Oligomer Formation by Amyloid-β42 in a Membrane-Mimicking Environment in Alzheimer’s Disease
Abstract
1. Introduction
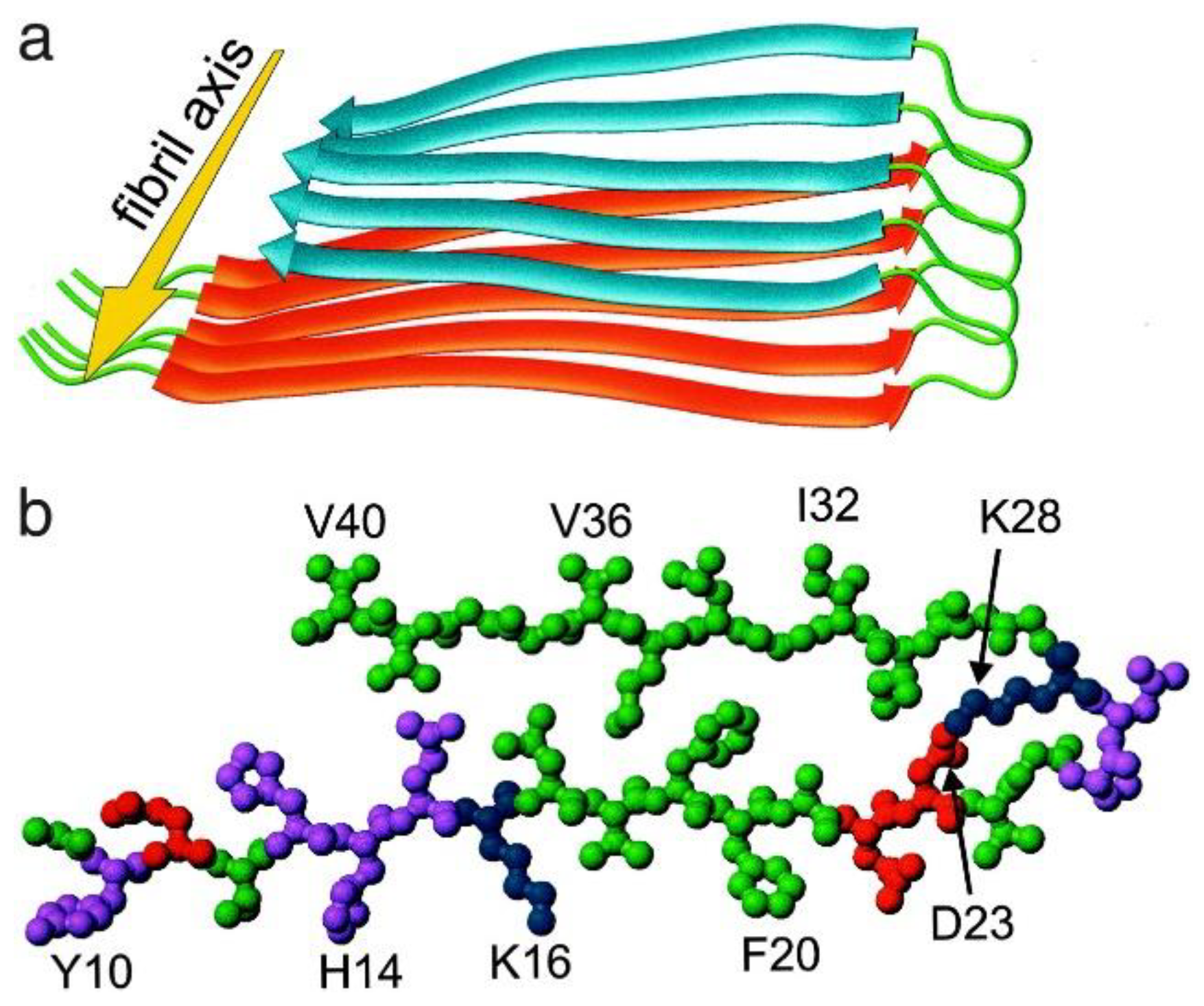
2. A Diverse Group of Amyloid-β Aggregates
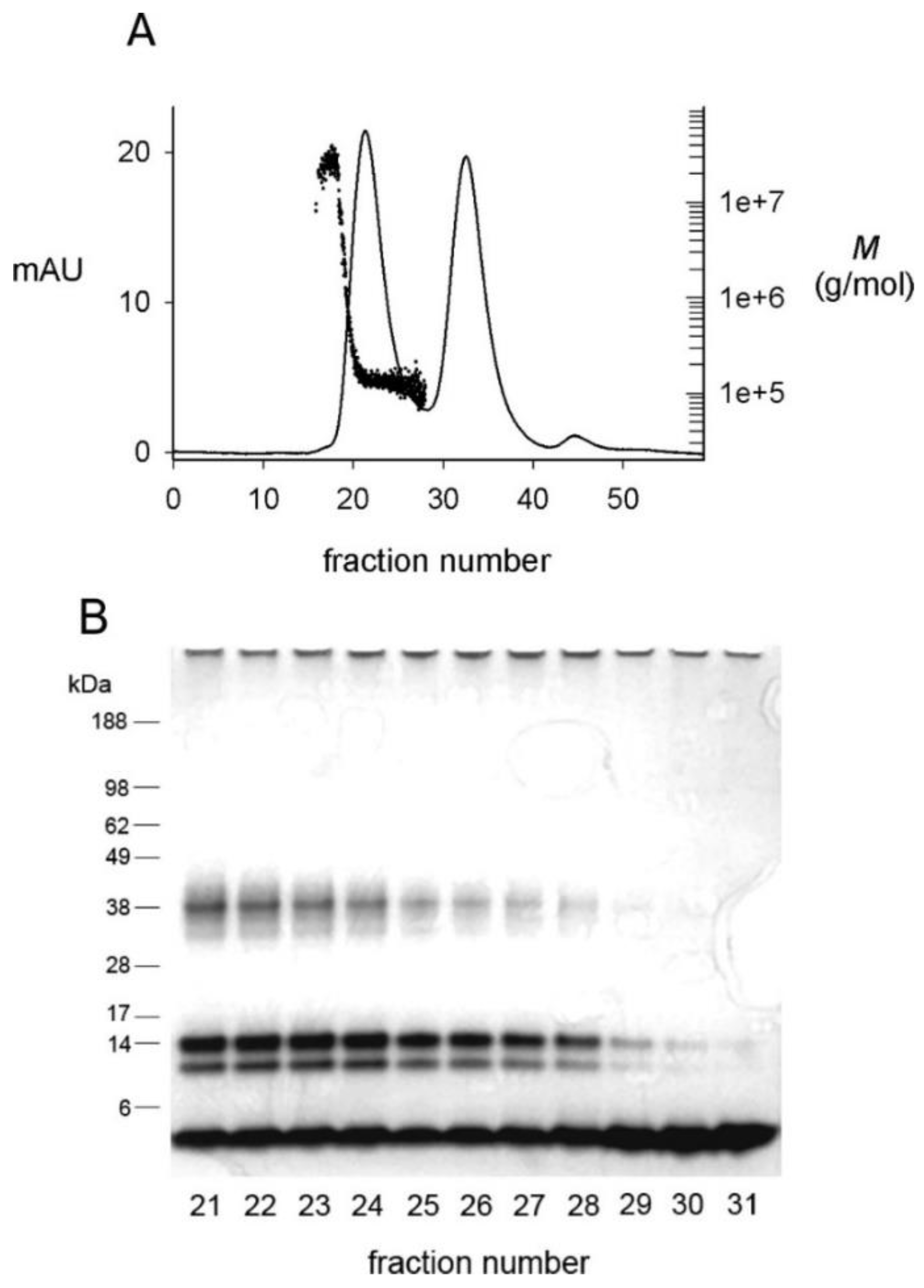
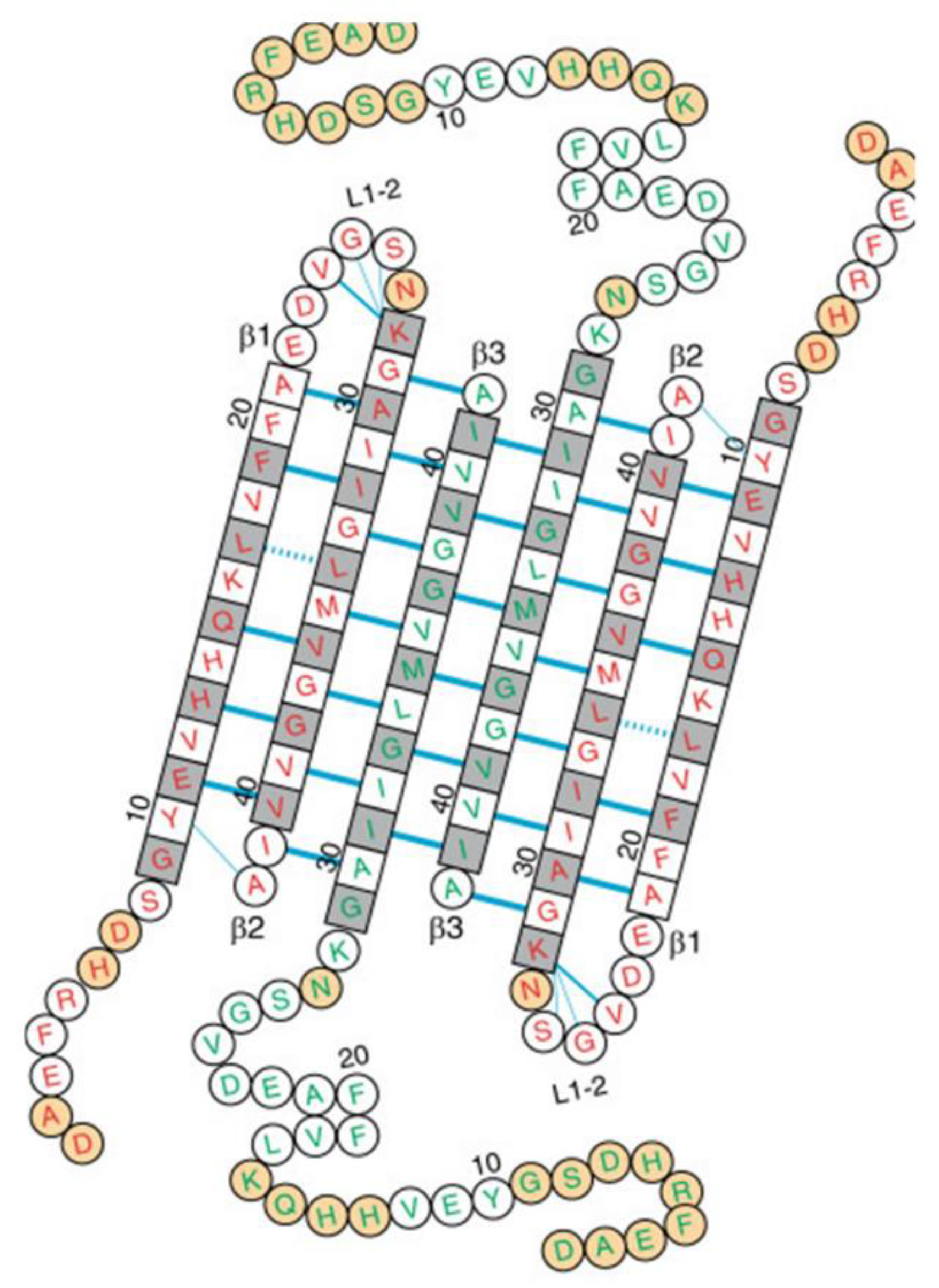
3. Analysis of 150-kDa Oligomers by Solid-State 2D NMR
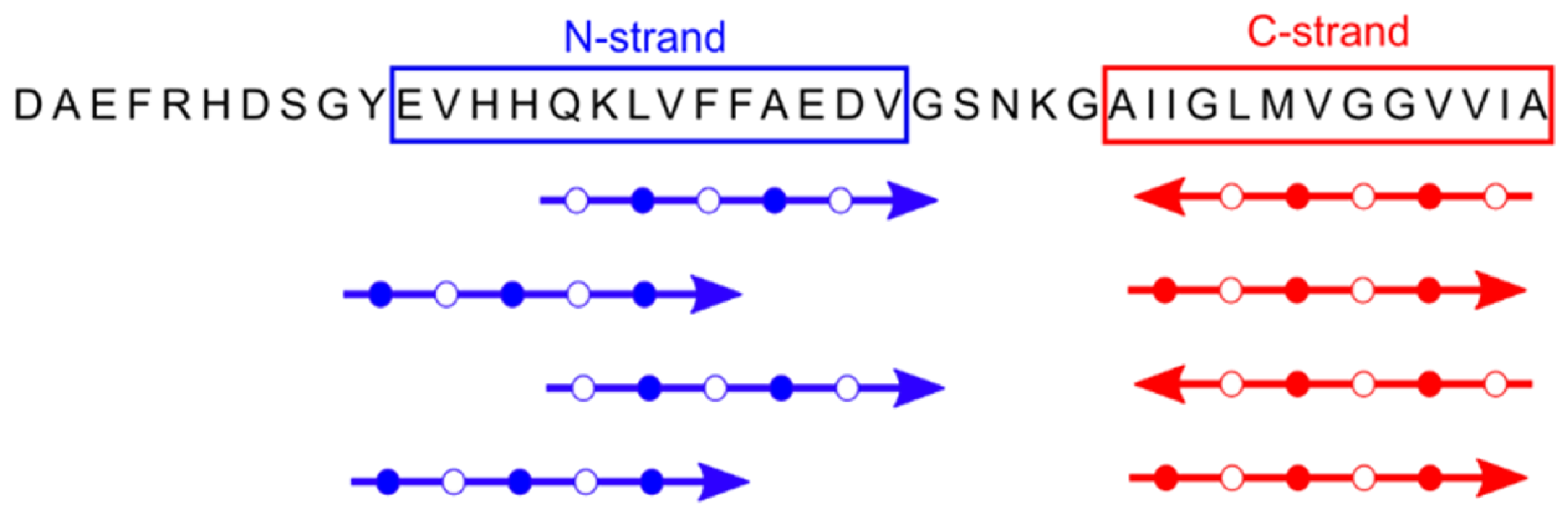
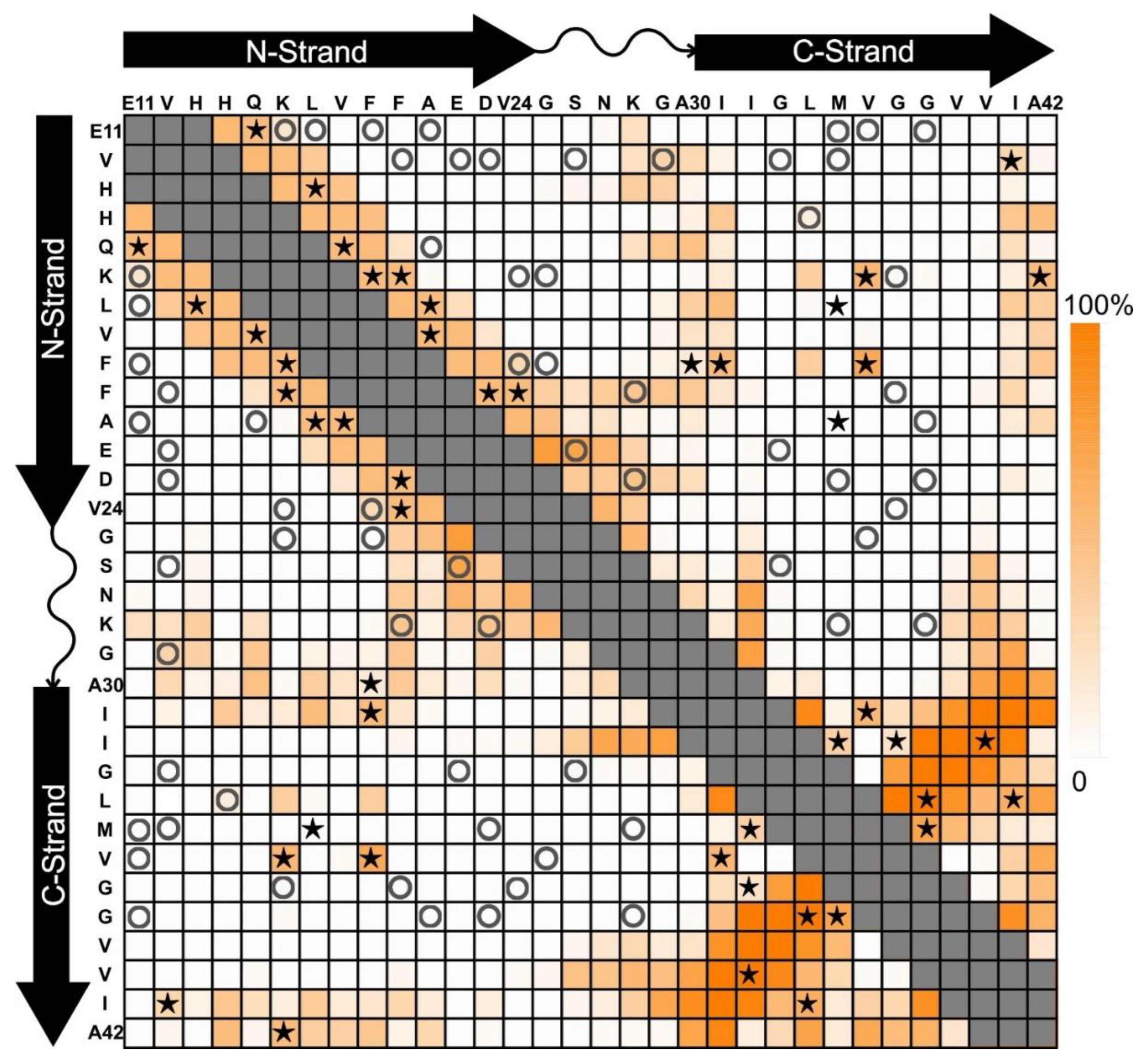
4. A Tetrameric Aβ42 Oligomer May Be an Intermediate to 150-kDa Oligomer Formation
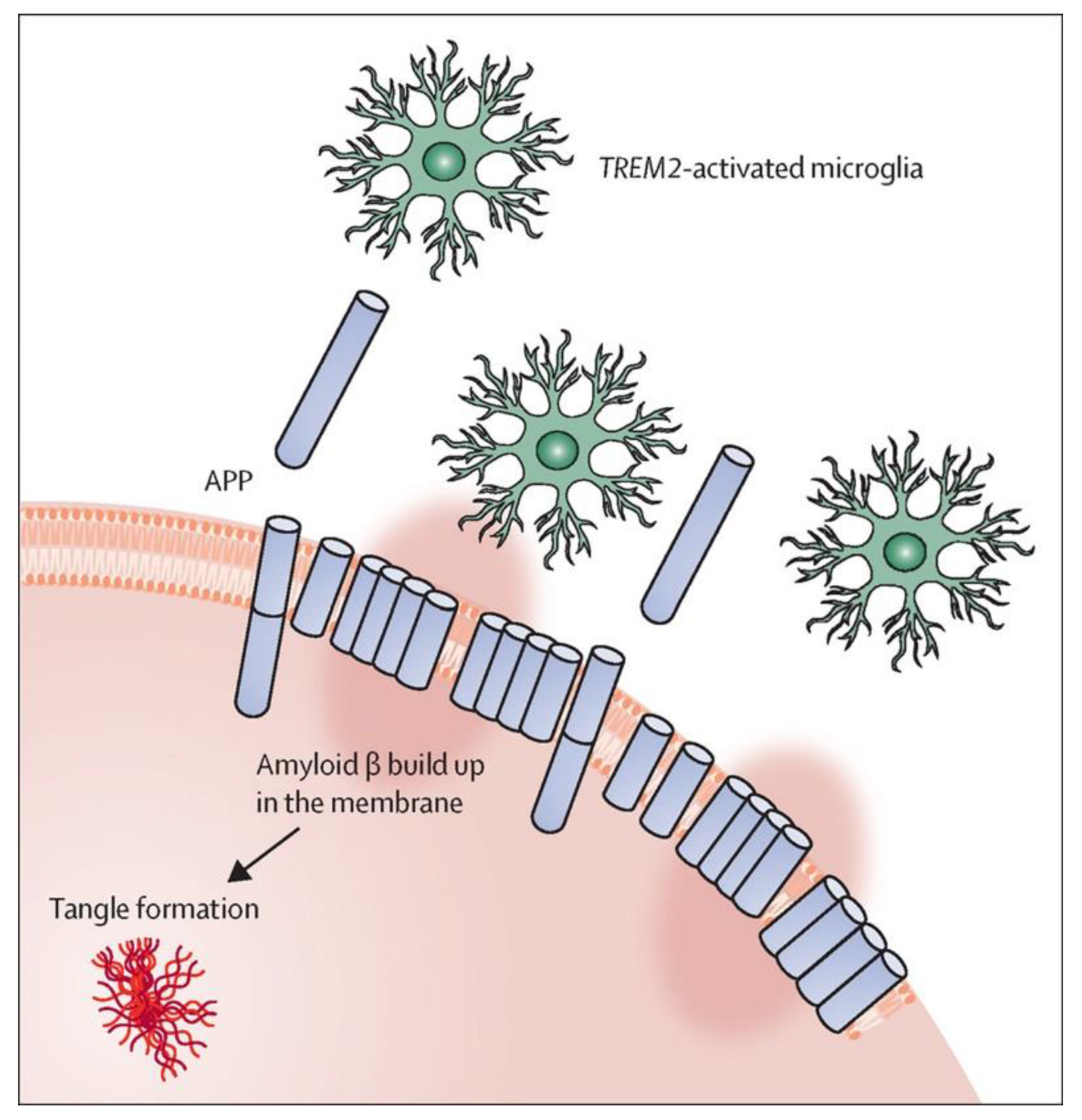
5. Aβ42 Interaction with Phospholipid Membranes
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Terry, R.D.; Gonatas, N.K.; Weiss, M. Ultrastructural studies in Alzheimer’s presenile dementia. Am. J. Pathol. 1964, 44, 269–297. [Google Scholar] [PubMed]
- Selkoe, D.J. The cell biology of b-amyloid precursor protein and presenilin in Alzheimer’s disease. Trends Cell Biol. 1998, 8, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Vassar, R.; Bennett, B.D.; Babu-Khan, S.; Kahn, S.; Mendiaz, E.A.; Denis, P.; Teplow, D.B.; Ross, S.; Amarante, P. b-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999, 286, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Funamoto, S.; Morishima-Kawashima, M.; Tanimura, Y.; Hirotani, N.; Saido, T.C.; Ihara, Y. Truncated carboxyl-terminal fragments of b-amyloid precursor protein are processed to amyloid b-proteins 40 and 42. Biochemistry 2004, 43, 13532–13540. [Google Scholar] [CrossRef]
- Takami, M.; Nagashima, Y.; Sano, Y.; Ishihara, S.; Morishima-Kawashima, M.; Funamoto, S.; Ihara, Y. g-Secretase: Successive tripeptide and tetrapeptide release from the transmembrane domain of b-carboxyl terminal fragment. J. Neurosci. 2009, 29, 13042–13052. [Google Scholar] [CrossRef] [PubMed]
- Bateman, R.J.; Aisen, P.S.; De Strooper, B.; Fox, N.C.; Lemere, C.A.; Ringman, J.M.; Salloway, S.; Sperling, R.A.; Windisch, M.; Xiong, C. Autosomal-dominant Alzheimer’s disease: A review and proposal for the prevention of Alzheimer’s disease. Alzheimer’s Res. Ther. 2011, 3, 1–13. [Google Scholar] [CrossRef]
- Annus, T.; Wilson, L.R.; Hong, Y.T.; Acosta-Cabronero, J.; Fryer, T.D.; Cardenas-Blanco, A.; Smith, R.; Boros, I.; Coles, J.P.; Aigbirhio, F.I.; et al. The pattern of amyloid accumulation in the brains of adults with Down syndrome. Alzheimer’s Dement. 2016, 12, 538–545. [Google Scholar] [CrossRef]
- Citron, M.; Oltersdorf, T.; Haass, C.; McConlogue, L.; Hung, A.Y.; Seubert, P.; Vigo-Pelfrey, C.; Lieberburg, I.; Selkoe, D.J. Mutation of the b-amyloid precursor protein in familial Alzheimer’s disease increases beta-protein production. Nature 1992, 360, 672–674. [Google Scholar] [CrossRef]
- De Jonghe, C.; Esselens, C.; Kumar-Singh, S.; Craessaerts, K.; Serneels, S.; Checler, F.; Annaert, W.; Van Broeckhoven, C.; De Strooper, B. Pathogenic APP mutations near the g-secretase cleavage site differentially affect Ab secretion and APP C-terminal fragment stability. Hum. Mol. Genet. 2001, 10, 1665–1671. [Google Scholar] [CrossRef]
- Murakami, K.; Irie, K.; Morimoto, A.; Ohigashi, H.; Shindo, M.; Nagao, M.; Shimizu, T.; Shirasawa, T. Synthesis, agregation, neurotoxicity, and secondary structure of various Ab 1-42 mutants of familial Alzheimer’s disease at positions 21–23. Biochem. Biophys. Res. Commun. 2002, 294, 5–10. [Google Scholar] [CrossRef]
- Borchelt, D.R.; Thinakaran, G.; Eckman, C.B.; Lee, M.K.; Davenport, F.; Ratovitsky, T.; Prada, C.M.; Kim, G.; Seekins, S.; Yager, D.; et al. Familial Alzheimer’s disease-linked presenilin 1 variants elevate Ab1-42/1-40 ratio in vitro and in vivo. Neuron 1996, 17, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Cline, E.N.; Bicca, M.A.; Viola, K.L.; Klein, W.L. The amyloid-β oligomer hypothesis: Beginning of the third decade. J. Alzheimer’s Dis. 2018, 64, 5567–5610. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.C. How early can we diagnose Alzheimer disease (and is it sufficient)? The 2017 Wartenberg lecture. Neurology 2018, 91, 395–402. [Google Scholar] [CrossRef]
- Jack, C.R.J.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 1, 119–128. [Google Scholar] [CrossRef]
- Petkova, A.T.; Ishii, Y.; Balbach, J.J.; Antzutkin, O.N.; Leapman, R.D.; Delaglio, F.; Tycko, R. A structural model for Alzheimer’s b-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA 2002, 99, 16742–16747. [Google Scholar] [CrossRef]
- Paravastu, A.K.; Qahwash, I.; Leapman, R.D.; Meredith, S.C.; Tycko, R. Seeded growth of b-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc. Natl. Acad. Sci. USA 2009, 106, 7443–7448. [Google Scholar] [CrossRef]
- Qiang, W.; Yau, W.M.; Lu, J.X.; Collinge, J.; Tycko, R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 2017, 541, 217–221. [Google Scholar] [CrossRef]
- Sarroukh, R.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. ATR-FTIR: A “rejuvenated” tool to investigate amyloid proteins. Biochim. Biophys. Acta. 2013, 1828, 2328–2338. [Google Scholar] [CrossRef]
- Lee, S.J.; Liyanage, U.; Bickel, P.E.; Xia, W.; Lansbury, P.T., Jr.; Kosik, K.S. A detergent-insoluble membrane compartment contains Ab in vivo. Nat. Med. 1998, 4, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Kawarabayashi, T.; Nakamura, T.; Sato, K.; Seino, Y.; Ichii, S.; Nakahata, N.; Takatama, M.; Westaway, D.; George-Hyslop, P.S.; Shoji, M. Lipid rafts act as a common platform for amyloid-β oligomer-induced Alzheimer’s disease pathology. J. Alzheimer’s Dis. 2022, 87, 1189–1203. [Google Scholar] [CrossRef] [PubMed]
- Serra-Batiste, M.; Ninot-Pedrosa, M.; Bayoumi, M.; Gairí, M.; Maglia, G.; Carulla, N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. USA 2016, 113, 10866–10871. [Google Scholar] [CrossRef]
- Rangachari, V.; Reed, D.K.; Moore, B.D.; Rosenberry, T.L. Secondary structure and interfacial aggregation of amyloid-b(1-40) on sodium dodecylsulfate micelles. Biochemistry 2006, 45, 8639–8648. [Google Scholar] [CrossRef]
- Rangachari, V.; Moore, B.D.; Reed, D.K.; Sonoda, L.K.; Bridges, A.W.; Conboy, E.; Hartigan, D.; Rosenberry, T.L. Amyloid-β(1-42) rapidly forms protofibrils and oligomers by distinct pathways in low concentrations of sodium dodecylsulfate. Biochemistry 2007, 46, 12451–12462. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.D.; Rangachari, V.; Tay, W.M.; Milkovic, N.M.; Rosenberry, T.L. Biophysical analyses of synthetic amyloid-β(1-42) aggregates before and after covalent cross-linking. Implications for deducing the structure of endogenous amyloid-β oligomers. Biochemistry 2009, 48, 11796–11806. [Google Scholar] [CrossRef]
- Barghorn, S.; Nimmrich, V.; Striebinger, A.; Krantz, C.; Keller, P.; Janson, B.; Bahr, M.; Schmidt, M.; Bitner, R.S.; Harlan, J.; et al. Globular amyloid b-peptide(1-42) oligomer—A homogenous and stable neuropathological protein in Alzheimer’s disease. J. Neurochem. 2005, 95, 834–847. [Google Scholar] [CrossRef]
- Tay, W.M.; Huang, D.; Rosenberry, T.L.; Paravastu, A.K. The Alzheimer’s amyloid-β(1-42) peptide forms off-pathway oligomers and fibrils that are distinguished structurally by intermolecular organization. J. Mol. Biol. 2013, 425, 2494–2508. [Google Scholar] [CrossRef]
- Huang, D.; Zimmerman, M.I.; Martin, P.K.; Nix, A.J.; Rosenberry, T.L.; Paravastu, A.K. Antiparallel β-sheet structure within the C-terminal region of 42-residue Alzheimer’s amyloid-β peptides when they form 150-kDa oligomers. J. Mol. Biol. 2015, 427, 2319–2328. [Google Scholar] [CrossRef]
- Gao, Y.; Guo, C.; Watzlawik, J.O.; Randolph, P.S.; Lee, E.J.; Huang, D.; Stagg, S.M.; Zhou, H.X.; Rosenberry, T.L.; Paravastu, A.K. Out-of-register parallel β-sheets and antiparallel β-sheets coexist in 150-kDa oligomers formed by amyloid-β(1-42). J. Mol. Biol. 2020, 432, 4388–4407. [Google Scholar] [CrossRef]
- Tycko, R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J. Chem. Phys. 2007, 126, 064506. [Google Scholar] [CrossRef] [PubMed]
- Morcombe, C.R.; Gaponenko, V.; Byrd, R.A.; Zilm, K.W. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J. Am. Chem. Soc. 2004, 126, 7196–7197. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Prasad, R.; Randolph, P.S.; Watzlawik, J.O.; Robang, A.S.; Guo, C.; Stagg, S.M.; Zhou, H.X.; Rosenberry, T.L.; Paravastu, A.K. Structural model for self-limiting β-strand arrangement within an Alzheimer’s amyloid-β oligomer. bioRxiv 2023. [Google Scholar] [CrossRef]
- Paravastu, A.K.; Leapman, R.D.; Yau, W.-M.; Tycko, R. Molecular structural basis of polymorphism in Alzheimer’s b-amyloid fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 18349–18354. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.R.; Cormier, A.R.; Pang, X.; Zimmerman, M.I.; Zhou, H.X.; Paravastu, A.K. Solid-state NMR evidence for β-hairpin structure within MAX8 designer peptide nanofibers. Biophys. J. 2013, 105, 222–230. [Google Scholar] [CrossRef]
- Ciudad, S.; Puig, E.; Botzanowski, T.; Meigooni, M.; Arango, A.S.; Do, J.; Mayze, M.; Bayoumi, M.; Chaignepain, S.; Maglia, G.; et al. Aβ(1-42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat. Commun. 2020, 11, 3014–3027. [Google Scholar] [CrossRef]
- McLaurin, J.; Chakrabartty, A. Membrane disruption by Alzheimer beta-amyloid peptides mediated through specific binding to either phospholipids or gangliosides. Implications for neurotoxicity. J. Biol. Chem. 1996, 271, 26482–26489. [Google Scholar] [CrossRef]
- Williams, T.L.; Serpell, L.C. Membrane and surface interactions of Alzheimer’s Aβ peptide--insights into the mechanism of cytotoxicity. FEBS J. 2011, 278, 3905–3917. [Google Scholar] [CrossRef]
- Tian, Y.; Liang, R.; Kumar, A.; Szwedziak, P.; Viles, J.H. 3D-visualization of amyloid-b oligomer interactions with lipid membranes by cryo-electron tomography. Chem. Sci. 2021, 12, 6896–6907. [Google Scholar] [CrossRef]
- Bode, D.C.; Baker, M.D.; Viles, J.H. Ion channel formation by amyloid-b42 oligomers but not amyloid-b40 in cellular membranes. J. Biol. Chem. 2017, 292, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S. In search of pathogenic amyloid β-peptide in familial Alzheimer’s disease. Prog. Mol. Biol. Transl. Sci. 2019, 168, 71–78. [Google Scholar] [PubMed]
- Hardy, J.; Salih, D. TREM2-mediated activation of microglia breaks link between amyloid and tau. Lancet Neurol. 2021, 20, 416–417. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Selkoe, D. If amyloid drives Alzheimer disease, why have anti-amyloid therapies not yet slowed cognitive decline? PLoS Biol. 2022, 20, e3001694. [Google Scholar] [CrossRef] [PubMed]
- Romero-Molina, C.; Garretti, F.; Andrews, S.J.; Marcora., E.; Goate, A.M. Microglial efferocytosis: Diving into the Alzheimer’s disease gene pool. Neuron 2022, 110, 3513–3533. [Google Scholar] [CrossRef]
- Lewcock, J.W.; Schlepckow, K.; Di Paolo, G.; Tahirovic, S.; Monroe, K.M.; Haass, C. Emerging microglia biology defines novel therapeutic approaches for Alzheimer’s disease. Neuron 2020, 108, 801–821. [Google Scholar] [CrossRef]
- Uhlmann, R.E.; Rother, C.; Rasmussen, J.; Schelle, J.; Bergmann, C.; Ullrich Gavilanes, E.M.; Fritschi, S.K.; Buehler, A.; Baumann, F.; Skodras, A.; et al. Acute targeting of pre-amyloid seeds in transgenic mice reduces Alzheimer-like pathology later in life. Nat. Neurosci. 2020, 23, 1580–1588. [Google Scholar] [CrossRef]
- Parhizkar, S.; Arzberger, T.; Brendel, M.; Kleinberger, G.; Deussing, M.; Focke, C.; Nuscher, B.; Xiong, M.; Ghasemigharagoz, A.; Katzmarski, N.; et al. Loss of TREM2 function increases amyloid seeding but reduces plaque-associated ApoE. Nat. Neurosci. 2019, 22, 191–204. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosenberry, T.L.; Zhou, H.-X.; Stagg, S.M.; Paravastu, A.K. Oligomer Formation by Amyloid-β42 in a Membrane-Mimicking Environment in Alzheimer’s Disease. Molecules 2022, 27, 8804. https://doi.org/10.3390/molecules27248804
Rosenberry TL, Zhou H-X, Stagg SM, Paravastu AK. Oligomer Formation by Amyloid-β42 in a Membrane-Mimicking Environment in Alzheimer’s Disease. Molecules. 2022; 27(24):8804. https://doi.org/10.3390/molecules27248804
Chicago/Turabian StyleRosenberry, Terrone L., Huan-Xiang Zhou, Scott M. Stagg, and Anant K. Paravastu. 2022. "Oligomer Formation by Amyloid-β42 in a Membrane-Mimicking Environment in Alzheimer’s Disease" Molecules 27, no. 24: 8804. https://doi.org/10.3390/molecules27248804
APA StyleRosenberry, T. L., Zhou, H.-X., Stagg, S. M., & Paravastu, A. K. (2022). Oligomer Formation by Amyloid-β42 in a Membrane-Mimicking Environment in Alzheimer’s Disease. Molecules, 27(24), 8804. https://doi.org/10.3390/molecules27248804




