Coordination Chemistry of Uranyl Ions with Surface-Immobilized Peptides: An XPS Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Evidence of (UO2) Uptake from X-ray Photoemission
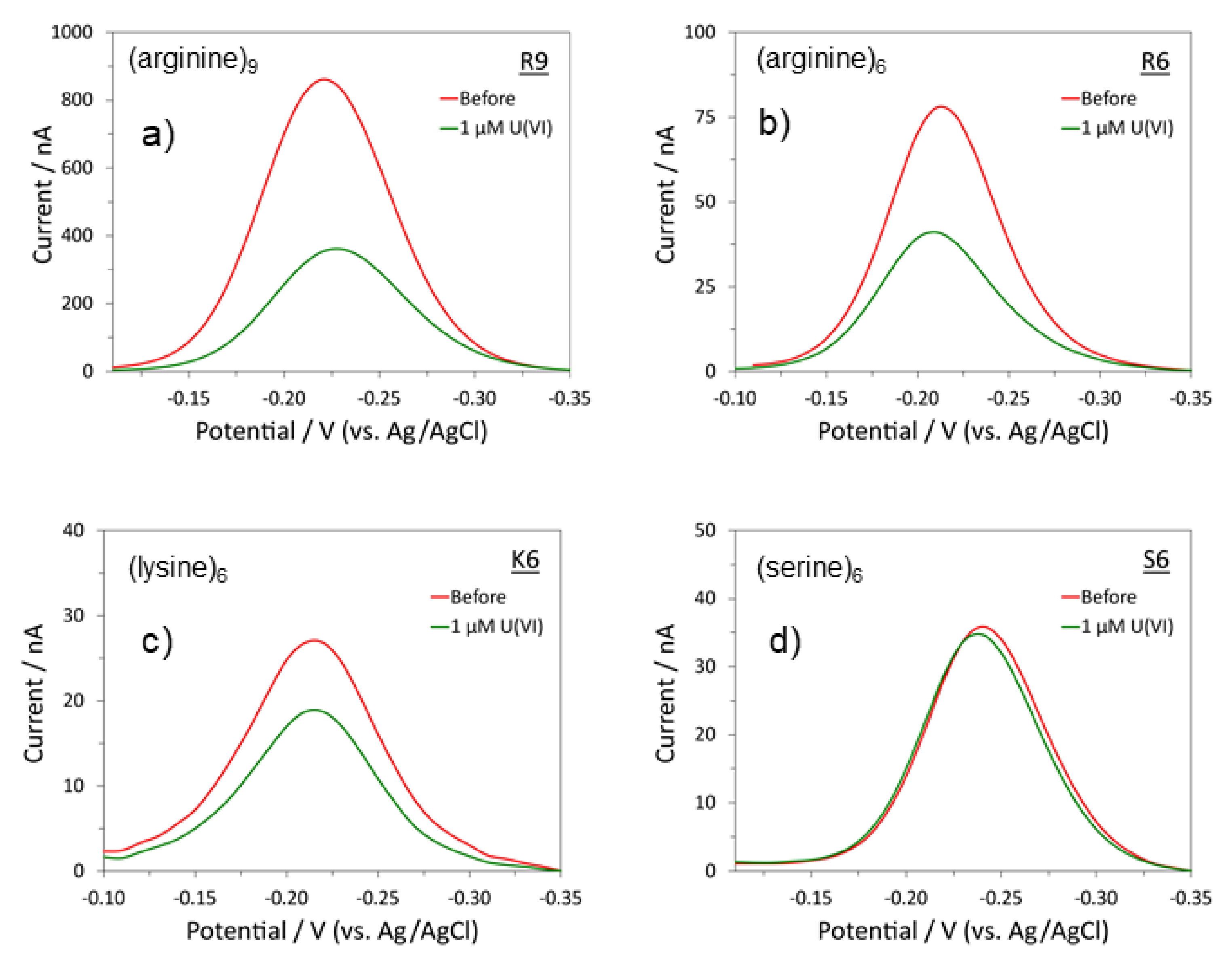
2.2. Modified Polypeptides as Sensors Characterization
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Domingo, J.L. Reproductive and developmental toxicity of natural and depleted uranium: A review. Reprod. Toxicol. 2001, 15, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Konietzka, R. Gastrointestinal absorption of uranium compounds—A review. Regul. Toxicol. Pharmacol. 2015, 71, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.; Oh, J.; Choung, S.; Cho, B.-W.; Lee, K.-S.; Yun, U.; Woo, N.-C.; Kim, H.K. Distribution and potential health risk of groundwater uranium in Korea. Chemosphere 2016, 163, 108–115. [Google Scholar] [CrossRef]
- Kurttio, P.; Auvinen, A.; Salonen, L.; Saha, H.; Pekkanen, J.; Mäkeläinen, I.; Väisänen, S.B.; Penttilä, I.M.; Komulainen, H. Renal effects of uranium in drinking water. Environ. Health Perspect. 2002, 110, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Sutton, M.; Burastero, S.R. Uranium (VI) solubility and speciation in simulated elemental human biological fluids. Chem. Res. Toxicol. 2004, 17, 1468–1480. [Google Scholar] [CrossRef]
- Yazzie, M.; Gamble, S.L.; Civitello, E.R.; Stearns, D.M. Uranyl acetate causes DNA single strand breaks in vitro in the presence of ascorbate (vitamin C). Chem. Res. Toxicol. 2003, 16, 524–530. [Google Scholar] [CrossRef]
- Hartsock, W.J.; Cohen, J.D.; Segal, D.J. Uranyl acetate as a direct inhibitor of DNA-binding proteins. Chem. Res. Toxicol. 2007, 20, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Cantaluppi, C.; Degetto, S. Civilian and military uses of depleted uranium: Environmental and health problems. Ann. Chim. 2000, 90, 665–676. [Google Scholar] [PubMed]
- Gao, N.; Huang, Z.; Liu, H.; Hou, J.; Liu, X. Advances on the toxicity of uranium to different organisms. Chemosphere 2019, 237, 124548. [Google Scholar] [CrossRef]
- Nolan, J.; Weber, K.A. Natural uranium contamination in major US aquifers linked to nitrate. Environ. Sci. Technol. Lett. 2015, 2, 215–220. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Chernikov, A.V.; Bruskov, V.I. Chemical and radiological toxicity of uranium compounds. Russ. J. Gen. Chem. 2016, 86, 1531–1538. [Google Scholar] [CrossRef]
- Ilangovan, R.; Daniel, D.; Krastanov, A.; Zachariah, C.; Elizabeth, R. Enzyme based biosensor for heavy metal ions determination. Biotechnol. Biotechnol. Equip. 2006, 20, 184–189. [Google Scholar] [CrossRef]
- Saidur, M.R.; Aziz, A.A.; Basirun, W.J. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: A review. Biosens. Bioelectron. 2017, 90, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Corbisier, P.; Van Der Lelie, D.; Borremans, B.; Provoost, A.; De Lorenzo, V.; Brown, N.L.; Lloyd, J.R.; Hobman, J.L.; Csöregi, E.; Johansson, G.; et al. Whole cell-and protein-based biosensors for the detection of bioavailable heavy metals in environmental samples. Anal. Chim. Acta 1999, 387, 235–244. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, J.; Boyd, B.J. Peptide-based biosensors. Talanta 2015, 136, 114–127. [Google Scholar] [CrossRef]
- Chow, E.; Gooding, J.J. Peptide modified electrodes as electrochemical metal ion sensors. Electroanalysis 2006, 18, 1437–1448. [Google Scholar] [CrossRef]
- Stellato, C.C.; Lai, R.Y. Engineering uranyl-chelating peptides from NikR for electrochemical peptide-based sensing applications. J. Electroanal. Chem. 2020, 858, 113698. [Google Scholar] [CrossRef]
- De la Rica, R.; Pejoux, C.; Matsui, H. Assemblies of functional peptides and their applications in building blocks for biosensors. Adv. Funct. Mater. 2011, 21, 1018–1026. [Google Scholar] [CrossRef]
- Miyake, R. Constructing multicomponent cooperative functional systems using metal complexes of short flexible peptides. Chem. Commun. 2021, 57, 7987–7996. [Google Scholar] [CrossRef]
- Garai, A.; Delangle, P. Recent advances in uranyl binding in proteins thanks to biomimetic peptides. J. Inorg. Biochem. 2020, 203, 110936. [Google Scholar] [CrossRef]
- Pardoux, R.; Sauge-Merle, S.; Lemaire, D.; Delangle, P.; Guilloreau, L.; Adriano, J.-M.; Berthomieu, C. Modulating uranium binding affinity in engineered calmodulin EF-hand peptides: Effect of phosphorylation. PLoS ONE 2012, 7, e41922. [Google Scholar] [CrossRef] [PubMed]
- Nourmand, M.; Meissami, N. Complex formation between uranium (VI) and thorium (IV) ions with some α-amino-acids. J. Chem. Soc. Dalton Trans. 1983, 1529–1533. [Google Scholar] [CrossRef]
- Wu, Q.-Y.; Wang, C.-Z.; Lan, J.-H.; Chai, Z.-F.; Shi, W.-Q. Theoretical insight into the binding affinity enhancement of serine with the uranyl ion through phosphorylation. RSC Adv. 2016, 6, 69773–69781. [Google Scholar] [CrossRef]
- Thompson, C.C.; Lai, R.Y. Threonine Phosphorylation of an Electrochemical Peptide-Based Sensor to Achieve Improved Uranyl Ion Binding Affinity. Biosensors 2022, 12, 961. [Google Scholar] [CrossRef]
- El-Bohy, M.N.; Abdel-Monem, Y.K.; Rabie, K.A.; Farag, N.M.; Mahfouz, M.G.; Galhoum, A.A.; Guibal, E. Grafting of arginine and glutamic acid onto cellulose for enhanced uranyl sorption. Cellulose 2017, 24, 1427–1443. [Google Scholar] [CrossRef]
- Lai, R.Y.; Walker, B.; Stormberg, K.; Zaitouna, A.J.; Yang, W. Electrochemical techniques for characterization of stem-loop probe and linear probe-based DNA sensors. Methods 2013, 64, 267–275. [Google Scholar] [CrossRef]
- Yang, W.; Gooding, J.J.; Hibbert, D.B. Characterisation of gold electrodes modified with self-assembled monolayers of L-cysteine for the adsorptive stripping analysis of copper. J. Electroanal. Chem. 2001, 516, 10–16. [Google Scholar] [CrossRef]
- Vallee, A.; Humblot, V.; Pradier, C.-M. Peptide interactions with metal and oxide surfaces. Acc. Chem. Res. 2010, 43, 1297–1306. [Google Scholar] [CrossRef]
- Mishra, E.; Majumder, S.; Varma, S.; Dowben, P.A. X-ray photoemission studies of the interaction of metals and metal ions with DNA. Z. Phys. Chem. 2022, 236, 439–480. [Google Scholar] [CrossRef]
- Schindler, M.; Hawthorne, F.C.; Freund, M.S.; Burns, P.C. XPS spectra of uranyl minerals and synthetic uranyl compounds. I: The U 4f spectrum. Geochim. Cosmochim. Acta 2009, 73, 2471–2487. [Google Scholar] [CrossRef]
- Allen, G.C.; Crofts, J.A.; Curtis, M.T.; Tucker, P.M.; Chadwick, D.; Hampson, P.J. X-ray photoelectron spectroscopy of some uranium oxide phases. J. Chem. Soc. Dalt. Trans. 1974, 1296–1301. [Google Scholar] [CrossRef]
- Ilton, E.S.; Bagus, P.S. XPS determination of uranium oxidation states. Surf. Interface Anal. 2011, 43, 1549–1560. [Google Scholar] [CrossRef]
- Raczyńska, E.D.; Cyrański, M.K.; Gutowski, M.; Rak, J.; Gal, J.-F.; Maria, P.-C.; Darowska, M.; Duczmal, K. Consequences of proton transfer in guanidine. J. Phys. Org. Chem. 2003, 16, 91–106. [Google Scholar] [CrossRef]
- Drozdov, F.V.; Kotovb, V.M. Guanidine: A Simple Molecule with Great Potential: From Catalysts to Biocides and Molecular Glues. J. Nesmeyanov Inst. Organoelement Compd. Russ. Acad. Sci. 2020, 3, 200–213. [Google Scholar] [CrossRef]
- Bain, C.D.; Whitesides, G.M. Attenuation lengths of photoelectrons in hydrocarbon films. J. Phys. Chem. 1989, 93, 1670–1673. [Google Scholar] [CrossRef]
- Ricci, F.; Lai, R.Y.; Heeger, A.J.; Plaxco, K.W.; Sumner, J.J. Effect of molecular crowding on the response of an electrochemical DNA sensor. Langmuir 2007, 23, 6827–6834. [Google Scholar] [CrossRef] [PubMed]
- Sumner, J.J.; Creager, S.E. Topological effects in bridge-mediated electron transfer between redox molecules and metal electrodes. J. Am. Chem. Soc. 2000, 122, 11914–11920. [Google Scholar] [CrossRef]
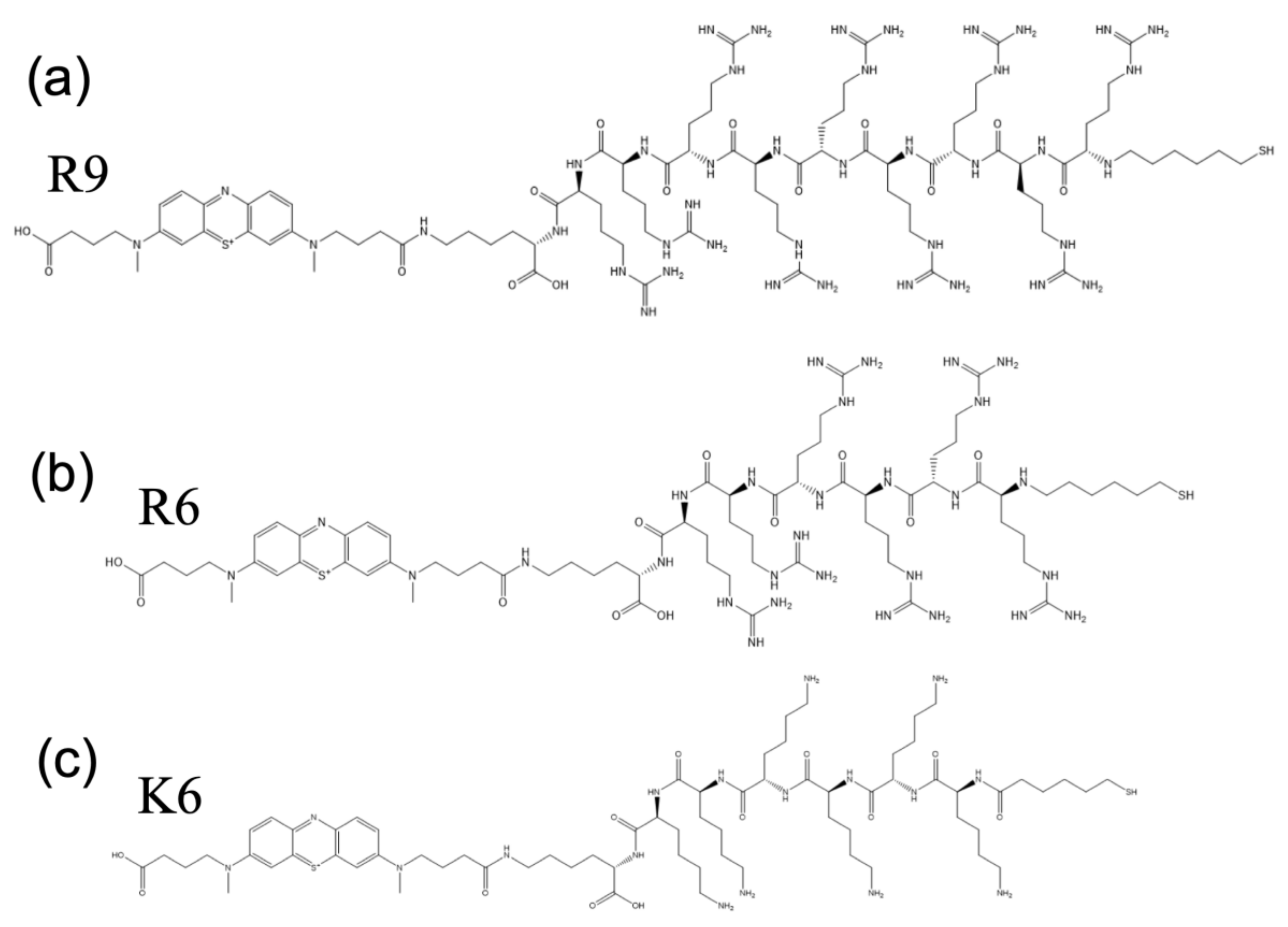
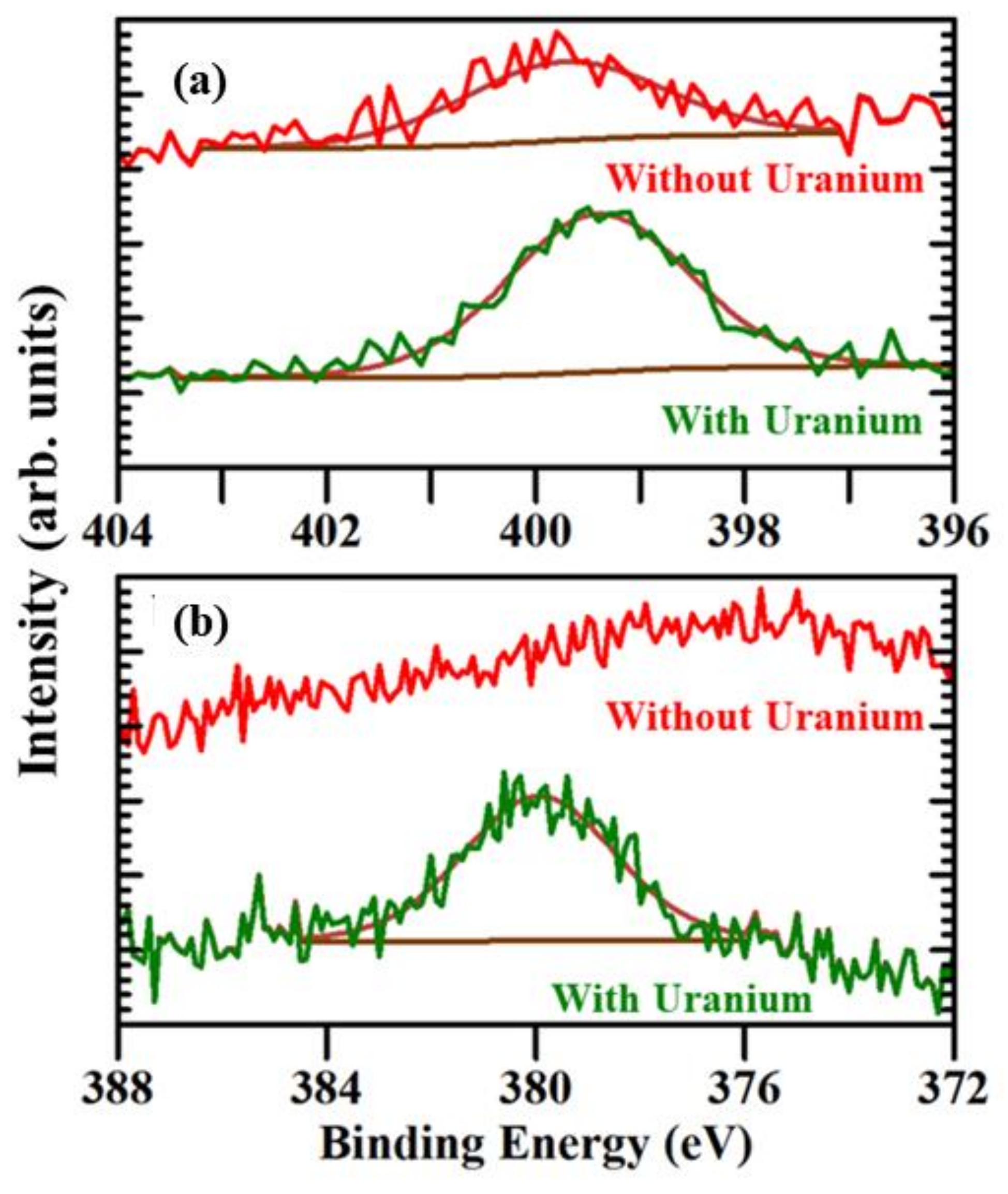
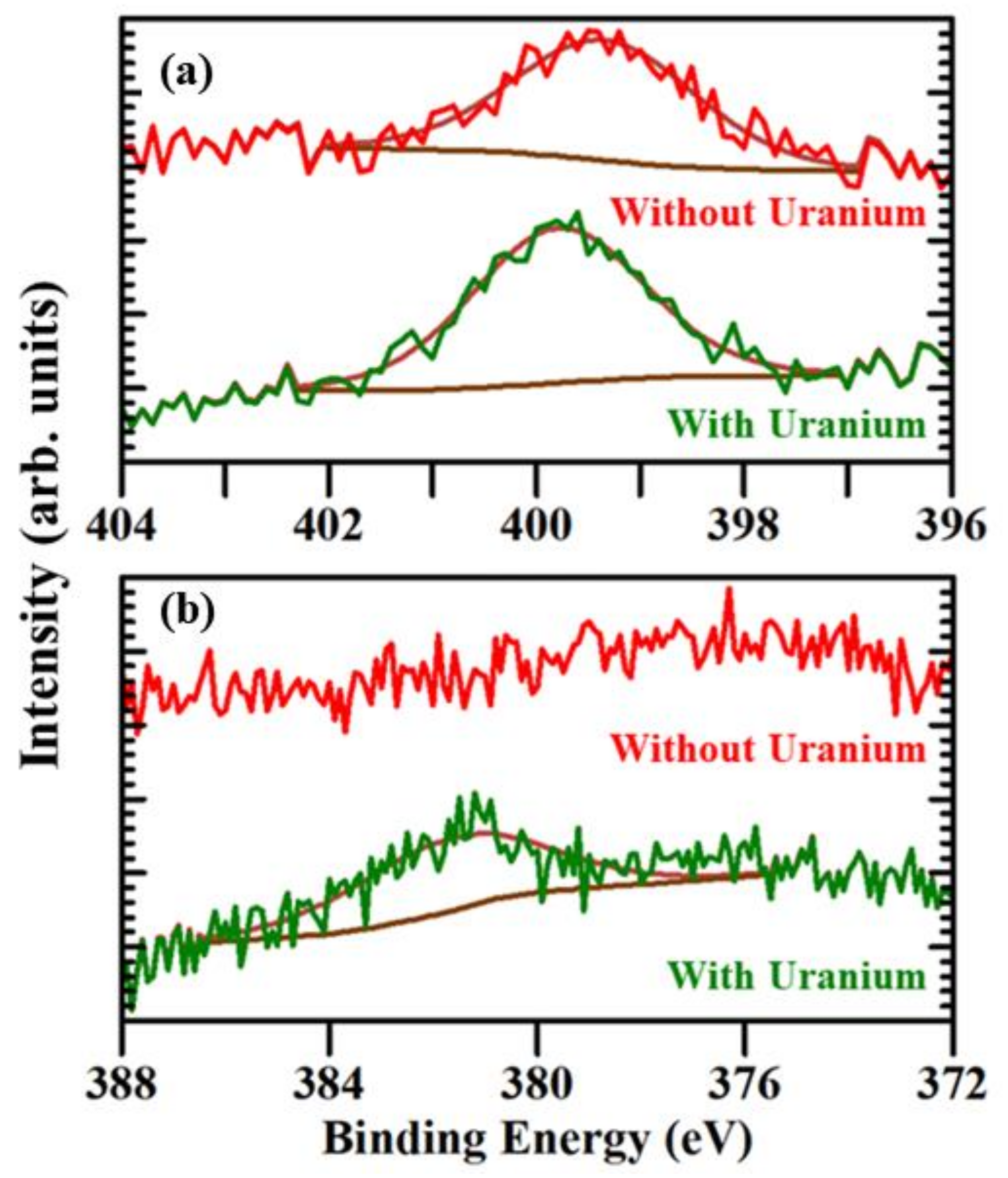
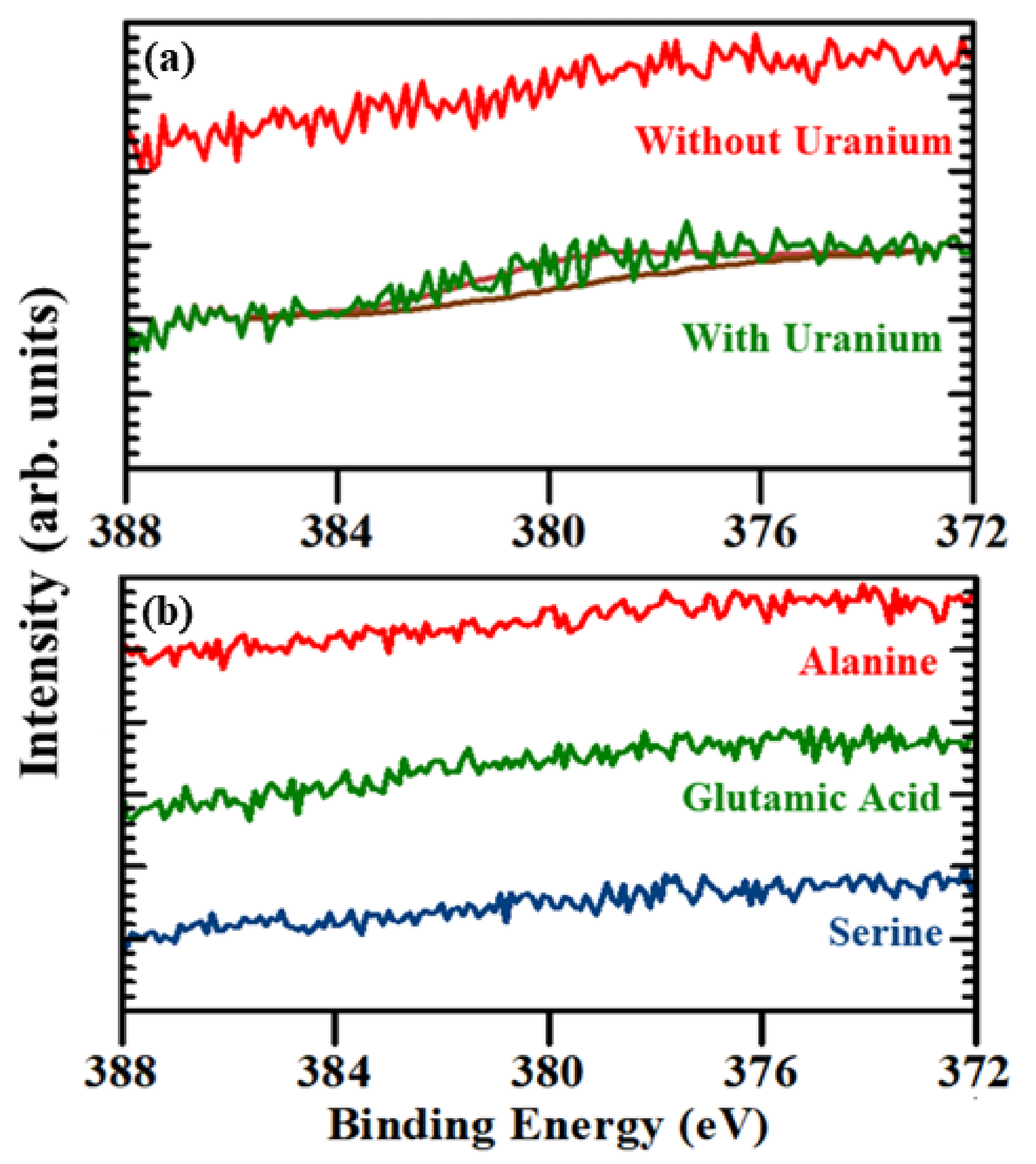
| Probe | Coverage (Teramolecules/cm2) |
|---|---|
| (arginine)6 (R6) | 1.37 |
| (arginine)9 (R9) | 8.89 |
| (lysine)6 (K6) | 0.15 |
| (alanine)6 (A6) | 9.20 |
| (serine)6 (S6) | 0.55 |
| (glutamic acid)6 (E6) | 1.70 |
| K-MB | 1.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, E.; Schultz, C.M.; Lai, R.Y.; Dowben, P.A. Coordination Chemistry of Uranyl Ions with Surface-Immobilized Peptides: An XPS Study. Molecules 2022, 27, 8960. https://doi.org/10.3390/molecules27248960
Mishra E, Schultz CM, Lai RY, Dowben PA. Coordination Chemistry of Uranyl Ions with Surface-Immobilized Peptides: An XPS Study. Molecules. 2022; 27(24):8960. https://doi.org/10.3390/molecules27248960
Chicago/Turabian StyleMishra, Esha, Cody M. Schultz, Rebecca Y. Lai, and Peter A. Dowben. 2022. "Coordination Chemistry of Uranyl Ions with Surface-Immobilized Peptides: An XPS Study" Molecules 27, no. 24: 8960. https://doi.org/10.3390/molecules27248960
APA StyleMishra, E., Schultz, C. M., Lai, R. Y., & Dowben, P. A. (2022). Coordination Chemistry of Uranyl Ions with Surface-Immobilized Peptides: An XPS Study. Molecules, 27(24), 8960. https://doi.org/10.3390/molecules27248960







