Modified Curcumins as Potential Drug Candidates for Breast Cancer: An Overview
Abstract
:1. Introduction
2. Materials and Methods
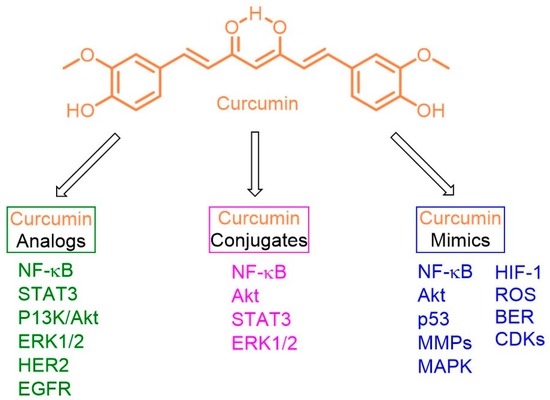
2.1. Anti-Breast Cancer Properties of Curcumin Analogs
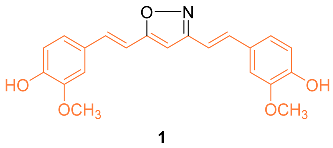
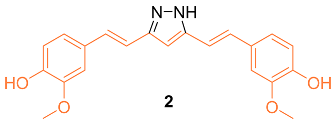
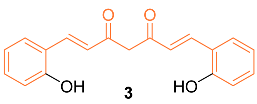
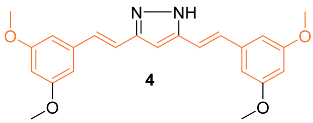


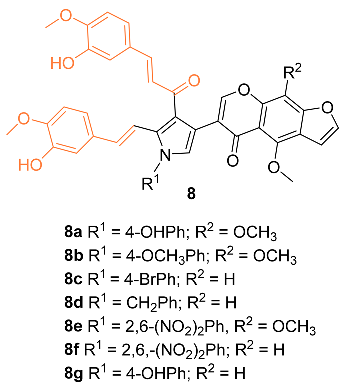

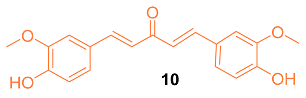
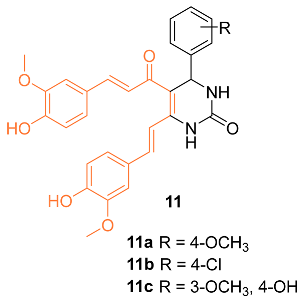
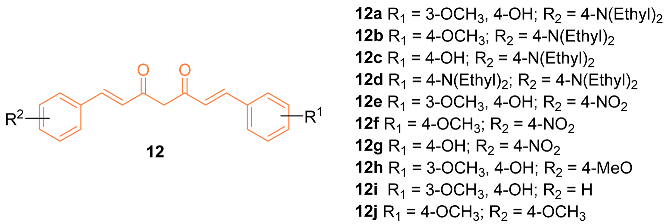
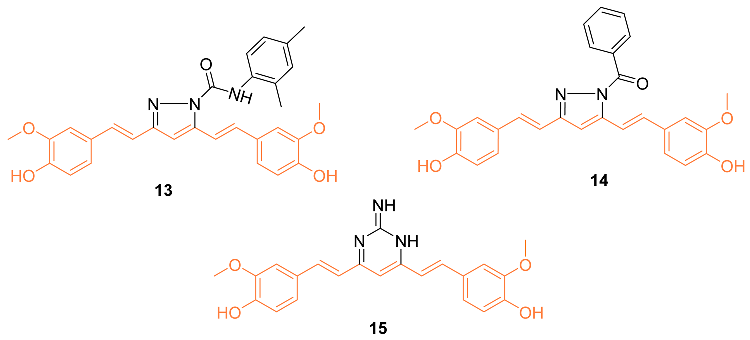
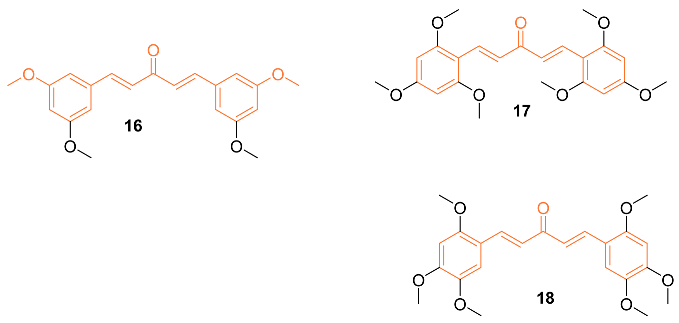
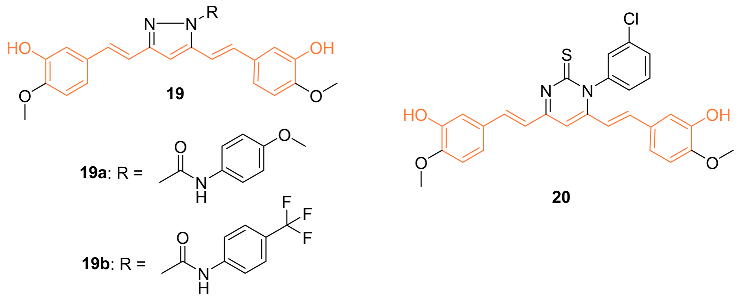
2.2. Anti-Breast Cancer Properties of Curcumin Conjugates
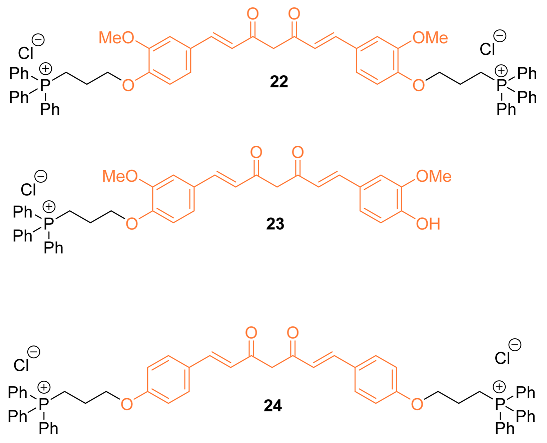
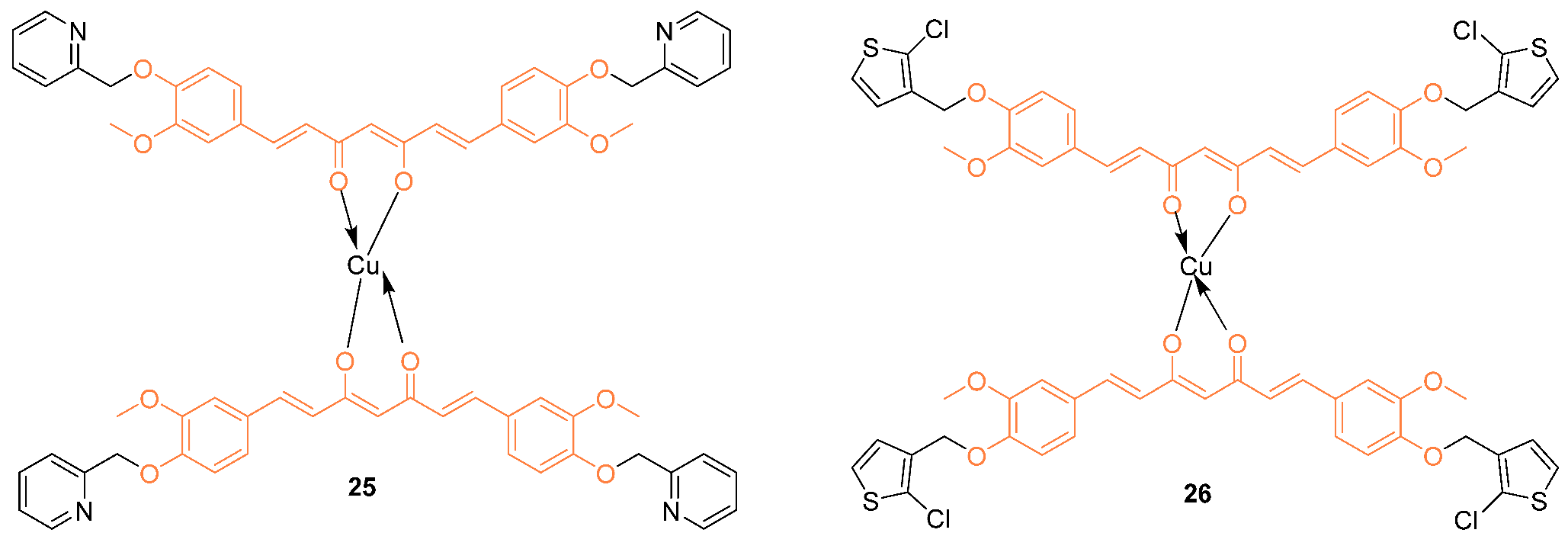
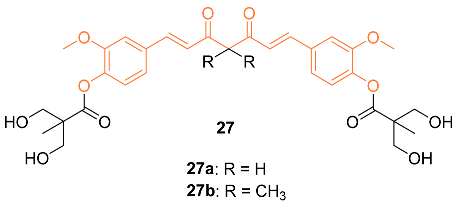

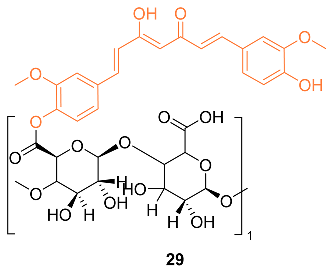
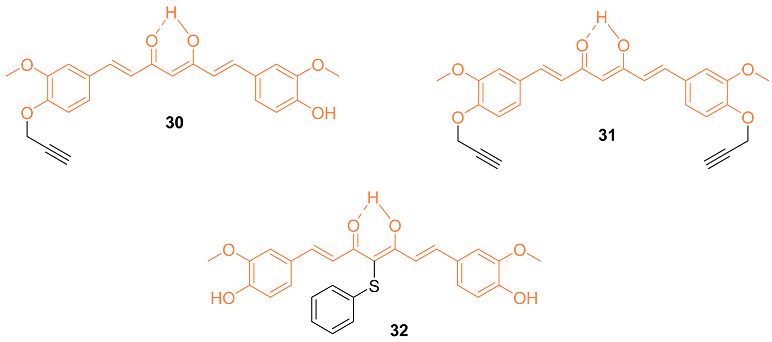




2.3. Anti-Breast Cancer Properties of Curcumin Mimic Conjugates
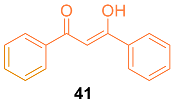
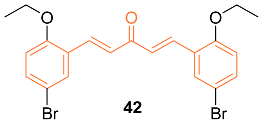
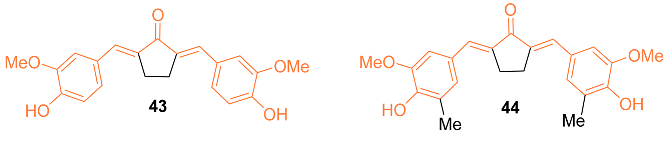
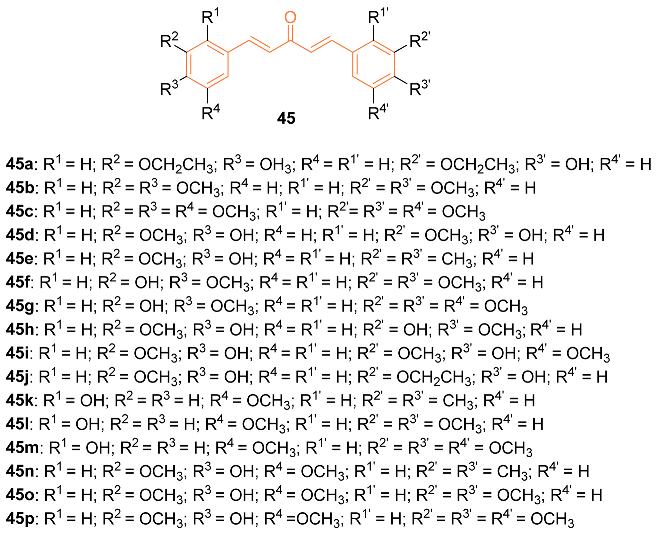
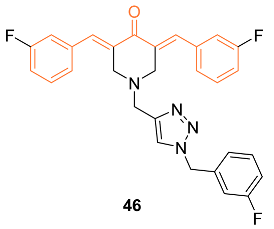
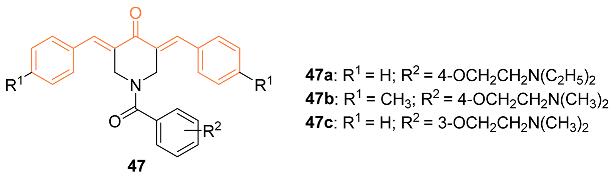
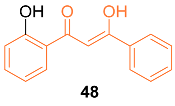


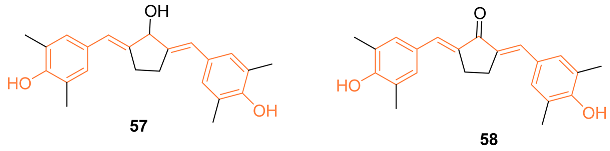
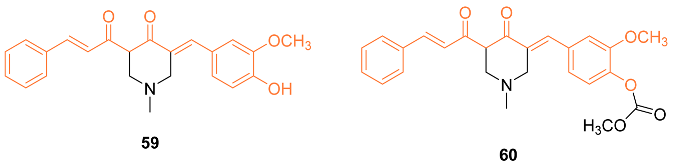
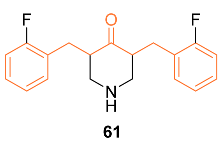
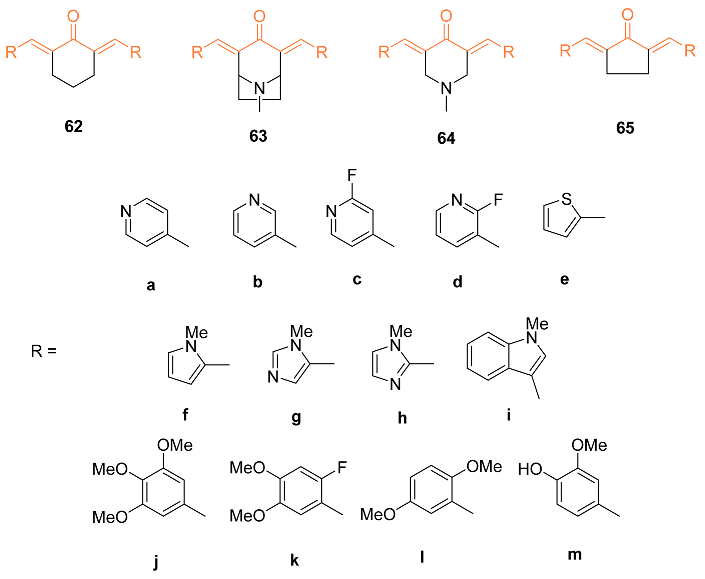
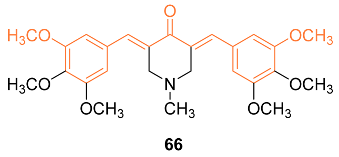
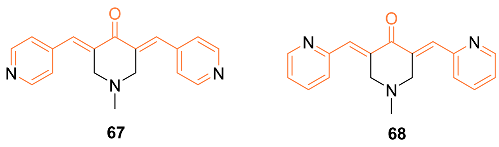

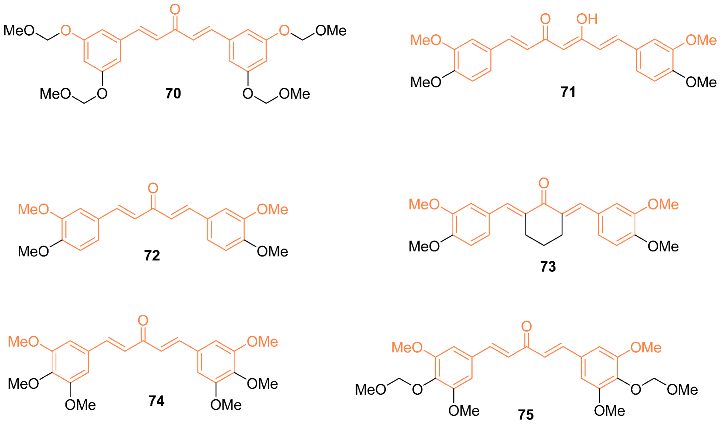

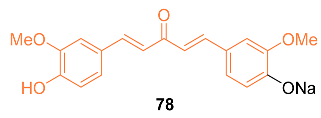
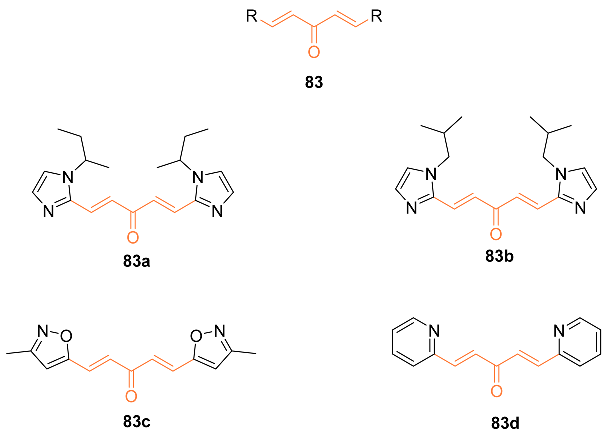
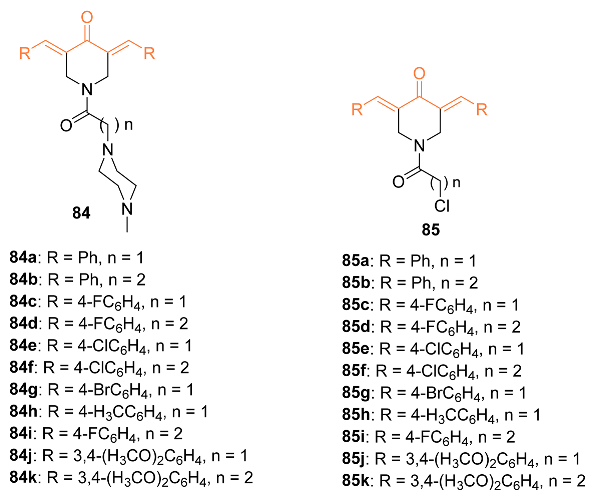
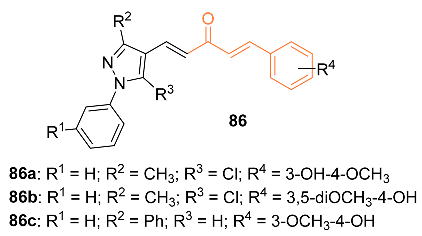

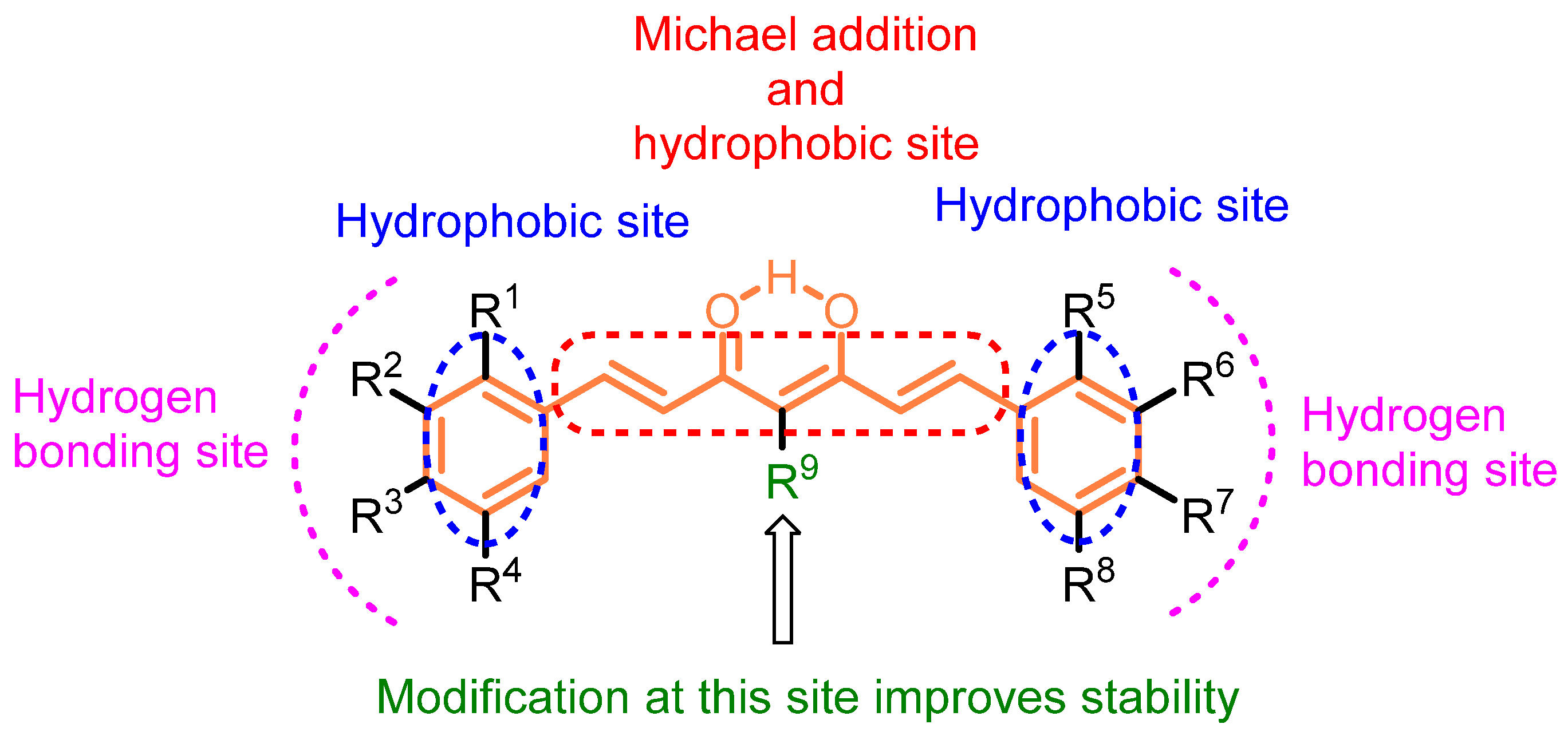
2.4. ADME Properties
3. Clinical Trials
4. Conclusions and Future Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Breast Cancer Facts and Statistics. Available online: https://www.breastcancer.org/facts-statistics (accessed on 11 October 2022).
- Basic Information About Breast Cancer. Available online: https://www.cdc.gov/cancer/breast/basic_info/index.htm#:~:text=About%2042%2C000%20women%20and%20500,breast%20cancer%20than%20White%20women (accessed on 11 October 2022).
- Chakraborty, S.; Rahman, T. The Difficulties in Cancer Treatment. Ecancermedicalscience 2012, 6, 1–5. [Google Scholar]
- Gordaliza, M. Natural Products as leads to anticancer drugs. Clin. Trans. Oncol. 2007, 9, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.S.; Girgis, A.S.; Thomas, S.J.; Capito, J.E.; George, R.F.; Salman, A.; El-Manawaty, M.A.; Samir, A. Synthesis, pharmacological profile and 2D-QSAR studies of curcuminamino acid conjugates as potential drug candidates. Eur. J. Med. Chem. 2020, 196, 112293. [Google Scholar] [CrossRef] [PubMed]
- Vyas, A.; Dandawate, P.; Padhye, S.; Ahmad, A.; Sarkar, F. Perspectives on New Synthetic Curcumin Analogs and their Potential Anticancer Properties. Curr. Pharm. Des. 2013, 19, 2047–2069. [Google Scholar]
- Yin, Y.; Tan, Y.; Wei, X.; Li, X.; Chen, H.; Yang, Z.; Tang, G.; Yao, X.; Mi, P.; Zheng, X. Recent Advances of Curcumin Derivatives in Breast Cancer. Chem. Biodivers. 2022, 19, e202200485. [Google Scholar] [CrossRef]
- Awasthi, M.; Singh, S.; Pandey, V.P.; Dwivedi, U. Curcumin: Structure-Activity Relationship Towards its Role as a Versatile Multi-Targeted Therapeutics. Mini Rev. Org. Chem. 2017, 14, 311–332. [Google Scholar] [CrossRef]
- Moreira, J.; Saraiva, L.; Pinto, M.M.; Cidade, H. Diarylpentanoids with antitumor activity: A critical review of structure-activity relationship studies. Eur. J. Med. Chem. 2020, 192, 112177. [Google Scholar] [CrossRef]
- Gupta, A.P.; Khan, S.; Manzoor, M.M.; Yadav, A.K.; Sharma, G.; Anand, R.; Gupta, S. Chapter 10—Anticancer Curcumin: Natural Analogues and Structure-Activity Relationship. Stud. Nat. Prod. 2017, 54, 355–401. [Google Scholar]
- Rodrigues, F.C.; Kumar, N.A.; Thakur, G. Developments in the anticancer activity of structurally modified curcumin: An up-to-date review. Eur. J. Med. Chem. 2019, 177, 76–104. [Google Scholar] [CrossRef]
- Shehzad, A.; Lee, J.; Lee, Y. Curcumin in various cancers. BioFactors 2013, 39, 56–68. [Google Scholar] [CrossRef]
- Sinha, D.; Biswas, J.; Sung, B.; Aggarwal, B.B.; Bishayee, A. Chemopreventive and chemotherapeutic potential of curcumin in breast cancer. Current Drug Targets 2012, 13, 1799–1819. [Google Scholar] [CrossRef] [PubMed]
- Mock, C.D.; Jordan, B.C.; Selvam, C. Recent advances of curcumin and its analogues in breast cancer prevention and treatment. RSC Adv. 2015, 5, 75575–75588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbese, Z.; Khwaza, V.; Aderibigbe, B.A. Curcumin and its derivatives as potential therapeutic agents in prostate, colon and breast cancers. Molecules 2019, 24, 4386. [Google Scholar] [CrossRef]
- Imran, M.; Ullah, A.; Saeed, F.; Nadeem, M.; Arshad, M.U.; Suleria, H.A.R. Cucurmin, anticancer, & antitumor perspectives: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1271–1293. [Google Scholar] [PubMed]
- Farghadani, R.; Naidu, R. Curcumin as an Enhancer of Therapeutic Efficiency of Chemotherapy Drugs in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 2144. [Google Scholar] [CrossRef] [PubMed]
- Poma, P.; Notarbartolo, M.; Labbozzetta, M.; Maurici, A.; Carina, V.; Alaimo, A.; Rizzi, M.; Simoni, D.; D’Alessandro, N. The antitumor activities of curcumin and of its isoxazole analogue are not affected by multiple gene expression changes in an MDR model of the MCF-7 breast cancer cell line: Analysis of the possible molecular basis. Int. J. Mol. Med. 2007, 20, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, Y.; Zhang, X.; Tian, W.; Feng, W.; Chen, T. The curcumin analogue hydrazinocurcumin exhibits potent suppressive activity on carcinogenicity of breast cancer cells via STAT3 inhibition. Int. J. Oncol. 2012, 40, 1189–1195. [Google Scholar] [CrossRef] [Green Version]
- Mohankumar, K.; Pajaniradje, S.; Sridharan, S.; Singh, V.K.; Ronsard, L.; Banerjea, A.C.; Benson, C.S.; Coumar, M.S.; Rukkumani, R. Mechanism of apoptotic induction in human breast cancer cell, MCF-7, by an analog of curcumin in comparison with curcumin—An in vitro and in silico approach. Chem. Biol. Interact. 2014, 210, 51–63. [Google Scholar] [CrossRef]
- Mohankumar, K.; Sridharan, S.; Pajaniradje, S.; Singh, V.K.; Ronsard, L.; Banerjea, A.C.; Somasundaram, D.B.; Coumar, M.S.; Periyasamy, L.; Rajagopalan, R. BDMC-A, an analog of curcumin, inhibits markers of invasion, angiogenesis, and metastasis in breast cancer cells via NF-kB pathway—A comparative study with curcumin. Biomed. Pharmacother. 2015, 74, 178–186. [Google Scholar] [CrossRef]
- Lien, J.C.; Hung, C.M.; Lin, Y.J.; Lin, H.C.; Ko, T.C.; Tseng, L.C.; Kuo, S.C.; Ho, C.T.; Lee, J.C.; Way, T.D. Pculin02H, a curcumin derivative, inhibits proliferation and clinical drug resistance of HER2-overexpressing cancer cells. Chem. Biol. Interact. 2015, 235, 17–26. [Google Scholar] [CrossRef]
- Elmegeed, G.A.; Yahya, S.M.M.; Abd-Elhalim, M.M.; Mohamed, M.S.; Mohareb, R.M.; Elsayed, G.H. Evaluation of heterocyclic steroids and curcumin derivatives as anti-breast cancer agents: Studying the effect on apoptosis in MCF-7 breast cancer cells. Steroids 2016, 115, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Bhuvaneswari, K.; Sivaguru, P.; Lalitha, A. Synthesis, Biological Evaluation and Molecular Docking of Novel Curcumin Derivatives as Bcl-2 Inhibitors Targeting Human Breast Cancer MCF-7 Cells. ChemistrySelect 2017, 2, 11552–11560. [Google Scholar] [CrossRef]
- Nagwa, M.F.; Sarhan, A.E.; Elhefny, E.A.; Nasef, A.M.; Aly, M.S. Synthesis, Cytotoxicity Evaluation, and Molecular Docking Studies of Novel Pyrrole Derivatives of Khellin and Visnagin via One-Pot Condensation Reaction with Curcumin. Russ. J. Bioorg. Chem. 2020, 46, 1117–1127. [Google Scholar] [CrossRef]
- Hong, J.; Meng, L.; Yu, P.; Zhou, C.; Zhang, Z.; Yu, Z.; Qin, F.; Zhao, Y. Novel drug isolated from mistletoe (1E,4E)-1,7-bis(4-hydroxyphenyl)hepta-1,4-dien-3-one for potential treatment of various cancers: Synthesis, pharmacokinetics and pharmacodynamics. RSC Adv. 2020, 10, 27794–27804. [Google Scholar] [CrossRef]
- Shen, H.; Shen, J.; Pan, H.; Xu, L.; Sheng, H.; Liu, B.; Yao, M. Curcumin analog B14 has high bioavailability and enhances the effect of anti-breast cancer cells in vitro and in vivo. Cancer Sci. 2021, 112, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Jadav, S.S.; Yasmin, S.; Bhatia, S.; Khalilullah, H.; Ahsan, M.J. Simple, efficient, and improved synthesis of Biginelli-type compounds of curcumin as anticancer agents. Med. Chem. Res. 2015, 24, 636–644. [Google Scholar] [CrossRef]
- Zhang, L.; Zong, H.; Lu, H.; Gong, J.; Ma, F. Discovery of novel anti-tumor curcumin analogues from the optimization of curcumin scaffold. Med. Chem. Res. 2017, 26, 2468–2476. [Google Scholar] [CrossRef]
- Ahsan, M.J.; Khalilullah, H.; Yasmin, S.; Jadav, S.S.; Govindasamy, J. Synthesis, Characterisation, and In Vitro Anticancer Activity of Curcumin Analogues Bearing Pyrazole/Pyrimidine Ring Targeting EGFR Tyrosine Kinase. BioMed Res. Int. 2013, 2013, 239354. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, J.R.; Pandit, B.; Bhasin, D.; Etter, J.P.; Regan, N.; Abdelhamid, D.; Li, C.; Lin, J.; Li, P. Structure-activity relationship studies of curcumin analogues. Bioorg. Med. Chem. Lett. 2009, 19, 2065–2069. [Google Scholar] [CrossRef]
- Ali, A.; Ali, A.; Tahir, A.; Bakht, A.M.; Salahuddin; Ahsan, M.J. Molecular Engineering of Curcumin, an Active Constituent of Curcuma longa L. (Turmeric) of the Family Zingiberaceae with Improved Antiproliferative Activity. Plants 2021, 10, 1559. [Google Scholar] [CrossRef]
- Basnet, P.; Hussain, H.; Tho, I.; Skalko-Basnet, N. Liposomal Delivery System Enhances Anti-Inflammatory Properties of Curcumin. J. Pharm. Sci. 2012, 101, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Mehranfar, F.; Bordbar, A.-K.; Fani, N.; Keyhanfar, M. Binding analysis for interaction of diacetylcurcumin with β-caseinnanoparticles by using fluorescence spectroscopy and molecular docking calculations. Spectrochim Acta A Mol. Biomol. Spectrosc. 2013, 115, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.A.; Somepalli, V.; Golakoti, T.; Kanugula, A.K.R.; Karnewar, S.; Rajendiran, K.; Vasagiri, N.; Prabhakar, S.; Kuppusamy, P.; Kotamraju, S.; et al. Mitochondrial-Targeted Curcuminoids: A Strategy to Enhance Bioavailability and Anticancer Efficacy of Curcumin. PLoS ONE 2014, 9, e89351/1–e89351/11. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Wang, J.; Si, G.; Wang, C.; Zhou, S. Synthesis, DNA-binding properties and cytotoxicity evaluation of two copper (II) complexes based on curcumin. Transit. Met. Chem. 2016, 41, 331–337. [Google Scholar] [CrossRef]
- Hsieh, M.T.; Chang, L.C.; Hung, H.Y.; Lin, H.Y.; Shih, M.H.; Tsai, C.H.; Kuo, S.C.; Lee, K.H. New bis(hydroxymethyl) alkanoate curcuminoid derivatives exhibit activity against triple-negative breast cancer in vitro and in vivo. Eur. J. Med. Chem. 2017, 131, 141–151. [Google Scholar] [CrossRef]
- Musib, D.; Pal, M.; Raza, K.M.; Roy, M. Photo-physical, theoretical and photo-cytotoxic evaluation of a new class of lanthanide (III)–curcumin/diketone complexes for PDT application. Dalton Trans. 2020, 49, 10786–10798. [Google Scholar] [CrossRef]
- Bai, F.; Diao, J.; Wang, Y.; Sun, S.; Zhang, H.; Liu, Y.; Wang, Y.; Cao, J. A New Water-Soluble Nano micelle Formed through Self-Assembly of Pectin-Curcumin Conjugates: Preparation, Characterization, and Anticancer Activity Evaluation. J. Agric. Food Chem. 2017, 65, 6840–6847. [Google Scholar] [CrossRef]
- Bonaccorsi, P.M.; Labbozzetta, M.; Barattucci, A.; Salerno, T.M.G.; Poma, P.; Notarbartolo, M. Synthesis of Curcumin Derivatives and Analysis of Their Antitumor Effects in Triple Negative Breast Cancer (TNBC) Cell Lines. Pharmaceuticals 2019, 12, 161. [Google Scholar] [CrossRef] [Green Version]
- Sertel, S.; Eichhorn, T.; Bauer, J.; Hock, K.; Plinkert, P.K.; Efferth, T. Pharmacogenomic determination of genes associated with sensitivity or resistance of tumor cells to curcumin and curcumin derivatives. J. Nutr. Biochem. 2012, 23, 875–884. [Google Scholar] [CrossRef]
- Kesharwani, R.K.; Srivastava, V.; Singh, P.; Rizvi, S.I.; Adeppa, K.; Misra, K. A Novel Approach for Overcoming Drug Resistance in Breast Cancer Chemotherapy by Targeting new Synthetic Curcumin Analogues Against Aldehyde Dehydrogenase 1 (ALDH1A1) and Glycogen SynthaseKinase-3β(GSK-3β). Appl. Biochem. Biotechnol. 2015, 176, 1996–2017. [Google Scholar] [CrossRef]
- Upadhyay, A.; Kundu, P.; Ramu, V.; Kondaiah, P.; Chakravarty, A.R. BODIPY-Tagged Platinum (II) Curcumin Complexes for Endoplasmic Reticulum-Targeted Red Light PDT. Inorg. Chem. 2022, 61, 1335–1348. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.S.; Tran, Q.L.; Rajpurohit, P.; Pillai, G.G.; Thomas, S.J.; Bridges, A.E.; Capito, J.E.; Thangaraju, M.; Lokeshwar, B.L. Design, Synthesis, and Molecular Docking Studies of Curcumin Hybrid Conjugates as Potential Therapeutics for Breast Cancer. Pharmaceuticals 2022, 15, 451. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Ho, C.T.; Huang, M.T. Chemoprevative Effects of Dibenzoylmethane on Mammary Tumorigenesis. ACS Symp. Ser. Am. Chem. Soc. 2008, 987, 281–292. [Google Scholar]
- Lin, C.; Wei, G.J.; Huang, M.T.; Ho, C.T. Metabolic pathway of dibenzoylmethane, a-diketone analogue of curcumin, by NADPH-dependent cytochrome P450 enzymes in of curcumin, by NADPH-dependent cytochrome P450 enzymes in mouse liver microsomes mouse liver microsomes. Yaowu Shipin Fenxi 2005, 13, 284–288. [Google Scholar]
- Liu, Z.; Sun, Y.; Ren, L.; Huang, Y.; Cai, Y.; Weng, Q.; Shen, X.; Li, X.; Liang, G.; Wang, Y. Evaluation of a curcumin analog as an anti-cancer agent inducing ER stress-mediated apoptosis in non-small cell lung cancer cells. BMC Cancer 2013, 13, 494. [Google Scholar] [CrossRef] [PubMed]
- Meiyanto, E.; Putri, D.D.P.; Susidarti, R.A.; Murwanti, R.; Sardjiman, S.; Fitriasari, A.; Husnaa, U.; Purnomo, H.; Kawaichi, M. Curcumin and its Analogues (PGV-0 and PGV-1) Enhance Sensitivity of Resistant MCF-7 Cells to Doxorubicin through Inhibition of HER2 and NF-kB Activation. APJCP 2014, 15, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Chen, J.; Luo, S.; Xu, J.; Huang, Q.; Liu, T. Synthesis and assessment of the antioxidant and antitumor properties of asymmetric curcumin analogues. Eur. J. Med. Chem. 2015, 93, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Mandalapu, D.; Saini, K.S.; Gupta, S.; Sharma, V.; Malik, M.Y.; Chaturvedi, S.; Bala, V.; Thakur, S.; Maikhuri, J.P.; Wahajuddin, M.; et al. Synthesis and biological evaluation of some novel triazole hybrids of curcumin mimics and their selective anticancer activity against breast and prostate cancer cell lines. Bioorg. Med. Chem. Lett. 2016, 26, 4223–4232. [Google Scholar] [CrossRef]
- Robles-Escajeda, E.; Das, U.; Ortega, N.M.; Parra, K.; Francia, G.; Dimmock, J.R.; Varela-Ramirez, A.; Aguilera, R.J. A novel curcumin-like dienone induces apoptosis in triple-negative breast cancer cells. Cell. Oncol. 2016, 39, 265–277. [Google Scholar] [CrossRef] [Green Version]
- Ali, N.M.; Yeap, S.K.; Abu, N.; Lim, K.L.; Ky, H.; Pauzi, A.Z.M.; Ho, W.Y.; Tan, S.W.; Alan-Ong, H.K.; Zareen, S.; et al. Synthetic curcumin derivative DK1 possessed G2/M arrest and induced apoptosis through accumulation of intracellular ROS in MCF-7 breast cancer cells. Cancer Cell Int. 2017, 17, 30/1–30/12. [Google Scholar] [CrossRef] [Green Version]
- Saini, K.S.; Ashraf, R.; Mandalapu, D.; Das, S.; Siddiqui, M.Q.; Dwivedi, S.; Sarkar, J.; Sharma, V.L.; Konwar, R. New Orally Active DNA Minor Groove Binding Small Molecule CT-1 Acts Against Breast Cancer by Targeting Tumor DNA Damage Leading top53-Dependent Apoptosis. Mol. Carcinog. 2017, 56, 1266–1280. [Google Scholar] [CrossRef] [PubMed]
- Badr, G.; Gul, H.I.; Yamali, C.; Mohamed, A.A.; Badr, B.M.; Gul, M.; Markeb, A.A.; El-Maali, N.A. Curcumin analogue 1,5-bis(4-hydroxy-3-((4-methylpiperazin-1-yl)methyl)phenyl)penta-1,4-dien-3-one mediates growth arrest and apoptosis by targeting the PI3K/AKT/mTOR and PKC-theta signaling pathways in human breast carcinoma cells. Bioorg. Chem. 2018, 78, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Zamrus, S.N.H.; Akhtar, M.N.; Yeap, S.K.; Quah, C.K.; Loh, W.-S.; Alitheen, N.B.; Zareen, S.; Tajuddin, S.N.; Hussin, Y.; Shah, S.A.A. Design, synthesis and cytotoxic effects of curcuminoids on HeLa, K562, MCF-7 and MDA-MB-231 cancer cell lines. Chem. Cent. J. 2018, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- De Freitas Silva, M.; Coelho, L.F.; Guirelli, I.M.; Pereira, R.M.; Ferreira-Silva, G.Á.; Graravelli, G.Y.; de Oliveira Horvath, R.; Caixeta, E.S.; Ionta, M.; Viegas, C. Synthetic resveratrol-curcumin hybrid derivative inhibits mitosis progression in estrogen positive MCF-7 breast cancer cells. Toxicol. In Vitro 2018, 50, 75–85. [Google Scholar] [CrossRef]
- Dhongade, S.R.; Chougale, U.B.; Chavan, H.V.; Deshmukh, S.M.; Kharade, P.R.; Kenwade, S.A.; Shetake, P.K. Synthesis and biological evaluation of pyrazole based curcuminanalogues as promising antimicrobial and anticancer agents. R. J. Life Sci. 2019, 5, 1164–1175. [Google Scholar]
- Lin, S.; Zhang, L.Y.; Zhang, X.; Yu, Z.; Huang, X.; Xu, J.; Liu, Y.; Chen, L.; Wu, L. Synthesis of novel dual target inhibitors of PARP and HSP90 and their antitumor activities. Bioorg. Med. Chem. 2020, 28, 115434. [Google Scholar] [CrossRef]
- Novitasari, D.; Jenie, R.I.; Utomo, R.Y.; Kato, J.; Meiyanto, E. CCA-1.1, A Novel Curcumin Analog, Exerts Cytotoxic anti-Migratory Activity toward TNBC and HER2-Enriched Breast Cancer Cells. Asian Pac. J. Cancer Prev. 2021, 22, 1827–1836. [Google Scholar] [CrossRef]
- Meiyanto, E.; Putri, H.; Larasati, Y.A.; Utomo, R.Y.; Jenie, R.I.; Ikawati, M.; Lestari, B.; Yoneda-Kato, N.; Nakamae, I.; Kawaichi, M.; et al. Anti-proliferative and Anti-metastatic Potential of Curcumin Analogue, Pentagamavunon-1 (PGV-1), Toward Highly Metastatic Breast Cancer Cells in Correlation with ROS Generation. Adv. Pharm. Bull. 2019, 9, 445–452. [Google Scholar] [CrossRef] [Green Version]
- Murwanti, R.; Rahmadani, A.; Ritmaleni, R.; Hermawan, A.; Sudarmanto, B.S.A. Curcumin Analogs Induce Apoptosis and G2/M Arrest In 4T1 Murine Triple-Negative Breast Cancer Cells. Indones. J. Pharm. 2020, 31, 11–18. [Google Scholar] [CrossRef]
- Meiyanto, E.; Husnaa, U.; Kastian, R.F.; Putri, H.; Larasati, Y.A.; Khumaira, A.; Pamungkas, D.D.P.; Jenie, R.I.; Kawaichi, M.; Lestari, B.; et al. The Target Differences of Anti-Tumorigenesis Potential of Curcumin and its Analogues Against HER-2 Positive and Triple-Negative Breast Cancer Cells. Adv. Pharm. Bull. 2021, 11, 188–196. [Google Scholar] [CrossRef]
- Kostrzewa, T.; Wolosewicz, K.; Jamrozik, M.; Drzezdzon, J.; Sieminska, J.; Jacewicz, D.; Magdalena, G.-P.; Marcin, K.; Lazny, R.; Kuban-Jankowska, A. Curcumin and Its New Derivatives: Correlation between Cytotoxicity against Breast Cancer Cell Lines, Degradation of PTP1B Phosphatase and ROS Generation. Int. J. Mol. Sci. 2021, 22, 10368. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.K.; Cai, J.; Armstrong, J.; Herold, M.; Lu, Y.J.; Sun, A.; Snyder, J.P.; Liotta, D.C.; Jones, D.P.; Shoji, M. EF24, A novel synthetic curcumin analog, induces apoptosis in cancer cells via a redox-dependent mechanism. Anti-Cancer Drugs 2005, 16, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.L.; Zhong, D.; Zhou, W.; Malik, S.; Liotta, D.; Snyder, J.P.; Hamel, E.; Giannakakou, P. EF24, a novel curcumin analog, disrupts the microtubule cytoskeleton and inhibits HIF-1. Cell Cycle 2008, 7, 2409–2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, B.; Taurin, S.; Rosengren, R.J.; Schumacher, M.; Diederich, M.; Somers-Edgar, T.J.; Larsen, L. Synthesis and cytotoxic potential of heterocyclic cyclohexanone analogues of curcumin. Bioorg. Med. Chem. 2010, 18, 6701–6707. [Google Scholar] [CrossRef]
- Yadav, B.; Taurin, S.; Larsen, L.; Rosengren, R.J. RL71, a second-generation curcumin analog, induces apoptosis and downregulates Akt in ER-negative breast cancer cells. Int. J. Oncol. 2012, 41, 1119–1127. [Google Scholar] [CrossRef] [Green Version]
- Yadav, B.; Taurin, S.; Larsen, L.; Rosengren, R.J. RL66 a second-generation curcumin analog has potent in vivo and in vitro anticancer activity in ER-negative breast cancer models. Int. J. Oncol. 2012, 41, 1723–1732. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Zhu, S.; Weitzmann, M.N.; Snyder, J.P.; Shoji, M. Curcumin analog UBS109 prevents bone marrow osteoblastogenesis and osteoclastogenesis disordered by coculture with breast cancer MDA-MB-231 bone metastatic cells in vitro. Mol. Cell. Biochem. 2015, 401, 1–10. [Google Scholar] [CrossRef]
- Nirgude, S.; Mahadeva, R.; Koroth, J.; Kumar, S.; Kumar, K.S.S.; Gopalakrishnan, V.; Karki, S.S.; Choudhary, B. ST09, A Novel Curcumin Derivative, Blocks Cell Migration by Inhibiting Matrix Metalloproteases in Breast Cancer Cells and Inhibits Tumor Progression in EAC Mouse Tumor Models. Molecules 2020, 25, 4499. [Google Scholar] [CrossRef]
- Ohori, H.; Yamakoshi, H.; Tomizawa, M.; Shibuya, M.; Kakudo, Y.; Takahashi, A.; Takahashi, S.; Kato, S.; Suzuki, T.; Ishioka, C.; et al. Synthesis and biological analysis of new curcumin analogues bearing an enhanced potential for the medicinal treatment of cancer. Mol. Cancer Ther. 2006, 5, 2563–2571. [Google Scholar] [CrossRef] [Green Version]
- Pandya, N.; Khan, E.; Jain, N.; Satham, L.; Singh, R.; Makde, R.D.; Mishra, A.; Kumar, A. Curcumin analogs exhibit anti-cancer activity by selectively targeting G-quadruplex forming c-myc promoter sequence. Biochimie 2021, 180, 205–221. [Google Scholar] [CrossRef]
- Al-Hujaily, E.M.; Mohamed, A.G.; Al-Sharif, I.; Youssef, K.M.; Manogaran, P.S.; Al-Otaibi, B.; Al-Haza’a, A.; Al-Jammaz, I.; Al-Hussein, K.; Aboussekhra, A. PAC, a novel curcumin analogue, has anti-breast cancer properties with higher efficiency on ER-negative cells. Breast Cancer Res. Treat. 2011, 128, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Faiao-Flores, F.; Suarez, J.A.Q.; Pardi, P.C.; Maria, D.A. DM-1, sodium 4-[5-(4-hydroxy-3-methoxyphenyl)-3-oxo-penta-1,4-dienyl]-2-methoxy-phenolate: A curcumin analog with a synergic effect in combination with paclitaxelin breast cancer treatment. Tumor Biol. 2012, 33, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Abd Razak, N.; Akhtar, M.N.; Abu, N.; Ho, W.Y.; Tan, S.W.; Zareen, S.; bin Taj-ud-din, S.N.; Long, K.; Alitheen, N.B.; Yeap, S.K. The in vivo anti-tumor effect of curcumin derivative(2E,6E)-2,6-bis(4-hydroxy-3-methoxybenzylidene)cyclohexanone (BHMC) on 4T1 breast cancer cells. RSC Adv. 2017, 7, 36185–36192. [Google Scholar] [CrossRef] [Green Version]
- Madan, E.; Parker, T.M.; Bauer, M.R.; Dhiman, A.; Pelham, C.J.; Nagane, M.; Kuppusamy, M.L.; Matti, H.; Thomas, R.H.; Shaik, K.; et al. The curcumin analog HO-3867 selectively kills cancer cells by converting mutant p53 protein to transcriptionally active wild type p53. J. Biol. Chem. 2018, 293, 4262–4276. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhao, P.; Yang, L.; Yang, D.; Hu, P.; Li, L.; Cheng, Y.; Yao, H. Reversal of P-glycoprotein-mediated multidrug resistance by novel curcumin analogues in paclitaxel-resistant human breast cancer cells. Biochem. Cell Biol. 2020, 98, 484–491. [Google Scholar] [CrossRef]
- Srour, A.M.; Panda, S.S.; Mostafa, A.; Fayad, W.; El-Manawaty, M.A.; Soliman, A.A.; Moatasim, Y.; El Taweel, A.; Abdelhameed, M.F.; Bekheit, M.S.; et al. Synthesis of aspirin-curcumin mimic conjugates of potential antitumor and anti-SARS-CoV-2 properties. Bioorg. Chem. 2021, 117, 105466. [Google Scholar] [CrossRef]
- Samaan, N.; Zhong, Q.; Fernandez, J.; Chen, G.; Hussain, A.M.; Zheng, S.; Wang, G.; Chen, Q. Design, Synthesis, and evaluation of novel heteroaromatic analogs of curcumin as anti-cancer agents. Eur. J. Med. Chem. 2014, 75, 123–131. [Google Scholar] [CrossRef]
- Youssef, M.A.; Panda, S.S.; Aboshouk, D.R.; Said, M.F.; El Taweel, A.; GabAllah, M.; Fayad, W.; Soliman, A.F.; Mostafa, A.; Fawzy, N.G.; et al. Novel Curcumin Mimics: Design, Synthesis, Biological Properties and Computational Studies of Piperidone-Piperazine Conjugates. ChemistrySelect 2022, 31, e202201406. [Google Scholar] [CrossRef]
- Doan, N.Q.; Nguyen, N.T.; Duong, V.B.; Nguyen, H.T.; Vong, L.B.; Duong, D.N.; Nguyen, N.T.T.; Nguyen, T.L.; Do, T.T.; Truong, T.N. Synthesis, Biological Evaluation, and Molecular Modeling Studies of 1-Aryl-1H-pyrazole-Fused Curcumin Analogues as Anticancer Agents. ACS Omega 2022, 7, 33963–33984. [Google Scholar] [CrossRef] [PubMed]
- Nirgude, S.; Desai, S.; Mahadeva, R.; Ravindran, F.; Choudhary, B. ST08 Altered NF-κB Pathway in Breast Cancer Cells In Vitro as Revealedby miRNA-mRNA Analysis and Enhanced the Effect of Cisplatin on Tumour Reduction in EAC Mouse Model. Front. Oncol. 2022, 12, 835027. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Gomis-Tena, J.; Brown, B.M.; Cano, J.; Trenor, B.; Yang, P.C.; Saiz, J.; Clancy, C.E.; Romero, L. When Does the IC50 Accurately Assess the Blocking Potency of a Drug? J. Chem. Inf. Model. 2020, 60, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Pardridge, W.M. Drug transport across the blood–brain barrier. J. Cereb. Blood Flow Metab. 2012, 32, 1959–1972. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, D.F.; Eggleston, W.D.; Wang, D. Validation and Clinical Utility of the hERG IC50:Cmax Ratio to Determine the Risk of Drug-Induced Torsades de Pointes: A Meta-Analysis. Pharmacotherapy 2018, 38, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://optibrium.com/stardrop/ (accessed on 7 December 2022).
- Salehi, B.; Stojanovic-Radic, Z.; Matejic, J.; Sharifi-Rad, M.; Anil Kumar, N.V.; Martins, N.; Sharifi-Rad, J. The therapeutic potential of curcumin: A review of clinical trials. Eur. J. Med. Chem. 2019, 163, 527–545. [Google Scholar] [CrossRef]
- Available online: https://clinicaltrials.gov (accessed on 7 December 2022).

| Compd. | Cell Lines Tested IC50 (µM) | Reference Molecules IC50 (µM) | Pathway/ Mechanism | Regulation | Cell Cycle Arrest (Phase) | [Ref] |
|---|---|---|---|---|---|---|
| 1 | MCF-7 13.1 ± 1.6 MCF-7R 12.0 ± 2.0 | MCF-7 (CUR) 29.3 ± 1.7 MCF-7R (CUR) 26.2 ± 1.6 | NF-κB STAT3 | Bcl-2↓ Bcl-XL↓ c-IAP-1↓ XIAP↓ NAIP↓ Survivin↓ COX-2↓ | S | [18] |
| 2 | MCF-7 2.56 MDA-MB-231 3.37 | MCF-7 (CUR) 21.22 MDA-MB-231 (CUR) 26.9 | STAT3 | MMP-9↓ MMP-2↓ Mcl-1↓ Cyclin D1↓ c-myc↓ Bcl-XL↓ Bcl-2↓ Survivin↓ VEGF↓ | G1 | [19] |
| 3 | MCF-7 30 | MCF-7 (CUR) 30 | NF-κB P13K/Akt | NF-κB p65↓ c-Rel↓ src↓ COX-2↓ MMP-9↓ TIMP-2↑ VEGF↓ TNFα↓ IL-1β↓ TGF-β↓ IL-8↓ | NA | [21] |
| 4 | NA | NA | HER2 P13K/Akt ERK1/2 | NA | G2/M | [22] |
| 5 | MCF-7 18 | MCF-7 (Tamoxifen) 4 | HER2 | Cyclin D1↓ Survivin↓ Bcl-2↓ CDC2↓ | NA | [23] |
| 6j | MCF-7 15 | NA | NA | Bcl-2↓ | NA | [24] |
| 7i | MCF-7 10 | NA | NA | Bcl-2↓ | NA | [24] |
| 8a | MCF-7 20 * | MCF-7 (DOX) 13 | NA | NA | G2/M | [25] |
| 9 | SKBR3 1.71 ± 0.10 MDA-MB-231 4.49 ± 0.46 MCF-7 9.09 ± 0.46 MDA-MB-453 10.42 ± 0.92 | SKBR3 (Cisplatin) 3.04 ± 0.17 MDA-MB-231 (Cisplatin) 12.81 ± 0.58 MCF-7 (Cisplatin) 11.16 ± 0.60 MDA-MB-453 (Cisplatin) 15.43 ± 0.83 | NA | NA | NA | [26] |
| 10 | MCF-7 8.84 MDA-MB-231 8.31 | MCF-7 (CUR) 16.85 MDA-MB-231 (CUR) 42.01 | NA | p21↑ Cyclin D1↓ CDK2↓ Bax↑ Cyto-C↑ Bcl-2↓ VEGF↓ TIMP1↑ TIMP2↑ | G1 | [27] |
| 12b | MDA-MB-231 4.99 ± 1.02 | MDA-MB-231 (Cisplatin) 6.18 ± 0.79 | NA | NA | NA | [29] |
| 13 | MCF-7 0.317 MDA-MB-231 0.473 HS 578T 0.295 BT-549 0.693 T-47D 0.79 MDA-MB-468 0.248 | NA | EGFR | NA | NA | [30] |
| 14 | MCF-7 0.162 MDA-MB-231 0.404 HS 578T 0.0779 BT-549 0.631 T-47D 0.793 MDA-MB-468 0.276 | NA | EGFR | NA | NA | [30] |
| 15 | MCF-7 1.81 MDA-MB-231 1.45 HS 578T 0.524 BT-549 1.3 T-47D 2.83 | NA | EGFR | NA | NA | [30] |
| 16 | MCF-7 2.7 ± 0.5 MDA-MB-231 1.5 ± 0.1 | MCF-7 (CUR) 21.5 ± 4.7 MDA-MB-231 (CUR) 25.6 ± 4.8 | NA | NA | NA | [31] |
| 17 | MCF-7 0.4 ± 0.1MDA-MB-231 0.6 ± 0.1 | MCF-7 (CUR) 21.5 ± 4.7 MDA-MB-231 (CUR) 25.6 ± 4.8 | NA | NA | NA | [31] |
| 18 | MCF-7 2.4 ± 1.0 MDA-MB-231 2.4 ± 0.4 | MCF-7 (CUR) 21.5 ± 4.7 MDA-MB-231 (CUR) 25.6 ± 4.8 | NA | NA | NA | [31] |
| 19b | MCF-7 1.97 ** MDA-MB-231 3.06 ** HS 578T 5.10 ** BT-549 1.96 ** T-47D 2.81 ** MDA-MB-468 2.30 ** | NA | EGFR | NA | NA | [32] |
| Compd. | Cell Lines Tested IC50 (µM) | Reference Molecules IC50 (µM) | Pathway/ Mechanism | Regulation | Cell Cycle Arrest (Phase) | [Ref] |
|---|---|---|---|---|---|---|
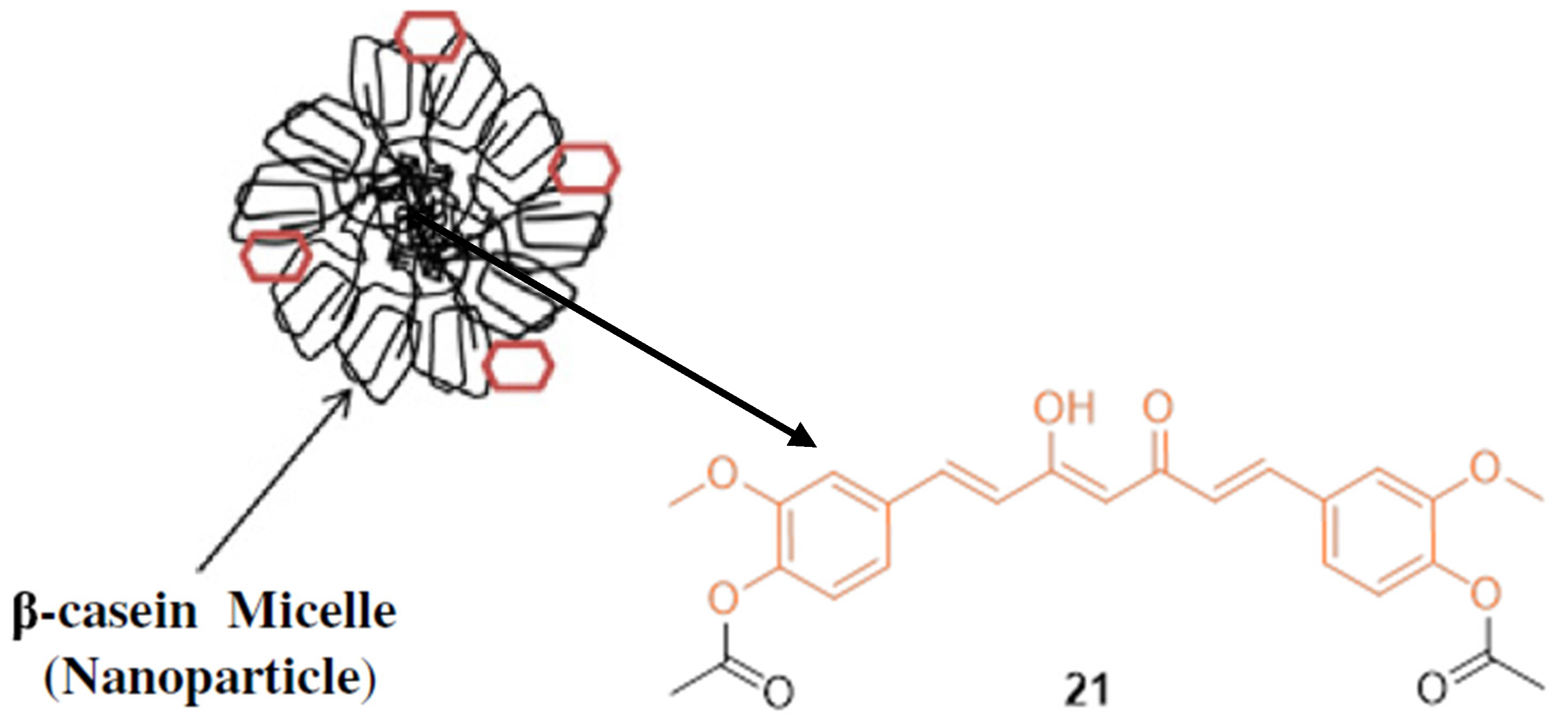
| 21 | MCF-7 22.5 | MCF-7 (DAC) 26.5 | NA | NA | NA | [34] |
| 22 | MCF-7 2.31 MDA-MB-231 5.1 | MCF-7 (CUR) 40.32 MDA-MB-231(CUR) 37.87 | Akt STAT3 ERK1/2 | Cyclin B1↓ Cyclin A↓ Cyclin D1↓ BNIP3↑ | G1/S G2/M | [35] |
| 25 | MCF-7 44.51± 1.74 (24 h) 42.43 ± 1.63 (48 h) | MCF-7 (Cisplatin) 56.24 ± 2.35 (24 h) 45.35 ± 1.59 (48 h) | NA | NA | NA | [36] |
| 26 | MCF-7 49.62 ± 2.23 (24 h) 46.32 ± 1.68 (48 h) | MCF-7 (Cisplatin) 56.24 ± 2.35 (24 h) 45.35 ± 1.59 (48 h) | NA | NA | NA | [36] |
| 27a | MCF-7 6.13 ± 0.51 T47D 19.28 ± 1.71 SKBR3 42.83 ± 1.47 BT474 30.14 ± 1.42 HS-578T 55.45 ± 1.39 MDA-MB-157 40.38 ± 3.26 MDA-MB-453 42.89 ± 2.35 | MCF-7(CUR) 42.89 ± 2.36 T47D(CUR) 47.91 ± 3.90 SKBR3(CUR) 6.39 ± 0.43 BT474(CUR) 6.15 ± 0.87 HS-578T(CUR) 7.96 ± 0.27 MDA-MB-157(CUR) 9.23 ± 0.11 MDA-MB-453(CUR) 6.13 ± 0.51 | NA | HO-1 | G2/M | [37] |
| 28 | MCF-7 >100 (dark) 1.9 ± 1.2 (light) | MCF-7 (Cisplatin) 28.0 ± 3.1 (Dark) ND (Light) | NA | NA | NA | [38] |
| 29 | MCF-7 12.0 ± 3.0 | MCF-7 (CUR) 48.3 ± 2.9 | NA | NA | NA | [39] |
| 30 | SUM 149 11.2 ± 1.30 MDA-MB-231 18.0 ± 0.41 | SUM 149 (CUR) 14.0 ± 0.29 MDA-MB-231 (CUR) 25.5 ± 0.35 | NF-κB | NF-κB Activity↓ | NA | [40] |
| 31 | SUM 149 13.2 ± 1.59 MDA-MB-231 20.0 ± 0.00 | SUM 149 (CUR) 14.0 ± 0.29 MDA-MB-231 (CUR) 25.5 ± 0.35 | NF-κB | NF-κB Activity↓ | NA | [40] |
| 32 | SUM 149 13.5 ± 0.88 MDA-MB-231 15.0 ± 0.85 | SUM 149 (CUR) 14.0 ± 0.29 MDA-MB-231 (CUR) 25.5 ± 0.35 | NF-κB | NF-κB Activity↓ | NA | [40] |
| 38a | MDA-MB-231 63.3 ± 0.1 (dark) 15.6 ± 0.5 (light) | NA | NA | NA | G1 | [43] |
| 38b | MDA-MB-231 57.3 ± 0.6 (dark) 11.3 ± 0.5 (light) | NA | NA | NA | G1 | [43] |
| 38c | MDA-MB-231 49.6 ± 0.8 (dark) 9.5 ± 0.3 (light) | NA | NA | NA | G1 | [43] |
| Compd | Cell Lines Tested IC50 (µM) | Reference Molecules IC50 (µM) | Pathway/ Mechanism | Regulation | Cell Cycle Arrest (Phase) | [Ref] |
|---|---|---|---|---|---|---|
| 41 | MCF-7 | NA | Cytochrome P450 | Bcl-2↓ c-myc↓ Ha-ras↓ hTERT↓ | NA | [46] |
| 42 | MCF-7 2.21 + 0.21 | MCF-7 (CUR) 8.98 ± 3.21 | NA | CHOP↑ Bcl-2↓ Cyclin D1↓ COX-2↓ | G1/S | [47] |
| 43 | MCF-7 60 ± 2.04 MCF-7/Dox 21 ± 0.008 | MCF-7 (CUR) 109 ± 1.915 MCF-7/Dox (CUR) 80 ± 2.39 MCF-7 (DOX) 0.4 MCF-7/Dox (DOX) 7 | NA | NA | NA | [48] |
| 44 | MCF-7 6 ± 2.02 MCF-7/Dox 82 ± 3.09 | MCF-7 (CUR) 109 ± 1.915 MCF-7/Dox (CUR) 80 ± 2.39 MCF-7 (DOX) 0.4 MCF-7/Dox (DOX) 7 | NF-κB | NF-κB↓ p65↓ HER2↓ | G2/M | [48] |
| 45e | MCF-7 22.53 | MCF-7 (CUR) 70.20 | NA | NA | NA | [49] |
| 45p | MCF-7 9.11 | MCF-7 (CUR) 70.20 | NA | NA | NA | [49] |
| 46 | MCF-7 6.0 ± 1.0 MDA-MB-231 10.0 ± 1.6 4T1 6.4 ± 1.9 | MCF-7 (CUR) 83.1 ± 4.4 MDA-MB-231 (CUR) 75.3 ± 2.8 4T1 (CUR) 49.4 ± 1.4 MCF-7 (DOX) 0.13 ± 0.22 MDA-MB-231 (DOX) 1.6 ± 0.23 4T1 (DOX) 0.99 ± 0.13 MCF-7 (5-FU) 15 ± 0.23 MDA-MB-231 (5-FU) 19 ± 2.51 4T1 (5-FU) 23 ± 3.52 MCF-7(Nocodazole) 0.42 ± 0.32 MDA-MB-231 (Nocodazole) 1.1 ± 1.2 4T1 (Nocodazole) 1.3 ± 0.23 | Akt | Akt Phosphorylation↓ PCNA↓ BAX↑ Bcl-2↓ | G0/G1 S | [50] |
| 47a | MDA-MB-231 0.91 MDA-MB-231-LM 0.90 HCC 1419 1.67 MCF-7 1.64 MCF-10A 41.03 | MDA-MB-231 (5-FU) 7.85 MDA-MB-231-LM (5-FU) 6.03 HCC 1419 (5-FU) N/D MCF-7(5-FU) 1.7 MCF-10A (5-FU) N/D | NA | NA | NA | [51] |
| 47b | MDA-MB-231 0.72 MDA-MB-231-LM 0.53 HCC 1419 1.7 MCF-7 4.4 MCF-10A 80.02 | MDA-MB-231 (5-FU) 7.85 MDA-MB-231-LM (5-FU) 6.03 HCC 1419 (5-FU) N/D MCF-7(5-FU) 1.7 MCF-10A (5-FU) N/D | NA | NA | NA | [51] |
| 47c | MDA-MB-231 0.88 MDA-MB-231-LM 1.11 HCC 1419 1.46 MCF-7 1.83 MCF-10A 79.65 | MDA-MB-231 (5-FU) 7.85 MDA-MB-231-LM (5 FU) 6.03 HCC 1419 (5-FU) N/D MCF-7(5-FU) 1.7 MCF-10A (5-FU) N/D | Intrinsic apoptotic pathway | Caspase-3↑ Phosphatidylserine externalization↑ Mitochondrial depolarization↑ | G2/M | [51] |
| 48 | MCF-7 25.00 ± 3.71 MDA-MB-231 37.50 ± 4.82 | MCF-7 (CUR) 30.15 ± 2.36 MDA-MB-231 (CUR) 21.72 ± 3.18 | ROS accumulation | ROS↑ p53↑ | G2/M | [52] |
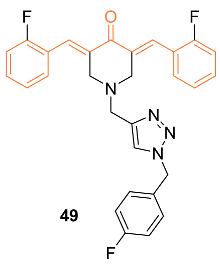
| 49 | MCF-7 2.1 LA-7 3.1 | NA | p53 | p53↑ Caspase-3↑ γH2AX foc↑ | S | [53] |
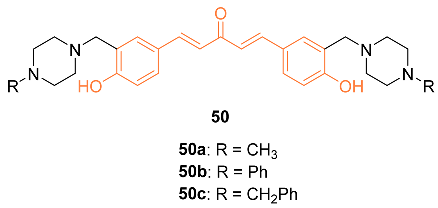
| 50a | MCF-7 10 ± 0 * MDA-MB-231 5 ± 0 * | NA | PI3K/Akt/mTOR NF-κB | Akt↓ mTOR↓ PKC-theta↓ Cyclin A↓ Cyclin D1↓ CDK2↓ Cyclin B1↑ | G2/M | [54] |
| 51d | MCF-7 8.70 ± 3.10 MDA-MB-231 2.30 ± 1.60 | MCF-7 (CUR) 22.50 ± 5.50 MDA-MB-231 (CUR) 26.50 ± 1.40 | NA | NA | NA | [55] |
| 52d | MCF-7 3.02 ± 1.20 MDA-MB-231 1.52 ± 0.60 | MCF-7 (CUR) 22.50 ± 5.50 MDA-MB-231 (CUR) 26.50 ± 1.40 | NA | NA | NA | [55] |
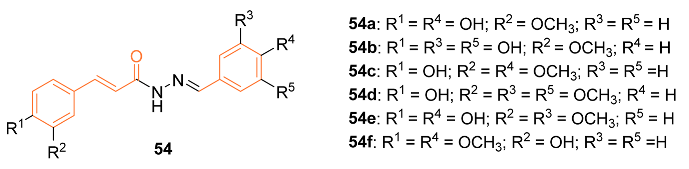
| 54c | MCF-7 40.49 ± 1.01 | MCF-7 (CUR) 68.25 ± 2.59 | CDKs | CCNB1↓ CDKN1A↑ p21↑ | G2/M | [56] |
| 55i | MCF-7 28.75 ± 0 | MCF-7 (Paclitaxel) 0.35 ± 0 | NA | NA | NA | [57] |
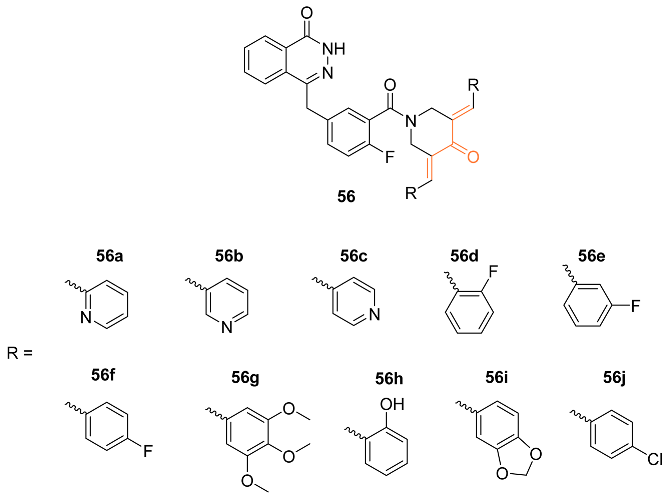
| 56a | MCF-7 1.47 ± 1.49 | MCF-7 (Olaparib) 232 ± 5.50 | BER | PARP↓ | NA | [58] |
| 56b | HCC1937 5.3 ± 3.21 | HCC1937 (Olaparib) 83.1 ± 8.99 | BER | PARP↓ | NA | [58] |
| 56c | MCF-7 0.97 ± 0.13 | MCF-7 (Olaparib) 232 ± 5.50 | BER | PARP↓ HSP90↓ BRCA↓ | NA | [58] |
| 57 | MCF-7/HER2+ 4.0 ± 0 4T1 1.0 ± 0 | MCF-7/HER2+ (58) 90 ± 0 4T1 (58) 2.0 ± 0 | MMPs | MMP-9↓ | NA | [59] |
| 58 | 4T1 38.21 ± 0 ** | 4T1 (CUR) 34.34 ± 0 ** | ROS | NA | G2/M | [61] |
| 59 | MCF-7 25.30 ± 2.54 MDA-MB-231 31.86 ± 1.07 | MCF-7 (CUR) 37.36 ± 1.88 MDA-MB-231 (CUR) 57.07 ± 6.23 | PTEN-Akt/pAkt ROS | NA | NA | [63] |
| 60 | MCF-7 30.51 ± 4.11 MDA-MB-231 11.80 ± 1.43 | MCF-7 (CUR) 37.36 ± 1.88 MDA-MB-231 (CUR) 57.07 ± 6.23 | PTEN-Akt/pAkt ROS | PTP1B↓ | NA | [63] |
| 61 | MDA-MB-231 | NA | HIF-1 | HIF-1α↓ | NA | [65] |
| 63a | MDA-MB-231 0.8 ± 0 *** MDA-MB-468 0.5 ± 0 SKBr3 0.6 ± 0 *** | MDA-MB-231 (CUR) 7.6 ± 0 *** MDA-MB-468 (CUR) 9.7 ± 0 *** SKBr3 (CUR) 2.4 ± 0 *** | NF-κB | NF-κB↓ | NA | [66] |
| 63j | MDA-MB-231 0.3 ± 0 *** MDA-MB-468 0.3 ± 0 SKBr3 0.4 ± 0 *** | MDA-MB-231 (CUR) 7.6 ± 0 *** MDA-MB-468 (CUR) 9.7 ± 0 *** SKBr3(CUR) 2.4 ± 0 *** | NF-κB | NF-κB↓ | NA | [66] |
| 64a | MDA-MB-231 1.1 ± 0 *** MDA-MB-468 0.6 ± 0 *** SKBr3 0.7 ± 0 *** | MDA-MB-231 (CUR) 7.6 ± 0 *** MDA-MB-468 (CUR) 9.7 ± 0 *** SKBr3 (CUR) 2.4 ± 0 *** | NF-κB | NF-κB↓ | NA | [66] |
| 66 | MDA-MB-231 0.3 ± 0 MDA-MB-468 0.3 ± 0 | NA | NF-κB MAPK p38/JNK | Akt↓ HER2/neu↓ p27↑ | G2/M | [67] |
| 67 | MDA-MB-231 0.8 ± 0 MDA-MB-468 0.5 ± 0 SKBr3 0.6 ± 0 | NA | NF-κB Akt MAPK | HER2/neu↓ p27↑ Akt↓ mTOR↓ NF-κB↓ | G2/M S | [68] |
| 68 | MDA-MB-231 | NA | MAPK/ERKNF-κB | TNF-α↑ | NA | [69] |
| 69 | MDA-MB-231 0.031 ± 0 MCF-7 0.093 ± 0 T47D 0.138 ± 0 | MDA-MB-231 (CUR) 10.53 ± 0 MCF-7 (CUR) 13.95 ± 0 T47D (CUR) 10.17 ± 0 | Intrinsic apoptotic pathway MMPs | MMP1,2↓ Apaf↑ Cytochrome C↑ BAX↑ BAD↑ Bcl-2↓ | G2/M | [70] |
| 72 | HCT116 MCF-7 | NA | Caspase-3-dependent pathway | NA | NA | [71] |
| 77 | MCF-7 MDA-MB231 MCF-10A T-47D | NA | NF-κB | Bcl-2↓ Cyclin D1↓ P21WAF1↑ | G2/M | [72] |
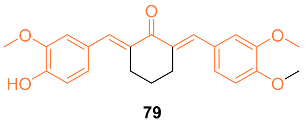
| 79 | 4T1 13.66 ± 3.24 | 4T1 (CUR) 27.15 ± 2.36 | p53 | NA | NA | [75] |
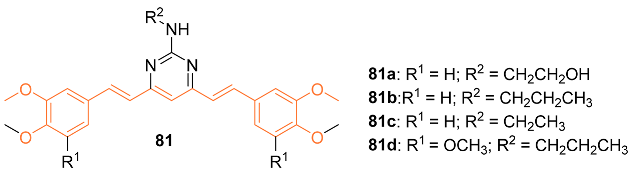
| 81c | MDA435/LCC6 70.5 ± 8.4 MDA435/LCC6MDR 59 ± 3.4 | MDA435/LCC6 (CUR) 19.8 ± 3.4 MDA435/LCC6MDR (CUR) 18.0 ± 2.2 | NA | NA | NA | [77] |
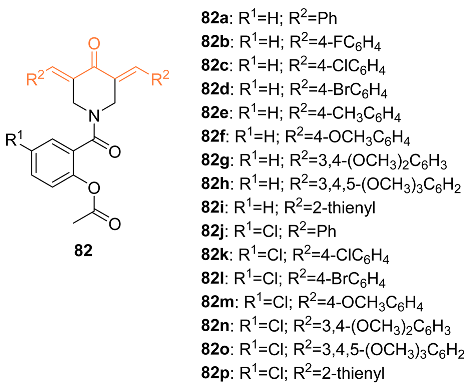
| 82c | MCF-7 2.806 ± 0.26 | MCF-7 (CUR) 16.00 ± 2.04 MCF-7 (Sunitinib) 3.97 ± 0.32 MCF-7 (5-FU) 3.15 ± 0.44 | NA | NA | G1 | [78] |
| 82o | MCF-7 2.653 ± 0.22 | MCF-7 (CUR) 16.00 ± 2.04 MCF-7 (Sunitinib) 3.97 ± 0.32 MCF-7 (5-FU) 3.15 ± 0.44 | NA | NA | S | [78] |
| 83a | MDA-MB-231 0.13 | MDA-MB-231 (CUR) 0.88 | NA | NA | NA | [79] |
| 83b | MDA-MB-231 0.15 | MDA-MB-231 (CUR) 0.88 | NA | NA | NA | [79] |
| 83c | MDA-MB-231 0.156 | MDA-MB-231 (CUR) 0.88 | NA | NA | NA | [79] |
| 83d | MDA-MB-231 0.097 | MDA-MB-231 (CUR) 0.88 | NA | NA | NA | [79] |
| 84e | MCF-7 1.148 ± 0.02 | MCF-7 (CUR) 16.00 ± 2.04 MCF-7 (5-FU) 3.15 ± 0.44 | NA | NA | G1/S | [80] |
| 86a | MDA-MB-231 5.61 ± 0.24 | MDA-MB-231 (CUR) 20.65 ± 0.80 MDA-MB-231 (Paclitaxel) 8.79 ± 0.96 | NA | NA | G2/M | [81] |
| 86b | MDA-MB-231 6.60 ± 0.16 | MDA-MB-231 (CUR) 20.65 ± 0.80 MDA-MB-231 (Paclitaxel) 8.79 ± 0.96 | NA | NA | G2/M | [81] |
| 86c | MDA-MB-231 5.50 ± 0.22 | MDA-MB-231 (CUR) 20.65 ± 0.80 MDA-MB-231 (Paclitaxel) 8.79 ± 0.96 | NA | NA | G2/M | [81] |
| 87 | MDA-MB-231 MCF-7 | MCF-7 (Cisplatin) 8.73 MCF-7 (Olaprib) 11.34 MCF-7 (DOX) 8.8 MDA-MB-231 (Cisplatin) 15.91 MDA-MB-231 (Olaprib) 11 MDA-MB-231 (DOX) 0.7 | NF-κB | TSG↑ | G2/M | [82] |
| Compound | BBB | hERG pIC50 | MW | HBA | HBD | Rotatable Bonds | TPSA | LogP |
|---|---|---|---|---|---|---|---|---|
| 1 | − | 5.360 | 365.4 | 6 | 2 | 6 | 84.95 | 3.501 |
| 2 | − | 4.893 | 364.4 | 6 | 3 | 6 | 87.60 | 2.561 |
| 3 | − | 4.732 | 308.3 | 4 | 2 | 6 | 74.60 | 1.873 |
| 4 | − | 4.865 | 392.4 | 6 | 1 | 8 | 65.60 | 3.877 |
| 6j | − | 5.149 | 568.6 | 10 | 4 | 10 | 173.30 | 3.733 |
| 7i | − | 5.303 | 583.6 | 9 | 3 | 11 | 146.30 | 5.248 |
| 8a | − | 6.298 | 727.7 | 12 | 3 | 11 | 163.00 | 5.107 |
| 9 | + | 5.446 | 294.3 | 3 | 2 | 6 | 57.53 | 3.904 |
| 10 | − | 4.853 | 326.3 | 5 | 2 | 6 | 75.99 | 2.931 |
| 11a | − | 4.546 | 528.6 | 9 | 4 | 9 | 126.30 | 3.892 |
| 12b | + | 5.690 | 377.5 | 4 | 0 | 10 | 46.61 | 3.173 |
| 13 | − | 5.679 | 511.6 | 8 | 3 | 9 | 105.8 | 4.547 |
| 14 | − | 5.669 | 468.5 | 7 | 2 | 8 | 93.81 | 3.986 |
| 15 | − | 5.169 | 391.4 | 7 | 4 | 6 | 111.5 | 2.837 |
| 16 | − | 4.792 | 354.4 | 5 | 0 | 8 | 53.99 | 3.724 |
| 17 | − | 4.835 | 414.4 | 7 | 0 | 10 | 72.45 | 3.092 |
| 18 | − | 5.113 | 414.4 | 7 | 0 | 10 | 72.45 | 3.051 |
| 19b | − | 6.025 | 551.5 | 8 | 3 | 10 | 105.80 | 4.566 |
| 20 | − | 5.877 | 519.0 | 6 | 2 | 7 | 76.74 | 5.504 |
| 22 | − | 7.561 | 977.1 | 6 | 0 | 24 | 71.06 | 9.130 |
| 23 | − | 6.396 | 672.7 | 6 | 1 | 16 | 82.06 | 5.806 |
| 24 | + | 7.823 | 917.1 | 4 | 0 | 22 | 52.60 | 9.534 |
| 27a | − | 3.786 | 600.6 | 12 | 4 | 18 | 186.10 | 1.031 |
| 30 | − | 4.946 | 406.4 | 6 | 2 | 10 | 85.22 | 4.180 |
| 31 | − | 5.088 | 444.5 | 6 | 1 | 13 | 74.22 | 3.840 |
| 32 | − | 5.033 | 476.5 | 6 | 3 | 9 | 96.22 | 4.954 |
| 33 | − | 5.647 | 915.9 | 18 | 6 | 24 | 244.40 | 2.960 |
| 34 | − | 5.520 | 860.0 | 17 | 3 | 24 | 226.50 | 2.589 |
| 35 | − | 5.081 | 862.0 | 18 | 3 | 24 | 235.70 | 2.491 |
| 36 | − | 5.965 | 969.3 | 11 | 2 | 24 | 146.30 | 5.476 |
| 37e | − | 4.652 | 340.3 | 6 | 4 | 6 | 115.10 | 1.204 |
| 39 | − | 5.113 | 596.3 | 8 | 1 | 16 | 108.40 | 4.142 |
| 40a | − | 3.768 | 710.4 | 12 | 3 | 22 | 166.60 | 2.543 |
| 41 | + | 4.533 | 224.3 | 2 | 1 | 3 | 37.30 | 3.845 |
| 42 | + | 5.647 | 480.2 | 3 | 0 | 8 | 35.53 | 6.265 |
| 43 | − | 5.084 | 352.4 | 5 | 2 | 4 | 75.99 | 3.335 |
| 44 | − | 4.944 | 380.4 | 5 | 2 | 4 | 75.99 | 4.032 |
| 45o | − | 4.859 | 370.4 | 6 | 1 | 8 | 74.22 | 3.093 |
| 46 | − | 7.147 | 500.5 | 5 | 0 | 6 | 51.02 | 4.921 |
| 47c | − | 6.049 | 452.5 | 5 | 0 | 7 | 49.85 | 4.230 |
| 48 | + | 4.280 | 240.3 | 3 | 2 | 3 | 57.53 | 4.056 |
| 49 | − | 7.342 | 500.5 | 5 | 0 | 6 | 51.02 | 4.921 |
| 50a | − | 7.332 | 490.6 | 7 | 2 | 8 | 70.49 | 2.440 |
| 52d | − | 5.120 | 354.4 | 5 | 0 | 8 | 53.99 | 3.660 |
| 54c | − | 4.387 | 326.3 | 6 | 2 | 7 | 80.15 | 2.385 |
| 55i | − | 5.220 | 438.9 | 6 | 0 | 8 | 62.58 | 4.713 |
| 56c | − | 5.919 | 557.6 | 8 | 1 | 6 | 108.90 | 3.110 |
| 57 | + | 5.156 | 350.5 | 3 | 3 | 2 | 60.69 | 4.712 |
| 58 | + | 4.901 | 348.4 | 3 | 2 | 2 | 57.53 | 5.092 |
| 59 | − | 5.341 | 377.4 | 5 | 1 | 5 | 66.84 | 1.800 |
| 60 | − | 5.299 | 435.5 | 7 | 0 | 8 | 82.14 | 1.829 |
| 61 | + | 5.599 | 315.4 | 2 | 1 | 4 | 29.10 | 3.650 |
| 63j | − | 5.494 | 495.6 | 8 | 0 | 8 | 75.69 | 3.279 |
| 66 | − | 5.338 | 469.5 | 8 | 0 | 8 | 75.69 | 2.592 |
| 67 | + | 4.980 | 291.3 | 4 | 0 | 2 | 46.09 | 1.628 |
| 68 | + | 5.001 | 291.3 | 4 | 0 | 2 | 46.09 | 1.628 |
| 69 | − | 5.903 | 742.5 | 6 | 0 | 7 | 74.76 | 6.241 |
| 71 | − | 4.918 | 396.4 | 6 | 0 | 10 | 71.06 | 2.089 |
| 72 | − | 5.000 | 354.4 | 5 | 0 | 8 | 53.99 | 3.724 |
| 77 | − | 5.437 | 381.4 | 6 | 2 | 4 | 79.23 | 2.570 |
| 78 | − | 4.911 | 348.3 | 5 | 1 | 7 | 64.99 | 3.084 |
| 79 | − | 5.264 | 380.4 | 5 | 1 | 5 | 64.99 | 3.962 |
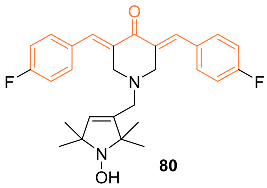
| 80 | + | 6.944 | 464.5 | 4 | 1 | 4 | 43.78 | 4.737 |
| 81c | − | 5.785 | 447.5 | 7 | 1 | 10 | 74.73 | 4.591 |
| 82c | − | 5.654 | 465.5 | 5 | 0 | 6 | 63.68 | 5.017 |
| 83a | − | 5.451 | 326.4 | 5 | 0 | 8 | 52.71 | 3.689 |
| 83b | − | 5.343 | 326.4 | 5 | 0 | 8 | 52.71 | 3.689 |
| 83c | − | 4.566 | 244.2 | 5 | 0 | 4 | 69.13 | 2.819 |
| 83d | + | 4.399 | 236.3 | 3 | 0 | 4 | 42.85 | 1.908 |
| 84e | + | 6.533 | 484.4 | 5 | 0 | 5 | 43.86 | 3.825 |
| 86b | − | 5.151 | 424.9 | 6 | 1 | 7 | 73.58 | 4.329 |
| 87 | − | 5.462 | 784.7 | 18 | 0 | 11 | 258.00 | 3.740 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flint, A.L.; Hansen, D.W.; Brown, L.D.; Stewart, L.E.; Ortiz, E.; Panda, S.S. Modified Curcumins as Potential Drug Candidates for Breast Cancer: An Overview. Molecules 2022, 27, 8891. https://doi.org/10.3390/molecules27248891
Flint AL, Hansen DW, Brown LD, Stewart LE, Ortiz E, Panda SS. Modified Curcumins as Potential Drug Candidates for Breast Cancer: An Overview. Molecules. 2022; 27(24):8891. https://doi.org/10.3390/molecules27248891
Chicago/Turabian StyleFlint, Abigail L., David W. Hansen, LaVauria D. Brown, Laura E. Stewart, Eduardo Ortiz, and Siva S. Panda. 2022. "Modified Curcumins as Potential Drug Candidates for Breast Cancer: An Overview" Molecules 27, no. 24: 8891. https://doi.org/10.3390/molecules27248891
APA StyleFlint, A. L., Hansen, D. W., Brown, L. D., Stewart, L. E., Ortiz, E., & Panda, S. S. (2022). Modified Curcumins as Potential Drug Candidates for Breast Cancer: An Overview. Molecules, 27(24), 8891. https://doi.org/10.3390/molecules27248891