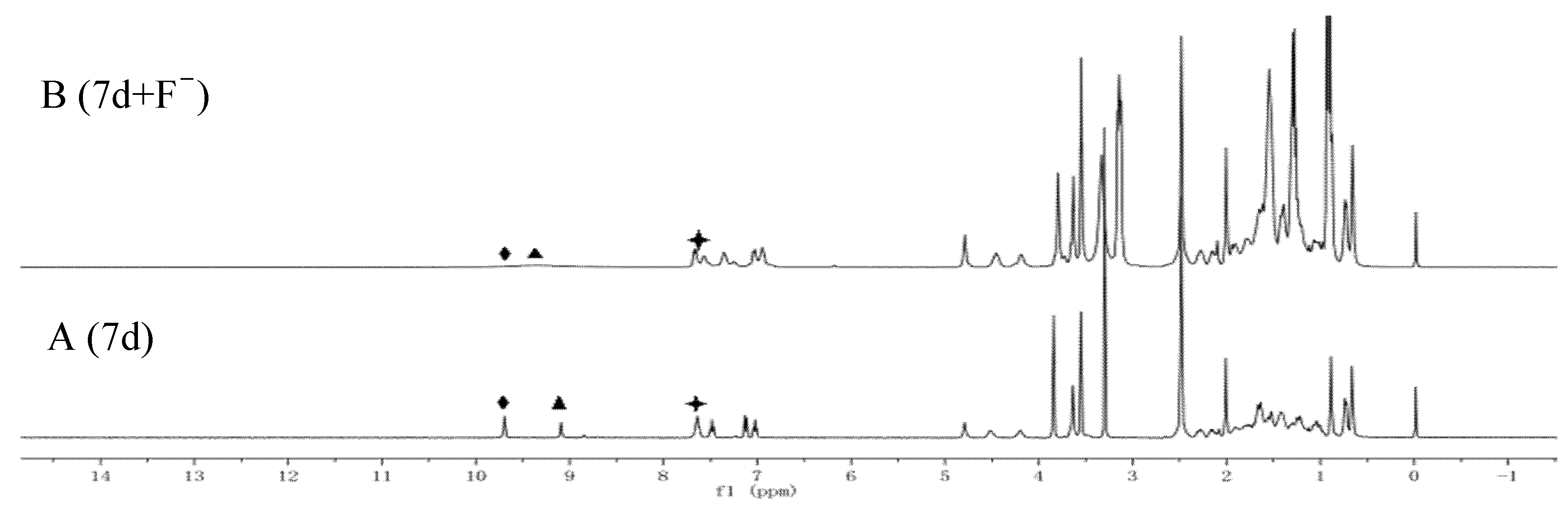
2.5. Recognition Mechanism
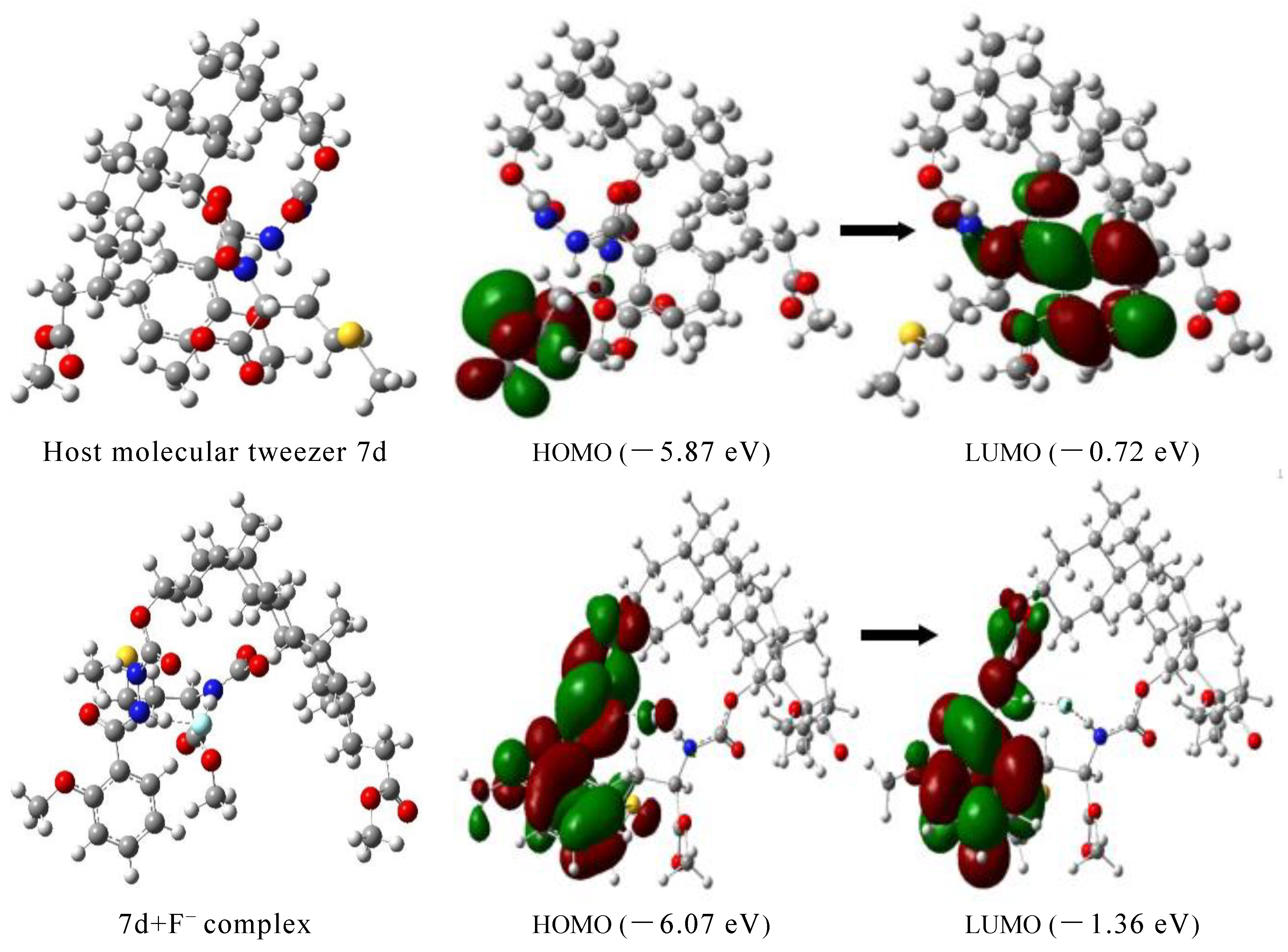
2.6. Theoretical Calculation
2.7. Quantitative Determination of F− in Actual Samples
3. Materials and Methods
3.1. Chemical Reagents and Instruments
3.2. Preparation of Cholesteric Molecular Tweezer 7d
3.3. Solution Preparation and Spectral Analysis
3.4. Selective Recognition of Anions by the Host Molecular Tweezer 7d
3.5. Interference Ability of Coexisting Ions and Response Time for Specific Recognition of F−
3.6. UV—Vis Spectral Titration and Job’s Plot Test for the Recognition of F− by the Host Molecular Tweezer 7d
3.7. Microstructure Observation and Energy Spectrum Imaging Analysis of the Complex
3.8. Recognition Mechanism of F− Ions by the Host Molecular Tweezer 7d
3.9. Detection of F− Ions in Actual Food
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Brown, A.; Beer, P.D. Halogen bonding anion recognition. Chem. Commun. 2016, 52, 8645–8658. [Google Scholar] [CrossRef]
- Kaur, K.; Saini, R.; Kumar, A.; Luxami, V.; Kaur, N.; Singh, P.; Kumar, S. Chemodosimeters: An approach for detection and estimation of biologically and medically relevant metal ions, anions and thiols. Coord. Chem. Rev. 2012, 256, 1992–2028. [Google Scholar] [CrossRef]
- Kumar, G.G.V.; Kesavan, M.P.; Sivaraman, G.; Rajesh, J. Colorimetric and NIR fluorescence receptors for F- ion detection in aqueous condition and its live cell imaging. Sens. Actuators B 2018, 255, 3194–3206. [Google Scholar] [CrossRef]
- Wu, H.B.; Wang, N.; Chen, Y.Z.; Wu, Q. Study on treatment of low-concentration fluoride wastewater by activated aluminum oxide. Appl. Chem. Ind. 2013, 42, 50–52. [Google Scholar]
- Wang, J.M.; Xu, H.M.; Yan, Z.P.; Yang, J.R.; Zhu, Y.Y.; Cheng, X.F.; Wang, J.D. Separation and identification of differential protein in rat’s bone with fluorosis and calcium supplementation intervention. Chin. J. Biotechnol. 2019, 6, 1097–1108. [Google Scholar]
- Wang, N.; Qi, P.; Chen, Y.Z.; Zeng, X.; Zhou, Q.Q.; Lin, Z.H.; Huang, M. Determination of fluoride in drinking water and health risk analysis. Light Ind. Sci. Technol. 2019, 5, 26–27. [Google Scholar]
- Li, Y.B.; Chai, L.H. Chronic effects of fluoride exposure on growth and development in Rana chensinensis Tadpoles. Asian J. Ecotoxicol. 2018, 13, 278–287. [Google Scholar]
- Chen, J.S.; Zhou, P.W.; Li, G.Y.; Chu, T.C.; He, G.Z. Fluoride anion sensing mechanism of 2-ureido-4 [1H]-pyrimidinone quadruple hydrogen-bonded supramolecular assembly: Photoinduced electron transfer and partial configuration change. J. Phys. Chem. B 2013, 117, 5212–5221. [Google Scholar] [CrossRef]
- GB/T5749-2006; Hygienic Standard for Drinking Water. National Standards of the People’s Republic of China; Ministry of Health: Beijing, China, 2006.
- Cheng, S.; Ding, M.S.; Wang, H.; Sun, S.N.; Chen, J.; Xue, D.J. Determination of total fluoride content in compound fertilizers with gas chromatography. Shandong Chem. Ind. 2021, 11, 93–94. [Google Scholar]
- Zhao, X.; Li, Y.; Carroll, K.C.; Li, F.F.; Qiu, L.C.; Huo, Z.L. Mesoporous goethite for rapid and high-capacity fluoride removal from drinking water. J. Environ. Chem. Eng. 2021, 9, 105278–105287. [Google Scholar] [CrossRef]
- Qiu, Z.W.; Liu, Y.L.; Li, Z.Q.; Ye, R.H.; Zhu, L.M.; Chen, Y.; Pan, W.L. Analysis of fluorinated surfactant in multicomponent systems by 19F NMR. Anal. Lab. 2015, 34, 549–553. [Google Scholar]
- Wei, B.H.; Huang, G.W.; Long, Z.K. Determination of 6-gingerol in toothpaste by HPLC. Oral Care Ind. 2009, 2, 40–42. [Google Scholar]
- Cao, C.; Qiu, M.H.; You, X.M.; Lu, B.B.; Ji, X.D. Syntheses of tripodal benzoyl thiourea ions receptors and their recognition properties. Chemia 2020, 83, 246–252. [Google Scholar]
- Wu, T.R.; Jian, R.C.; Chen, X.L.; Tang, X.W.; Li, Y.L.; Sheng, Z.J.; Wu, P.P.; Hong, W.Q. Synthesis and biological activity of novel amides. Chin. J. Synth. Chem. 2021, 1, 65–70. [Google Scholar]
- Song, X.W.; Liu, J.B.; Fu, M. Synthesis of lactam impurities in sacubitril. Shandong Chem. Ind. 2021, 17, 63–64. [Google Scholar]
- Yan, X.Q.; Zhuo, J.B.; Wang, J.C.; Xie, L.L.; Yuan, Y.F. Design, synthesis and anion recognition of benzocoumarin-based thiourea receptors. Chin. J. Org. Chem. 2015, 35, 2184–2190. [Google Scholar] [CrossRef]
- Ji, X.D.; Wang, Y.F.; An, P.; You, X.M.; Cao, C. Anions recognition properties of long-chain alkoxy substituted benzoyl thiourea. Chem. Res. Appl. 2020, 6, 1007–1013. [Google Scholar]
- Qian, H.R.; Wang, B. A Study of recognition to multiple anions based on Brominated N,N′-di ( 2-naphthalene) Azopyridines salts. J. China West Norm. Univ. 2019, 40, 65–68. [Google Scholar]
- Han, F.; Wang, T.; Feng, B.; Xu, Q. N,N-Dimethylformamide(DMF)-Promoted Specific N-Alkylation of Hydroxyl N-Heterocycles with Organohalides:A Direct and Efficient Method for Synthesis of Pyridone Derivatives. Chin. J. Org. Chem. 2021, 7, 2831–2838. [Google Scholar]
- Zhang, D.Q.; Liu, X.; Pang, X.J.; Liu, H.M.; Zhang, Q.R. Design, synthesis and anticancer activity studies of novel indole-pyrimidine biaryl derivatives. Chin. J. Org. Chem. 2021, 1, 267–275. [Google Scholar] [CrossRef]
- Yu, R.R.; Fang, X.J. Synthesis of 3-methyl indoles via catellani reaction. Chin. J. Org. Chem. 2021, 6, 2532–2533. [Google Scholar] [CrossRef]
- Mao, S.W.; Tang, J.; Yang, F. Recent progress on biological activities of bile acid derivatives. Chin. Bull. Life Sci. 2017, 29, 90–96. [Google Scholar]
- Liu, X.L.; Zhao, Z.G. Design and synthesis of novel deoxycholic acid clamp anion receptor and its recognition performance for anions. Chem. Res. Appl. 2007, 19, 670–674. [Google Scholar]
- Chahar, M.; Pandey, P.S. Design of steroid-based imidazolium receptors for fluoride ion recognition. Tetrahedron 2008, 64, 6488–6493. [Google Scholar] [CrossRef]
- Ye, Y.; Suo, Y.R.; Yang, Y.J.; Yang, F.; Han, L.J. Application of UV spectrophotometry-NMR methodology in research on recognition properties of receptors based on deoxycholic acid for phenolic compounds. Anal. Test. Technol. Instrum. 2015, 21, 1–6. [Google Scholar]
- Liu, Y.H.; Wang, K.; Zhang, M.; Liu, G. Synthesis of HBBP-P-nitrophenylhydrazone and its Anion recognition. Chin. J. Inorg. Anal. Chem. 2020, 10, 64–68. [Google Scholar]
- Cao, C.; Cao, T.; Yan, P.J.; Wang, Q.Y.; Yue, G.R.; Ji, X.D. High selectivity of enol-keto tautomers imine derivative for the recognition of Co2+. Chem. J. Chin. Univ. 2020, 41, 1544–1551. [Google Scholar]
- Zheng, X.; Zhu, W.; Liu, D.; Ai, H.; Huang, Y.; Lu, Z.Y. Highly Selective Colorimetric/Fluorometric Dual-Channel Fluoride Ion Probe, and Its Capability of Differentiating Cancer Cells. ACS Appl. Mater. Interfaces 2014, 6, 7996–8000. [Google Scholar] [CrossRef]
- Zhang, Y.R.; Xing, F.F.; Zhu, S.R. Structures and chromogenic ion-pair recognition of a catechol-functionalized 1,8-anthraquinone macrocycle in dimethyl sulfoxide. Inorg. Chem. 2021, 60, 5042–5053. [Google Scholar] [CrossRef]
- Misra, A.; Shahid, M.; Dwivedi, P.; Srivastava, P.; Razi, S.S. A simple naphthalimide-based receptor for selective recognition of fluoride anion. Arkivoc Online J. Org. Chem. 2012, 2, 133–145. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.Y.; Liu, R.; Zhu, X.L.; Cai, X.; Zhu, H.J. A highly efficient and selective probe for F detection based on 1H-imidazo [4,5-b] phenazine derivative. Chin. Chem. Lett. 2015, 26, 339–342. [Google Scholar] [CrossRef]
- Sharma, S.; Hundal, M.S.; Hundal, G. Selective recognition of fluoride ions through fluorimetric and colorimetric response of a first mesitylene based dipodal sensor employing thiosemicarbazones. Tetrahedron Lett. 2013, 54, 2423–2427. [Google Scholar] [CrossRef]
- Liu, R.Y.; Gao, Y.; Zhang, Q.B.; Yang, X.D.; Lu, X.W.; Ke, Z.Y.; Zhou, W.Q.; Qu, J.Q. A fluorescent probe based on hydroxylnaphthalene 2-cyanoacrylate: Fluoride ion detection and its bio-imaging in live cells. New J. Chem. 2014, 38, 2941–2945. [Google Scholar] [CrossRef]
- Xu, J.; Sun, S.; Li, Q.; Yue, Y.; Li, Y.; Shao, S. A novel “Turn-On” fluorescent probe for F detection in aqueous solution and its application in live-cell imaging. Anal. Chim. Acta 2014, 849, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.C.; Yuan, F.; Li, R.X.; Li, Y.M.; Wei, Q.; Ma, Z.M.; Du, B.; Zhang, X.L. A highly selective colorimetric and ratiometric fluorescent chemodosimeter for imaging fluoride ions in living cells. Chem. Commun. 2011, 47, 7098–7100. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Wan, W.L.; Qiu, J.K.; Li, Z.Y.; Wang, H.Y.; Pei, Y.C.; Zhao, Y.; Wang, J.J. Molecular dynamic simulation for the microstructure of ordered molecular aggregates in ionic liquid solutions. Scientia Sinica Chimica 2021, 51, 1406–1414. [Google Scholar] [CrossRef]
- Choi, Y.W.; Park, G.J.; Na, Y.J.; Jo, H.Y.; Lee, S.A.; You, G.R.; Kim, C. A single schiff base molecule for recognizing multiple metal ions: A fluorescence sensor for Zn(Ⅱ) and Al(Ⅲ) and colorimetric sensor for Fe(Ⅱ) and Fe(Ⅲ). Sens. Actuators B 2014, 194, 343–352. [Google Scholar] [CrossRef]
- Fu, Y.L.; Fan, C.B.; Liu, G.; Pu, S.Z. A Colorimetric and Fluorescent Sensor for Cu2+ and F- based on a Diarylethene with a 1,8-Naphthalimide Schiff Base Unit. Sens. Actuators B Chem. 2017, 239, 295–303. [Google Scholar] [CrossRef]
- Sohn, D.H.; Park, J.; Cho, S.J.; Kang, J. Novel anion receptors for selective recognition of dimethyl phosphinate and carboxylate. Tetrahedron 2017, 73, 212–221. [Google Scholar] [CrossRef]
- Dwivedi, S.K.; Razi, S.S.; Misra, A. Sensitive colorimetric detection of CN- and AcO- anions in a semi-aqueous environment through a coumarin–naphthalene conjugate azo dye. New J. Chem. 2019, 43, 5126–5132. [Google Scholar] [CrossRef]
- Liu, M.; Wang, X.H.; Bi, Q.W.; Zhao, Z.G. Microwave-assisted synthesis and recognition properties of chiral molecular tweezers based on deoxycholic acid. J. Chem. Res. 2013, 37, 394–397. [Google Scholar] [CrossRef]
- Song, X.; Chen, M.; Sun, L. Study on synthesis of amino acid methyl ester hydrochloride in DMSO—Methanol solution. Chem. Reag. 2007, 29, 687–688. [Google Scholar]
- Ye, Y.; Suo, Y.R.; Yang, F.; Han, L.J. Microwave-assisted synthesis of novel chiral receptors derived from deoxycholic acid and their molecular recognition properties. Chem. Lett. 2014, 43, 1812–1814. [Google Scholar] [CrossRef]
- Ji, X.D.; Li, S.B.; Cao, C.; Ru, T.; Yang, R.F.; Yue, G.R. Selective Single Recognition of F- by Long Chain Alkoxyphenylhydrazone Derivatives. Fine Chem. 2021, 38, 323–418. [Google Scholar]
- Wu, G.; Zhang, Z.F.; Song, F.; Liu, X.; Ma, H.C.; Pang, X.M.; Zhang, J.L.; Liu, Y.Y. Determination of total nitrogen in plant fruits by graphite digestion apparatus and automatic azotometer. Rock Min. Anal. 2020, 39, 311–317. [Google Scholar]

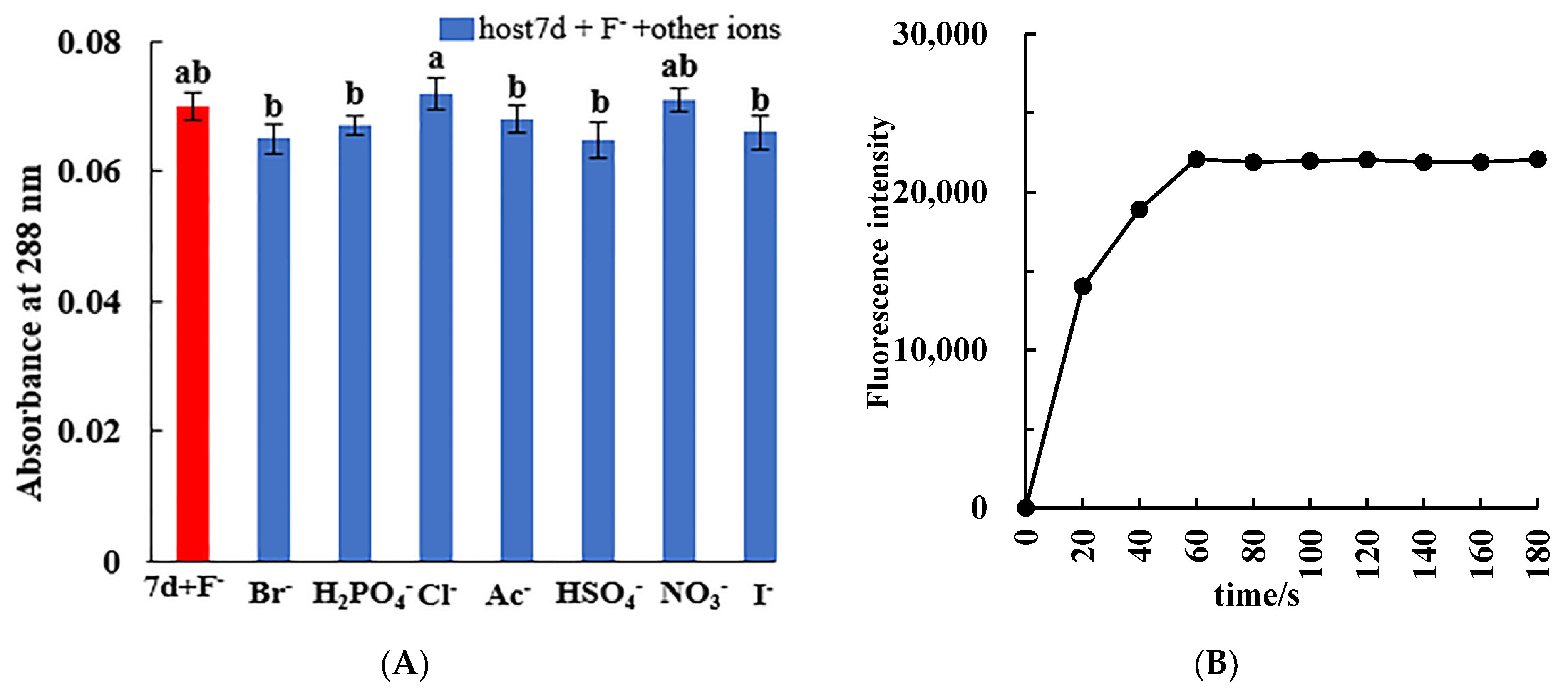


| Coexisting Ion | CCoexisting Ion (mol/L) | (A−A0)/A0 (%) |
|---|---|---|
| Br− | 8.33 × 10−4 | −7.14 |
| H2PO4− | 8.33 × 10−4 | −4.29 |
| Cl− | 8.33 × 10−4 | 2.86 |
| Ac− | 8.33 × 10−4 | −2.86 |
| HSO4− | 8.33 × 10−4 | −7.43 |
| NO3− | 8.33 × 10−4 | 1.43 |
| I− | 8.33 × 10−4 | −5.71 |
| Samples | New Method (mg/kg) | RSD (%) | Selective Electrode Method (mg/kg) | Relativeerror (%) | p |
|---|---|---|---|---|---|
| Quinoa | 2.45 ± 0.029 | 1.17 | 2.41 ± 0.013 | 1.67 | 0.83 |
| Ribes stenocarpum Maxim | 4.68 ± 0.032 | 0.68 | 4.50 ± 0.010 | 4.02 | 0.94 |
| Chinese wolfberry | 2.57 ± 0.042 | 1.63 | 2.50 ± 0.006 | 2.65 | 0.63 |
| Milk powder | 2.23 ± 0.016 | 0.71 | 2.18 ± 0.008 | 2.20 | 0.88 |
| Wheat flour | 1.77 ± 0.028 | 1.57 | 1.70 ± 0.006 | 3.83 | 0.33 |
| Samples | Add (mg/Kg) | This Method (mg/kg) | Recovery (%) | RSD (%, n = 3) | Selective Electrode Method (mg/kg) | Recovery (%) | RSD (%, n = 3) | p |
|---|---|---|---|---|---|---|---|---|
| Quinoa | 2 | 4.66 ± 0.071 | 105 | 1.53 | 4.51 ± 0.009 | 98 | 2.29 | 0.62 |
| 5 | 7.61 ± 0.073 | 102 | 0.95 | 7.27 ± 0.008 | 102 | 3.86 | 0.60 | |
| Chinese wolfberry | 2 | 4.61 ± 0.041 | 101 | 0.88 | 4.77 ± 0.015 | 106 | 1.66 | 0.26 |
| 5 | 7.57 ± 0.028 | 100 | 0.38 | 7.61 ± 0.030 | 102 | 4.59 | 0.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wang, H.; Chen, Z.; Ye, Y.; Wang, Y. Cholesteric Chiral Molecular Tweezer for Rapid Detection of F− in Food Samples. Molecules 2022, 27, 1098. https://doi.org/10.3390/molecules27031098
Liu Z, Wang H, Chen Z, Ye Y, Wang Y. Cholesteric Chiral Molecular Tweezer for Rapid Detection of F− in Food Samples. Molecules. 2022; 27(3):1098. https://doi.org/10.3390/molecules27031098
Chicago/Turabian StyleLiu, Zhe, Hong Wang, Zhu Chen, Ying Ye, and Yuwei Wang. 2022. "Cholesteric Chiral Molecular Tweezer for Rapid Detection of F− in Food Samples" Molecules 27, no. 3: 1098. https://doi.org/10.3390/molecules27031098
APA StyleLiu, Z., Wang, H., Chen, Z., Ye, Y., & Wang, Y. (2022). Cholesteric Chiral Molecular Tweezer for Rapid Detection of F− in Food Samples. Molecules, 27(3), 1098. https://doi.org/10.3390/molecules27031098