Quantitative Analysis of Isoniazid and Its Four Primary Metabolites in Plasma of Tuberculosis Patients Using LC-MS/MS
Abstract
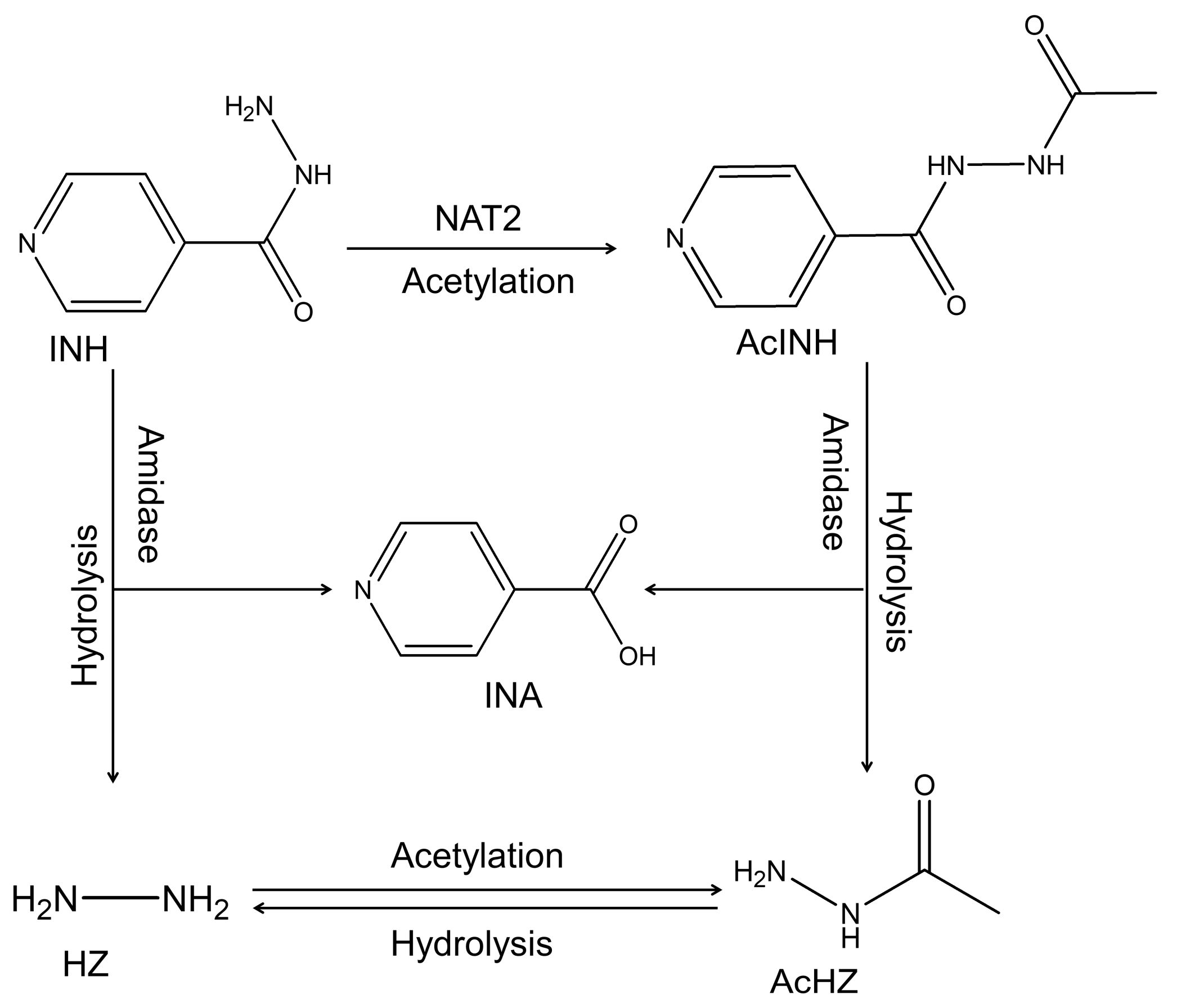
1. Introduction
2. Results and Discussion
2.1. Method Optimization for LC and MS Conditions
2.2. Method Validations
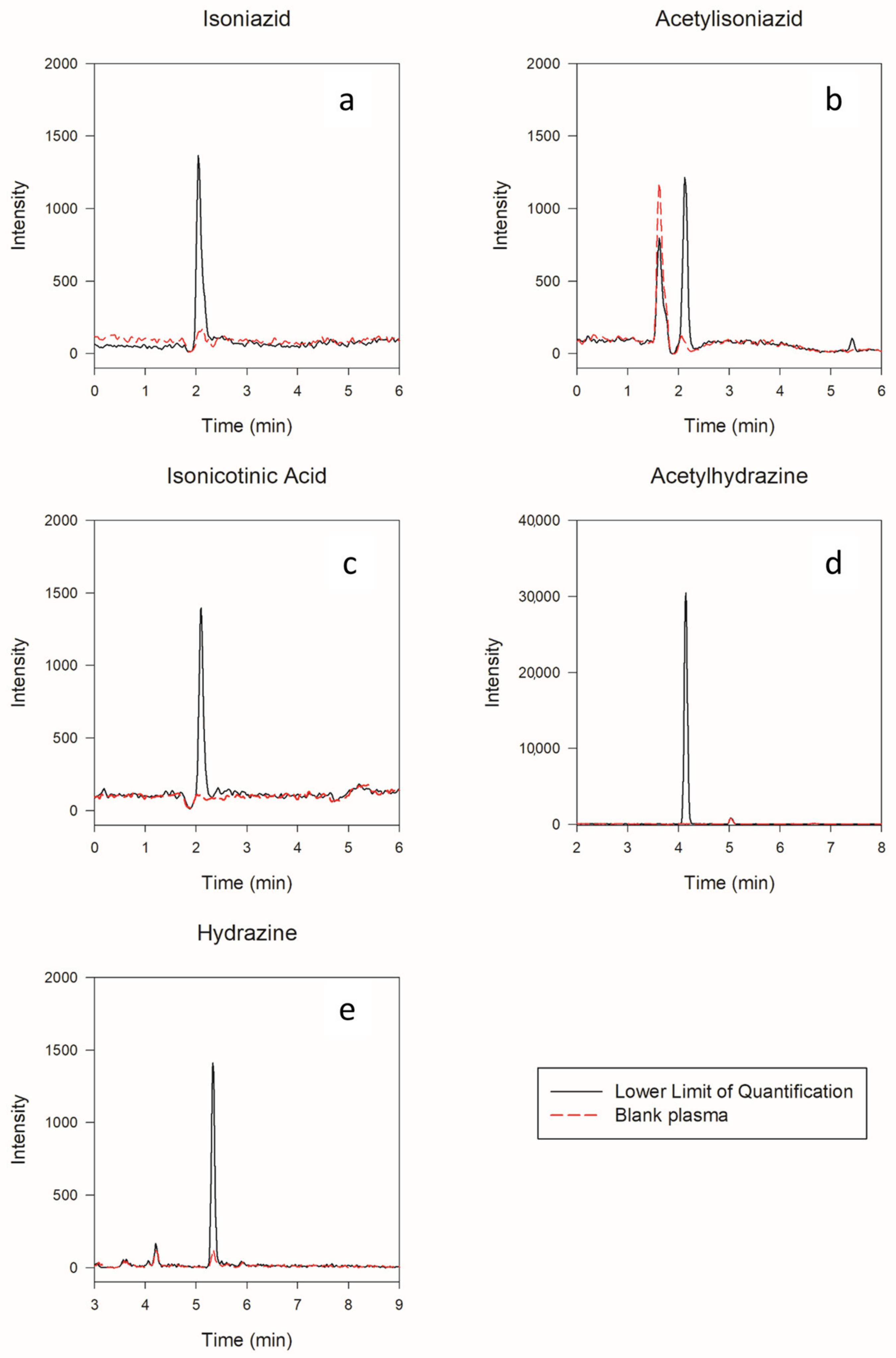
2.2.1. Selectivity, Linearity, and Carryover
2.2.2. Accuracy and Precision
2.2.3. Matrix Effects and Recovery
2.2.4. Stability
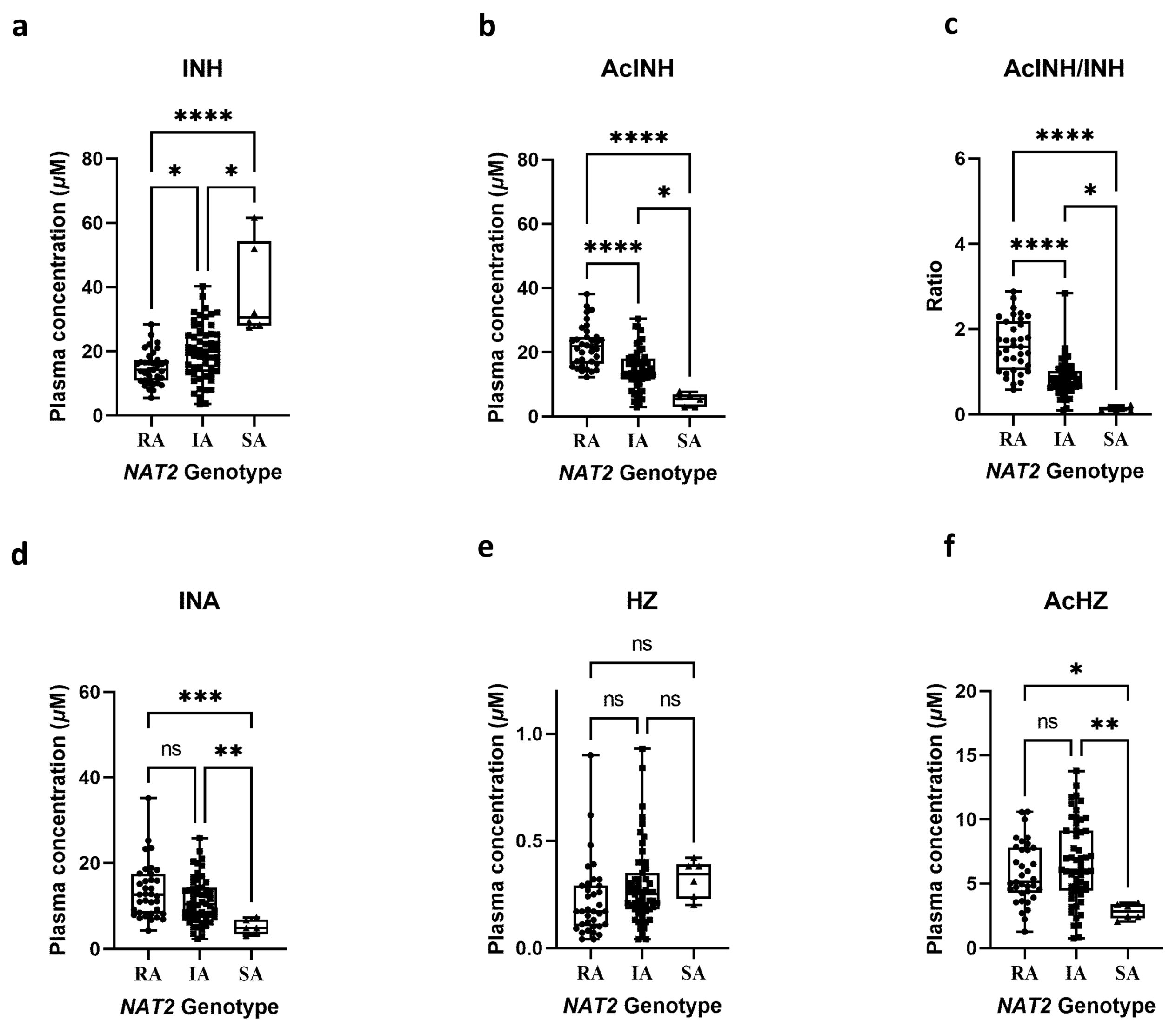
2.3. Application to Clinical Research
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Preparation of Standards and Quality Controls Samples
3.3. Sample Preparation
3.4. Liquid Chromatography–Tandem Mass Spectrometry
3.5. Method Validation
3.5.1. Selectivity and Carryover
3.5.2. Linearity and Lower Limit of Quantification
3.5.3. Accuracy and Precision
3.5.4. Matrix Effect and Recovery
3.5.5. Stability
3.6. Clinical Application
3.7. Determination of NAT2 Genotype
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- World Health Organization. Global Tuberculosis Report 2021; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Forget, E.J.; Menzies, D. Adverse reactions to first-line antituberculosis drugs. Expert Opin. Drug Saf. 2006, 5, 231–249. [Google Scholar] [CrossRef] [PubMed]
- Nolan, C.M.; Goldberg, S.V.; Buskin, S.E. Hepatotoxicity associated with isoniazid preventive therapy: A 7-year survey from a public health tuberculosis clinic. JAMA 1999, 281, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M. Drug-induced hepatotoxicity. N. Engl. J. Med. 2003, 349, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.R.; Zimmerman, H.J.; Ishak, K.G.; Thorgeirsson, U.P.; Timbrell, J.A.; Snodgrass, W.R.; Nelson, S.D. Isoniazid liver injury: Clinical spectrum, pathology, and probable pathogenesis. Ann. Intern. Med. 1976, 84, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Sarich, T.C.; Youssefi, M.; Zhou, T.; Adams, S.P.; Wall, R.A.; Wright, J.M. Role of hydrazine in the mechanism of isoniazid hepatotoxicity in rabbits. Arch. Toxicol. 1996, 70, 835–840. [Google Scholar] [CrossRef]
- Metushi, I.; Uetrecht, J.; Phillips, E. Mechanism of isoniazid-induced hepatotoxicity: Then and now. Br. J. Clin. Pharmacol. 2016, 81, 1030–1036. [Google Scholar] [CrossRef]
- Metushi, I.G.; Cai, P.; Zhu, X.; Nakagawa, T.; Uetrecht, J.P. A fresh look at the mechanism of isoniazid-induced hepatotoxicity. Clin. Pharmacol. Ther. 2011, 89, 911–914. [Google Scholar] [CrossRef]
- Metushi, I.G.; Sanders, C.; Acute Liver Study, G.; Lee, W.M.; Uetrecht, J. Detection of anti-isoniazid and anti-cytochrome P450 antibodies in patients with isoniazid-induced liver failure. Hepatology 2014, 59, 1084–1093. [Google Scholar] [CrossRef]
- Seng, K.Y.; Hee, K.H.; Soon, G.H.; Chew, N.; Khoo, S.H.; Lee, L.S. Population pharmacokinetic analysis of isoniazid, acetylisoniazid, and isonicotinic acid in healthy volunteers. Antimicrob. Agents Chemother. 2015, 59, 6791–6799. [Google Scholar] [CrossRef]
- Sundell, J.; Bienvenu, E.; Janzen, D.; Birgersson, S.; Abelo, A.; Ashton, M. Model-Based Assessment of Variability in Isoniazid Pharmacokinetics and Metabolism in Patients Co-Infected With Tuberculosis and HIV: Implications for a Novel Dosing Strategy. Clin. Pharmacol. Ther. 2020, 108, 73–80. [Google Scholar] [CrossRef]
- Wilkins, J.J.; Langdon, G.; McIlleron, H.; Pillai, G.; Smith, P.J.; Simonsson, U.S. Variability in the population pharmacokinetics of isoniazid in South African tuberculosis patients. Br. J. Clin. Pharmacol. 2011, 72, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.S.; Jang, T.W.; Kim, H.J.; Oh, J.Y.; Lee, H.K.; Park, H.K.; Ghim, J.L.; Long, N.P.; Park, Y.; Choi, Y.K.; et al. Isoniazid Population Pharmacokinetics and Dose Recommendation for Korean Patients With Tuberculosis Based on Target Attainment Analysis. J. Clin. Pharmacol. 2021, 61, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.W.; Shin, A.Y.; Ha, J.H.; Ahn, J.H.; Kang, H.S.; Kim, J.S. Effect of serum isoniazid level on treatment outcomes among tuberculosis patients with slow response—A retrospective cohort study. J. Infect. Chemother. 2021, 27, 1555–1561. [Google Scholar] [CrossRef] [PubMed]
- Preziosi, P. Isoniazid: Metabolic aspects and toxicological correlates. Curr. Drug Metab. 2007, 8, 839–851. [Google Scholar] [CrossRef]
- Erwin, E.R.; Addison, A.P.; John, S.F.; Olaleye, O.A.; Rosell, R.C. Pharmacokinetics of isoniazid: The good, the bad, and the alternatives. Tuberculosis 2019, 116S, S66–S70. [Google Scholar] [CrossRef]
- Hee, K.H.; Seo, J.J.; Lee, L.S. Development and validation of liquid chromatography tandem mass spectrometry method for simultaneous quantification of first line tuberculosis drugs and metabolites in human plasma and its application in clinical study. J. Pharm. Biomed. Anal. 2015, 102, 253–260. [Google Scholar] [CrossRef]
- Sundell, J.; Bienvenu, E.; Birgersson, S.; Abelo, A.; Ashton, M.; Hoffmann, K.J. Simultaneous quantification of four first line antitubercular drugs and metabolites in human plasma by hydrophilic interaction chromatography and tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1105, 129–135. [Google Scholar] [CrossRef]
- Kivrane, A.; Grinberga, S.; Sevostjanovs, E.; Igumnova, V.; Pole, I.; Viksna, A.; Bandere, D.; Krams, A.; Cirule, A.; Pugovics, O.; et al. LC-MS/MS method for simultaneous quantification of the first-line anti-tuberculosis drugs and six primary metabolites in patient plasma: Implications for therapeutic drug monitoring. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1185, 122986. [Google Scholar] [CrossRef]
- Song, L.; Gao, D.; Li, S.; Wang, Y.; Liu, H.; Jiang, Y. Simultaneous quantitation of hydrazine and acetylhydrazine in human plasma by high performance liquid chromatography-tandem mass spectrometry after derivatization with p-tolualdehyde. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1063, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Song, S.H.; Jun, S.H.; Park, K.U.; Yoon, Y.; Lee, J.H.; Kim, J.Q.; Song, J. Simultaneous determination of first-line anti-tuberculosis drugs and their major metabolic ratios by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 1331–1338. [Google Scholar] [CrossRef]
- Fachi, M.M.; Vilhena, R.O.; Boger, B.; Domingos, E.L.; Dos Santos, J.; Junkert, A.M.; de Fatima Cobre, A.; Momade, D.R.O.; Beraldi-Magalhaes, F.; de Liz, M.V.; et al. LC-QToF-MS method for quantification of ethambutol, isoniazid, pyrazinamide and rifampicin in human plasma and its application. Biomed. Chromatogr. 2020, 34, e4812. [Google Scholar] [CrossRef] [PubMed]
- Zuma, P.; Joubert, A.; van der Merwe, M.; Norman, J.; Waitt, C.; Court, R.; Loveday, M.; Castel, S.; Wiesner, L. Validation and application of a quantitative LC-MS/MS assay for the analysis of first-line anti-tuberculosis drugs, rifabutin and their metabolites in human breast milk. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2022, 1211, 123489. [Google Scholar] [CrossRef] [PubMed]
- FDA. Guidance for Industry: Bioanalytical Method Validation; FDA: Silver Spring, MA, USA, 2018. [Google Scholar]
- Kim, H.J.; Seo, K.A.; Kim, H.M.; Jeong, E.S.; Ghim, J.L.; Lee, S.H.; Lee, Y.M.; Kim, D.H.; Shin, J.G. Simple and accurate quantitative analysis of 20 anti-tuberculosis drugs in human plasma using liquid chromatography-electrospray ionization-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2015, 102, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wang, Z.; Xie, X.; You, C.; Yang, Y.; Xi, Y.; Chen, W. Rapid and sensitive method for simultaneous determination of first-line anti-tuberculosis drugs in human plasma by HPLC-MS/MS: Application to therapeutic drug monitoring. Tuberculosis 2018, 109, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, M.; Kishikawa, Y.; Takekuma, Y.; Matsuura, M.; Narahara, K.; Inoue, T.; Hamdy, S.I.; Endo, N.; Goto, J.; Mizugaki, M. Genotyping of the N-acetyltransferase2 polymorphism in the prediction of adverse drug reactions to isoniazid in Japanese patients. Drug Metab. Pharmacokinet. 2002, 17, 357–362. [Google Scholar] [CrossRef]
- Sotsuka, T.; Sasaki, Y.; Hirai, S.; Yamagishi, F.; Ueno, K. Association of isoniazid-metabolizing enzyme genotypes and isoniazid-induced hepatotoxicity in tuberculosis patients. In Vivo 2011, 25, 803–812. [Google Scholar]
- Fukino, K.; Sasaki, Y.; Hirai, S.; Nakamura, T.; Hashimoto, M.; Yamagishi, F.; Ueno, K. Effects of N-acetyltransferase 2 (NAT2), CYP2E1 and Glutathione-S-transferase (GST) genotypes on the serum concentrations of isoniazid and metabolites in tuberculosis patients. J. Toxicol. Sci. 2008, 33, 187–195. [Google Scholar] [CrossRef]
- Parkin, D.P.; Vandenplas, S.; Botha, F.J.; Vandenplas, M.L.; Seifart, H.I.; van Helden, P.D.; van der Walt, B.J.; Donald, P.R.; van Jaarsveld, P.P. Trimodality of isoniazid elimination: Phenotype and genotype in patients with tuberculosis. Am. J. Respir. Crit. Care Med. 1997, 155, 1717–1722. [Google Scholar] [CrossRef]
- Donald, P.R.; Sirgel, F.A.; Venter, A.; Parkin, D.P.; Seifart, H.I.; van de Wal, B.W.; Werely, C.; van Helden, P.D.; Maritz, J.S. The influence of human N-acetyltransferase genotype on the early bactericidal activity of isoniazid. Clin. Infect. Dis. 2004, 39, 1425–1430. [Google Scholar] [CrossRef]
- Park, J.S.; Lee, J.Y.; Lee, Y.J.; Kim, S.J.; Cho, Y.J.; Yoon, H.I.; Lee, C.T.; Song, J.; Lee, J.H. Serum Levels of Antituberculosis Drugs and Their Effect on Tuberculosis Treatment Outcome. Antimicrob. Agents Chemother. 2016, 60, 92–98. [Google Scholar] [CrossRef]
- Wang, P.; Pradhan, K.; Zhong, X.B.; Ma, X. Isoniazid metabolism and hepatotoxicity. Acta Pharm. Sin. B 2016, 6, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Azuma, J.; Ohno, M.; Kubota, R.; Yokota, S.; Nagai, T.; Tsuyuguchi, K.; Okuda, Y.; Takashima, T.; Kamimura, S.; Fujio, Y.; et al. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: A randomized controlled trial for pharmacogenetics-based therapy. Eur. J. Clin. Pharmacol. 2013, 69, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Kinzig-Schippers, M.; Tomalik-Scharte, D.; Jetter, A.; Scheidel, B.; Jakob, V.; Rodamer, M.; Cascorbi, I.; Doroshyenko, O.; Sorgel, F.; Fuhr, U. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob. Agents Chemother. 2005, 49, 1733–1738. [Google Scholar] [CrossRef]
- Kuznetsov, I.B.; McDuffie, M.; Moslehi, R. A web server for inferring the human N-acetyltransferase-2 (NAT2) enzymatic phenotype from NAT2 genotype. Bioinformatics 2009, 25, 1185–1186. [Google Scholar] [CrossRef] [PubMed]
- Conte, J.E., Jr.; Golden, J.A.; McQuitty, M.; Kipps, J.; Duncan, S.; McKenna, E.; Zurlinden, E. Effects of gender, AIDS, and acetylator status on intrapulmonary concentrations of isoniazid. Antimicrob. Agents Chemother. 2002, 46, 2358–2364. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nguyen, V.L.; Ahn, S.; Hoa, P.Q.; Long, N.P.; Ahn, S.; Cho, Y.S.; Shin, J.G. Center for Personalized Precision Medicine for Tuberculosis: Smart Research and Development Workstation. Healthc. Inform. Res. 2022, 28, 176–180. [Google Scholar] [CrossRef]
| Analytes | Transition (m/z) | DP (V) | CE (eV) | Calibration Range (ng/mL) | Correlation Coefficient | Internal Standard |
|---|---|---|---|---|---|---|
| INH | 138 → 121 | 50 | 30 | 80–10,000 | 0.9970 | Phenacetin |
| AcINH | 180 → 121 | 60 | 30 | 80–10,000 | 0.9958 | Phenacetin |
| INA | 124 → 80 | 40 | 25 | 80–10,000 | 0.9947 | Phenacetin |
| HZ | 237.2 → 120 | 80 | 25 | 1–30 | 0.9975 | Sulfamethazine |
| AcHZ | 177 → 118 | 50 | 19 | 40–1200 | 0.9993 | Sulfamethazine |
| Analytes | Nominal Conc. (ng/mL) | Intra-Day (n = 5) | Inter-Day (n = 3) * | ||
|---|---|---|---|---|---|
| RSD (%) | Accuracy (%) | RSD (%) | Accuracy (%) | ||
| INH | 80 | 4.41 | 104.43 | 4.78 | 104.34 |
| 240 | 3.92 | 107.78 | 5.29 | 103.84 | |
| 2000 | 5.21 | 106.16 | 3.01 | 103.10 | |
| 8000 | 7.93 | 105.84 | 3.30 | 101.69 | |
| AcINH | 80 | 3.34 | 92.85 | 7.10 | 97.46 |
| 240 | 11.57 | 95.38 | 4.67 | 98.74 | |
| 2000 | 3.73 | 99.10 | 4.93 | 99.25 | |
| 8000 | 4.59 | 97.20 | 5.43 | 99.89 | |
| INA | 80 | 13.43 | 102.07 | 8.51 | 108.78 |
| 240 | 10.54 | 91.63 | 9.69 | 103.01 | |
| 2000 | 9.31 | 96.00 | 2.87 | 97.89 | |
| 8000 | 8.74 | 100.94 | 8.95 | 100.59 | |
| HZ | 1 | 5.58 | 112.00 | 9.36 | 106.26 |
| 3 | 1.86 | 109.00 | 3.73 | 107.74 | |
| 12 | 6.28 | 98.10 | 6.49 | 100.36 | |
| 24 | 6.90 | 100.32 | 2.49 | 102.13 | |
| AcHZ | 40 | 6.54 | 106.00 | 9.19 | 99.19 |
| 120 | 1.12 | 114.00 | 8.22 | 106.07 | |
| 480 | 11.30 | 97.68 | 5.32 | 100.26 | |
| 960 | 9.00 | 100.30 | 5.60 | 100.67 | |
| Analytes | Nominal Conc. (ng/mL) | Recovery (n = 6) | Matrix Effect (n = 6) | ||
|---|---|---|---|---|---|
| Mean (%) ± SD | RSD (%) | Mean (%) ± SD | RSD (%) | ||
| INH | 240 | 92.38 ± 3.56 | 3.86 | 36.42 ± 2.55 | 7.01 |
| 8000 | 96.92 ± 1.97 | 2.03 | 43.46 ± 3.08 | 7.09 | |
| AcINH | 240 | 110.76 ± 3.05 | 2.75 | 53.88 ± 1.70 | 3.15 |
| 8000 | 96.65 ± 1.23 | 1.27 | 54.08 ± 2.95 | 5.45 | |
| INA | 240 | 66.94 ± 6.27 | 9.36 | 40.65 ± 1.67 | 4.12 |
| 8000 | 78.17 ± 0.84 | 1.07 | 39.68 ± 2.97 | 7.50 | |
| HZ | 1.5 | 109.67 ± 7.65 | 6.98 | NA | NA |
| 24 | 70.35 ± 1.36 | 1.94 | NA | NA | |
| AcHZ | 120 | 105.12 ± 2.42 | 2.30 | NA | NA |
| 960 | 112.19 ± 1.64 | 1.46 | NA | NA | |
| Analytes | Nominal Conc. (ng/mL) | Stability (%) | ||
|---|---|---|---|---|
| Bench-Top at RT (4 h) | Autosampler at 4 °C (*) | Freeze–Thaw (3 Cycles) | ||
| INH | 240 | 103.82 | 104.46 | 77.36 |
| 8000 | 93.84 | 93.55 | 92.83 | |
| AcINH | 240 | 94.10 | 97.44 | 97.97 |
| 8000 | 95.76 | 98.33 | 106.54 | |
| INA | 240 | 92.93 | 92.98 | 79.44 |
| 8000 | 108.23 | 92.86 | 113.96 | |
| HZ | 3 | 86.65 | 91.07 | 87.13 |
| 24 | 97.77 | 83.83 | 86.83 | |
| AcHZ | 120 | 113.67 | 111.33 | 112.33 |
| 960 | 112.00 | 101.00 | 107.00 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ky Anh, N.; My Tung, P.; Kim, M.J.; Phuoc Long, N.; Cho, Y.-S.; Kim, D.-H.; Shin, J.-G. Quantitative Analysis of Isoniazid and Its Four Primary Metabolites in Plasma of Tuberculosis Patients Using LC-MS/MS. Molecules 2022, 27, 8607. https://doi.org/10.3390/molecules27238607
Ky Anh N, My Tung P, Kim MJ, Phuoc Long N, Cho Y-S, Kim D-H, Shin J-G. Quantitative Analysis of Isoniazid and Its Four Primary Metabolites in Plasma of Tuberculosis Patients Using LC-MS/MS. Molecules. 2022; 27(23):8607. https://doi.org/10.3390/molecules27238607
Chicago/Turabian StyleKy Anh, Nguyen, Pham My Tung, Min Jung Kim, Nguyen Phuoc Long, Yong-Soon Cho, Dong-Hyun Kim, and Jae-Gook Shin. 2022. "Quantitative Analysis of Isoniazid and Its Four Primary Metabolites in Plasma of Tuberculosis Patients Using LC-MS/MS" Molecules 27, no. 23: 8607. https://doi.org/10.3390/molecules27238607
APA StyleKy Anh, N., My Tung, P., Kim, M. J., Phuoc Long, N., Cho, Y.-S., Kim, D.-H., & Shin, J.-G. (2022). Quantitative Analysis of Isoniazid and Its Four Primary Metabolites in Plasma of Tuberculosis Patients Using LC-MS/MS. Molecules, 27(23), 8607. https://doi.org/10.3390/molecules27238607








