Multi-Dimensional Elimination of β-Lactams in the Rural Wetland: Molecule Design and Screening for More Antibacterial and Degradable Substitutes
Abstract
1. Introduction
2. Results and Discussion
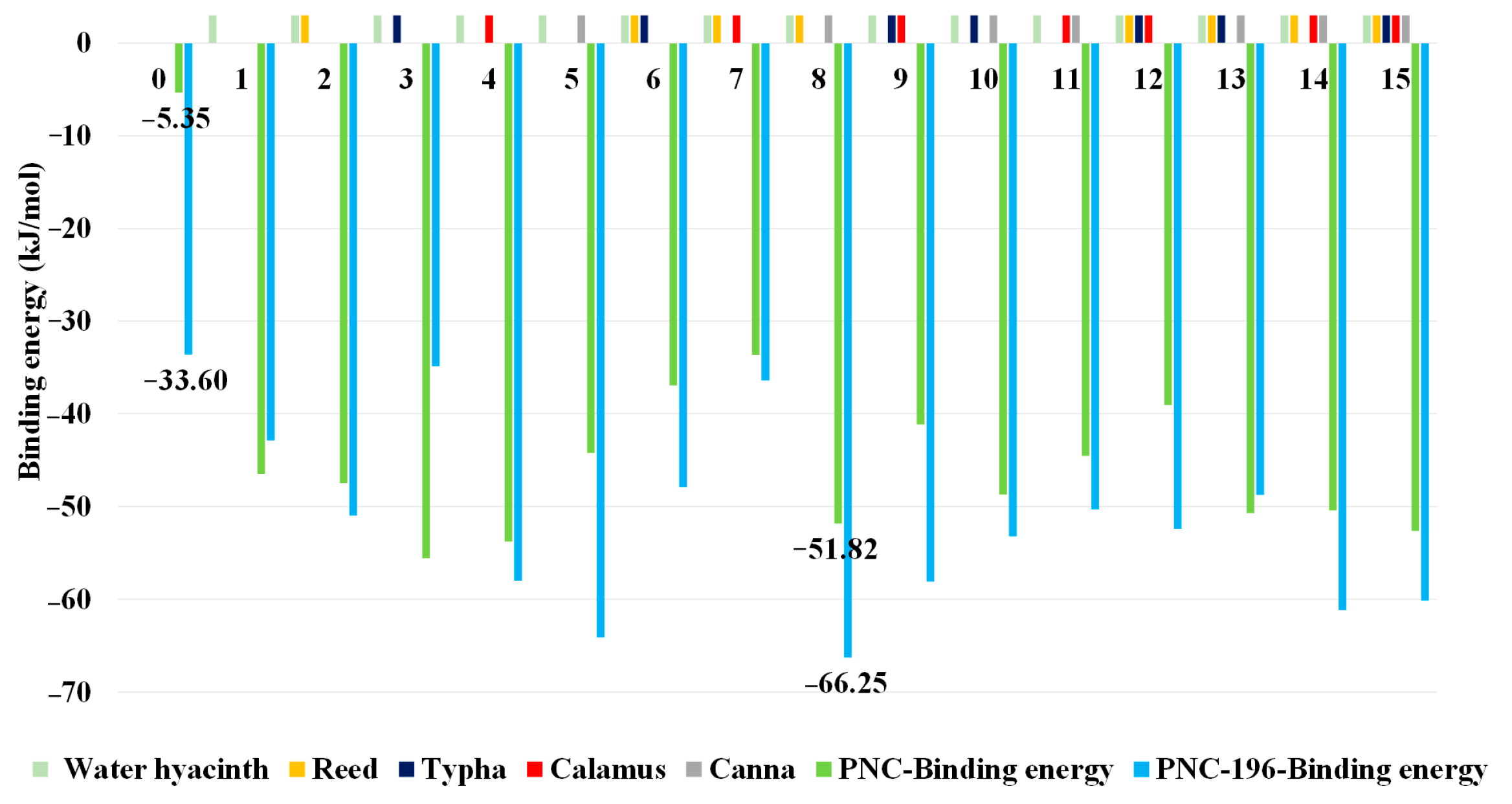
2.1. Acquisition of the Comprehensive Evaluation Value of Combined Biodegradability of β-Lactams Based on CM
2.2. Molecular Modification and Evaluation of High-Performance Combined Biodegradation of β-Lactams Based on the 3D-QSAR Model
2.2.1. 3D-QSAR Model Evaluation of High-Performance Combined Biodegradability of β-Lactams
2.2.2. Molecular Modification of High-Performance Combined Biodegradation β-Lactam Substitutes
2.2.3. Evaluation of Environmental Friendliness of β-Lactam Substitutes and Validation of the 3D-QSAR Model
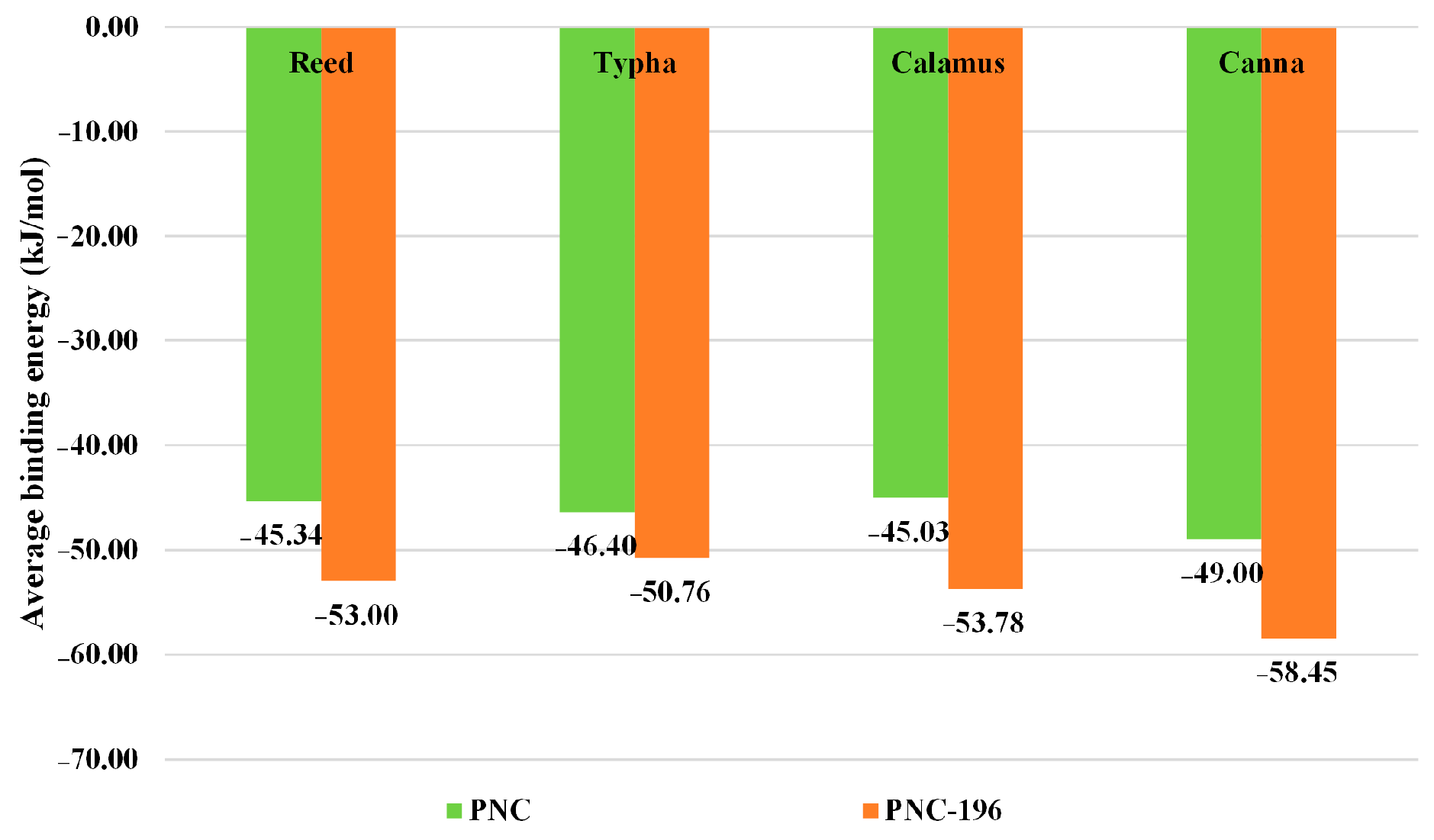
2.3. Screening of Antibacterial Resistance of β-Lactam Substitutes
2.3.1. Construction of Drug Resistant Mutations of E. coli PBP
2.3.2. Evaluation and Screening of Antibacterial Resistant Properties of β-Lactam Substitutes
2.3.3. Expression of Drug Resistance in E. coli and Analysis of the Mechanism of Resistance to β-Lactam Substitute
2.4. Construction, Screening, and Characterization of CW Based on Molecular Dynamics Simulation to Eliminate the Effects of the β-Lactam System
3. Materials and Methods
3.1. Principles and Sources of Receptor Protein Selection for Characterizing the Combined Biodegradability of β-Lactams
3.2. Combined Biodegradation of β-Lactams Characterization
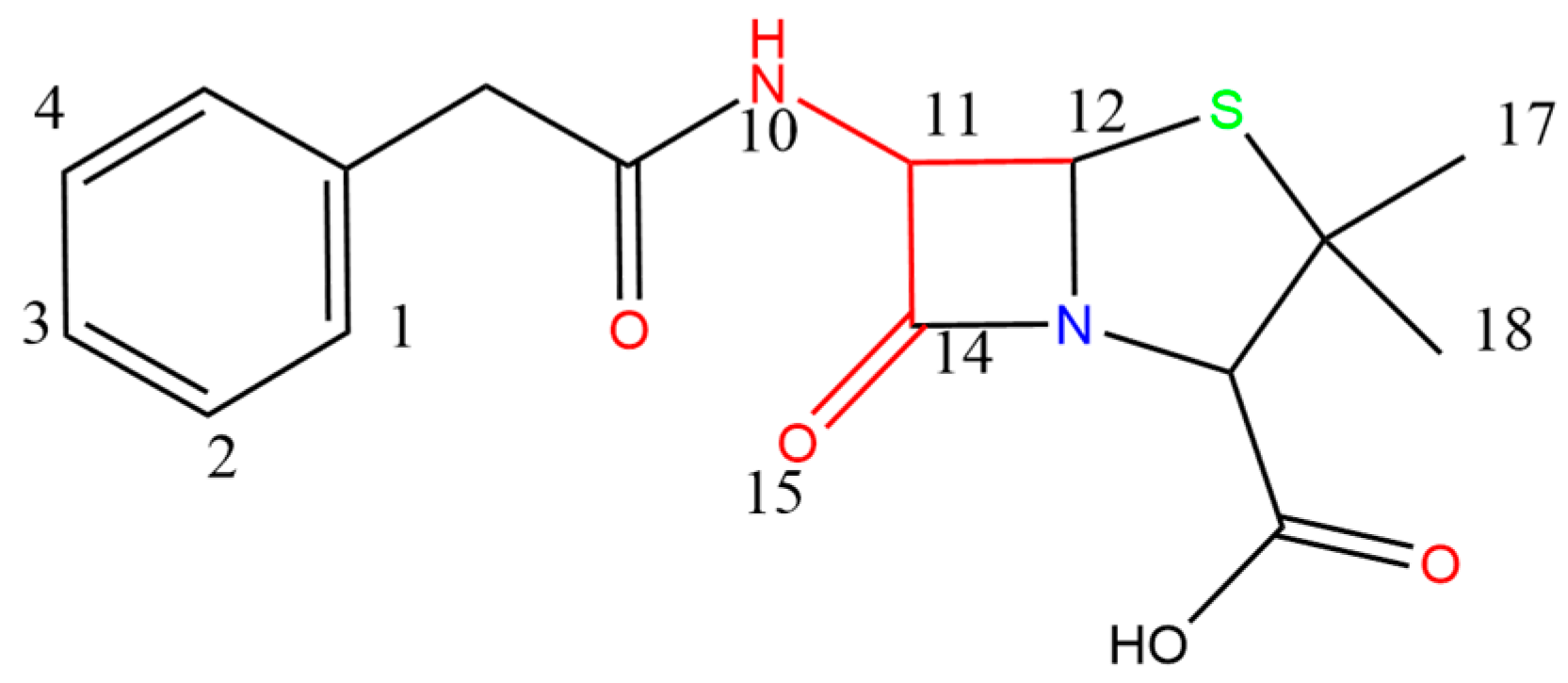
3.2.1. Characterization of Biodegradability of β-Lactams—Molecular Docking Method
3.2.2. Characterization of β-Lactams Combined Biodegradation—Comprehensive Matrix Scoring Method with Skewed Weights (CM)
3.3. Molecular Modification of β-Lactams with High Performance Combined Biodegradation Properties Using Quantitative Structure–Activity Relationship (QSAR) Model
3.4. Construction of Resistant Mutations of β-Lactam Target Receptor Proteins
3.4.1. Drug Resistant Mutations in E. coli PBP Using Sequence Manipulation Suite
3.4.2. Construction of Drug Resistant Mutations of E. coli PBP Using Homology Modeling Algorithms
3.5. System Construction for Integrated Elimination of the Effects of β-Lactams in CW Using Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirsch, R.; Ternes, T.; Haberer, K.; Kratz, K.L. Occurrence of Antibiotics in the Aquatic Environment. Sci. Total Environ. 1999, 225, 109–118. [Google Scholar] [CrossRef]
- Kumar, M.; Jaiswal, S.; Sodhi, K.K.; Shree, P.; Singh, D.K.; Agrawal, P.K.; Shukla, P. Antibiotics Bioremediation: Perspectives on Its Ecotoxicity and Resistance. Environ. Int. 2019, 124, 448–461. [Google Scholar] [CrossRef]
- Chen, Q.W.; Guo, X.; Hua, G.F.; Li, G.L.; Feng, R.R.; Liu, X.L. Migration and Degradation of Swine Farm Tetracyclines at the River Catchment Scale: Can the Multi-Pond System Mitigate Pollution Risk to Receiving Rivers? Environ. Pollut. 2017, 220, 1301–1310. [Google Scholar] [CrossRef]
- Camara, M.; Gallego-Picó, A.; Garcinuno, R.M.; Fernández-Hernando, P.; Durand-Alegría, J.S.; Sánchez, P.J. An HPLC-DAD Method for the Simultaneous Determination of Nine β-lactams Antibiotics in Ewe Milk. Food Chem. 2013, 141, 829–834. [Google Scholar] [CrossRef]
- Shao, Y.T.; Wang, Y.P.; Yuan, Y.W.; Xie, Y.J. A Systematic Review on Antibiotics Misuse in Livestock and Aquaculture and Regulation Implications in China. Sci. Total Environ. 2021, 798, 149205. [Google Scholar] [CrossRef]
- Chen, N.; Chen, S.; Chao, L.; Sun, L.; Zheng, D.M.; Liu, Q.; Zhang, Q. Study on Plant-Microbial Remediation of Antibiotic and Heavy Metal Contaminated Soil. Appl. Mech. Mater. 2014, 587, 816–819. [Google Scholar] [CrossRef]
- Gu, W.W.; Zhao, Y.Y.; Li, Q.; Li, Y. Plant-Microorganism Combined Remediation of Polychlorinated Naphthalenes Contaminated Soils Based on Molecular Directed Transformation and Taguchi Experimental Design-Assisted Dynamics Simulation. J. Hazard. Mater. 2020, 396, 122753. [Google Scholar] [CrossRef]
- Zhou, H.D.; Zhang, J.Y.; Cui, J.Y.; Li, D.Y.; Huang, L.P. Membrane Combined with Artificial Floating Ecosystems for the Removal of Antibiotics and Antibiotic-resistance Genes from Urban Rivers. J. Environ. Eng. 2021, 9, 106070. [Google Scholar] [CrossRef]
- Al-Ahmad, A.; Daschner, F.D.; Kümmerer, K. Biodegradability of Cefotiam, Ciprofloxacin, Meropenem, Penicillin G, and Sulfamethoxazole and Inhibition of Wastewater Bacteria. Arch. Environ. Contam. Toxicol. 1999, 37, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.H.; Ge, Q.P. Detection and Degradation of Antibiotics in Ecological Environment: A Review. Chin. Agric. Sci. Bull. 2021, 37, 59–64. [Google Scholar]
- Yu, F.; Han, S.; Ma, J. Adsorptive removal of antibiotics from aqueous solution using carbon materials. Chemosphere 2016, 153, 365–385. [Google Scholar] [CrossRef] [PubMed]
- Bartolomeu, M.; Neves, M.G.P.M.S.; Faustino, M.A.F.; Almeida, A. Wastewater chemical contaminants: Remediation by advanced oxidation processes. Photochem. Photobiol. Sci. 2018, 17, 1573–1589. [Google Scholar] [CrossRef] [PubMed]
- Rozas, O.; Contreras, D.; Mondaca, M.A.; Pérez-Moya, M.; Mansilla, H. Experimental design of fenton and photo-fenton reactions for the treatment of ampicillin solutions. J. Hazard. Mater. 2010, 177, 1025–1030. [Google Scholar] [CrossRef]
- Alvarino, T.; Suarez, S.; Lema, J.; Omil, F. Understanding the sorption and biotransformation of organic micropollutants in innovative biological wastewater treatment technologies. Sci. Total Environ. 2018, 615, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.M.; Ullman, J.L.; Teel, A.L.; Watts, R.J. pH and temperature effects on the hydrolysis of three β-lactam antibiotics: Ampicillin, cefalotin and cefoxitin. Sci. Total Environ. 2014, 466, 547–555. [Google Scholar] [CrossRef]
- Mabey, W.; Mill, T. Critical review of hydrolysis of organic compounds in water under environmental conditions. J. Phys. Chem. Ref. Data 1978, 7, 383–415. [Google Scholar] [CrossRef]
- Sheng, F. Transformation and the Associated Toxicity of Penicillin Antibiotics in Natural Water and Soil Mineral Environments. Ph.D. Thesis, Nanjing University, Nanjing, China, 2019. [Google Scholar]
- Hijosa-Valsero, M.; Fink, G.; Schlüsener, M.P.; Sidrach-Cardona, R.; Martín-Villacorta, J.; Ternes, T. Removal of Antibiotics from Urban Wastewater by Constructed Wetland Optimization. Chemosphere 2011, 83, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, Y.H.; Wang, Z.; Liu, C.X.; Huang, X.; Zhu, G.F. Behavior of Tetracycline and Sulfamethazine with Corresponding Resistance Genes from Swine Wastewater in Pilot-Scale Constructed Wetlands. J. Hazard. Mater. 2014, 278, 304–310. [Google Scholar] [CrossRef]
- Yang, J.W.; Zhao, Y.Y.; Li, M.H.; Du, M.J.; Li, X.X.; Li, Y. A Review of a class of emerging contaminants: The classification, distribution, intensity of consumption, synthesis routes, environmental effects and expectation of pollution abatement to organophosphate flame retardants (OPFRs). Int. J. Mol. Sci. 2019, 20, 2874. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, C.X.; Li, K.; Su, J.Q.; Zhu, G.F. Performance of Vertical up-flow Constructed Wetlands on Swine Wastewater Containing Tetracyclines and Tet Genes. Water Res. 2015, 70, 109–117. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, L.H.; Zoh, K.D. Removal Characteristics and Mechanism of Antibiotics Using Constructed Wetlands. Ecol. Eng. 2016, 91, 85–92. [Google Scholar] [CrossRef]
- Dorival-García, N.; Zafra-Gómez, A.; Navalón, A.; González-López, J.; Hontoria, E.; Vílchez, J.L. Removal and Degradation Characteristics of Quinolone Antibiotics in Laboratory-Scale Activated Sludge Reactors under Aerobic, Nitrifying and Anoxic Conditions. J. Environ. Manag. 2013, 120, 75–83. [Google Scholar] [CrossRef]
- Li, X.X.; Gu, W.W.; Chen, B.; Zhu, Z.W.; Zhang, B. Functional Modification of HHCB: Strategy for Obtaining Environmentally Friendly Derivatives. J. Hazard. Mater. 2021, 416, 126116. [Google Scholar] [CrossRef]
- He, Y.J.; Zhou, K.P.; Rao, Y.X.; Ji, R. Environmental Risks of Antibiotics in Soil and the Related Bioremediation Technologies. Chin. J. Biotechnol. 2021, 37, 3487–3504. [Google Scholar] [CrossRef]
- Lee, S.C.; Harner, T.; Pozo, K.; Shoeib, M.; Wania, F.; Muir, D.C.G.; Barrie, L.A.; Jones, K.C. Polychlorinated Naphthalenes in the Global Atmospheric Passive Sampling (GAPS) Study. Environ. Sci. Technol. 2007, 41, 2680–2687. [Google Scholar] [CrossRef] [PubMed]
- Mottl, H.; Nieland, P.; De, K.G.; Wierenga, J.J.; Keck, W. Deletion of an Additional Domain Located Between Sxxk and Sxn Active-site Fingerprints in Penicillin Binding Protein 4 from Escherichia Coli. J. Bacteriol. 1992, 174, 3261–3269. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chu, Z.H.; Li, Y. Designing Modified Polybrominated Diphenyl Ether BDE-47, BDE-99, BDE-100, BDE-183, and BDE-209 Molecules with Decreased Estrogenic Activities Using 3D-QSAR, Pharmacophore Models Coupled with Resolution V of the 210-3 Fractional Factorial Design and Molecular Docking. J. Hazard. Mater. 2019, 364, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Shi, M. Inhibition on Coliphage of Plants Root Exudates. Ph.D. Thesis, Ocean University of China, Qingdao, China, 2014. [Google Scholar]
- Wu, H.L.; Wang, X.Z.; He, X.J. Effects of Selected Root Exudate Components on Nitrogen Removal and Development of Denitrifying Bacteria in Constructed Wetlands. Water 2017, 9, 430. [Google Scholar] [CrossRef]
- Yi, L.L. Experimental Study on Townlet Domestic Sewage Treatment with Integrated Vertical Flow Constructed Wetland. Ph.D. Thesis, Henan University, Kaifeng, China, 2016. [Google Scholar]
- Kumar, K.; Gupta, S.C.; Baidoo, S.K.; Chander, Y.; Rosen, C.J. Antibiotic Uptake by Plants from Soil Fertilized with Animal Manure. J. Environ. Qual. 2005, 34, 2082–2085. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.X.; Xu, H.H.; Wang, Y.W.; Li, Y.; Han, S.; Ren, J.B. Combined Toxicity Characteristics and Regulation of Residual Quinolone Antibiotics in Aquatic Environment. Chemosphere 2021, 263, 128301. [Google Scholar] [CrossRef] [PubMed]
- Berger, W.A.; Mattina, M.I.; White, J.C. Effect of Hydrogen Peroxide on the Uptake of Chlordane by Cucurbita Pepo. Plant Soil 2012, 360, 135–144. [Google Scholar] [CrossRef]
- Gu, W.W.; Li, X.X.; Li, Q.; Hou, Y.L.; Zheng, M.S.; Li, Y. Combined Remediation of Polychlorinated Naphthalene-Contaminated Soil under Multiple Scenarios: An Integrated Method of Genetic Engineering and Environmental Remediation Technology. J. Hazard. Mater. 2020, 405, 124139. [Google Scholar] [CrossRef]
- Zhou, P.C. Removal of Antibiotics in Constructed Wetland and Its Effect on Plant Growth and Sewage Treatment. M.D. thesis, South China Agricultural University, Guangzhou, China, 2019. [Google Scholar]
- Li, X.X.; Gu, W.W.; Zhang, B.Y.; Xin, X.Y.; Kang, Q.; Yang, M.; Chen, B.; Li, Y. Insights into Toxicity of Polychlorinated Naphthalenes to Multiple Human Endocrine Receptors: Mechanism and Health Risk Analysis. Environ. Int. 2022, 165, 107291. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Hou, Y.L.; Li, Y. Multi-Directional Selective Toxicity Effects on Farmland Ecosystems: A Novel Design of Green Substitutes for Neonicotinoid Insecticides. J. Clean. Prod. 2020, 272, 122715. [Google Scholar] [CrossRef]
- Riedel, S.L.; Pitz, G.F. Utilization-Oriented Evaluation of Decision Support Systems. IEEE Trans. Syst. Man Cybern. 1986, 16, 980–996. [Google Scholar] [CrossRef]
- Feng, S.; Xu, L.D. Decision Support for Fuzzy Comprehensive Evaluation of Urban Development. Fuzzy Sets Syst. 1999, 105, 1–12. [Google Scholar] [CrossRef]
- Ren, Z.X.; Zhao, Y.Y.; Han, S.; Li, X. Regulatory Strategies for Inhibiting Horizontal Gene Transfer of ARGs in Paddy and Dryland Soil Through Computer-based Methods. Sci. Total Environ. 2023, 856, 159096. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, X.Y.; Jiang, L.; Li, Y. Prediction of Octanol-Air Partition Coefficients for Polychlorinated Biphenyls (PCBs) Using 3D-QSAR Models. Ecotoxicol. Environ. Saf. 2016, 124, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.L.; Qu, R.J.; Feng, M.B.; Chen, J.; Wang, L.S.; Wang, Z.Y. Photodegradation of Polyfluorinated Dibenzo-p-Dioxins (PFDDs) in Organic Solvents: Experimental and Theoretical Studies. Environ. Sci. Technol. 2016, 50, 8128–8134. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Li, Y. Modified Neonicotinoid Insecticide with Bi-Directional Selective Toxicity and Drug Resistance. Ecotoxicol. Environ. Saf. 2018, 164, 467–473. [Google Scholar] [CrossRef]
- Qu, R.J.; Liu, H.X.; Feng, M.B.; Yang, X.; Wang, Z.Y. Investigation on Intramolecular Hydrogen Bond and Some Thermodynamic Properties of Polyhydroxylated Anthraquinones. J. Chem. Eng. Data 2012, 57, 2442–2455. [Google Scholar] [CrossRef]
- Ren, Z.X.; Wang, S.; Liu, D.; Yu, J.; Zhang, X.Y.; Zhao, P.N.; Sun, Y.X.; Han, S. Control Strategies for the Vertical Gene Transfer of Quinolone ARGs in Escherichia Coli Through Molecular Modification and Molecular Dynamics. J. Hazard. Mater. 2021, 420, 126667. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.Z.; Liu, M. A Double-Activity (Green Algae Toxicity and Bacterial Genotoxicity) 3D-QSAR Model Based on the Comprehensive Index Method and Its Application in Fluoroquinolones’ Modification. Int. J. Environ. Res. Public Health 2020, 17, 942. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gehring, R.; Riviere, J.E.; Lin, Z.M. Development and Application of a Population Physiologically Based Pharmacokinetic Model for Penicillin G in Swine and Cattle for Food Safety Assessment. Food Chem. Toxicol. 2017, 107, 74–87. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A Web-Based Environment for Protein Structure Homology Modelling. Bioinformatics 2006, 2, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling Protein Quaternary Structure of Homo-and Hetero-oligomers Beyond Binary Interactions by Homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; De Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new Features and Functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated Comparative Protein Structure Modeling with SWISS-MODEL and Swiss-PdbViewer: A Historical Perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Schwede, T. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance Constraints Applied on Model Quality Estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Rainey, P.B. Adaptation of Pseudomonas Fluorescens to the Plant Rhizosphere. Environ. Microbiol. 1999, 1, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Li, Y. Design of Environmentally Friendly Neonicotinoid Insecticides with Bioconcentration Tuning and Bi-Directional Selective Toxic Effects. J. Clean. Prod. 2019, 221, 113–121. [Google Scholar] [CrossRef]
- Ren, Z.X.; Zhao, Y.Y.; Huang, J.; Han, S.; Wang, Y.W. Validation and Inhibition Study for Toxic Expression of Quinolone Antibiotic Resistance Genes in Agricultural Soils of Eastern China. Ecotoxicol. Environ. Saf. 2022, 241, 113806. [Google Scholar] [CrossRef]
- Deng, Z.Y.; Ren, Z.X.; Sun, S.H.; Wang, Y.J. Theoretical Design and Process Control of Neonicotinoids Insecticides Suitable for Synergistic Degradation with the Rubisco Enzyme from Rhizobia and Carbon-fixing Bacteria in Soil. Environ. Sci. Pollut. Res. 2022, 29, 12355–12376. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Zhao, Y.Y.; Chen, B.; Zhu, Z.W.; Kang, Q.; Husain, T.; Zhang, B.Y. Inhalation and Ingestion of Synthetic Musks in Pregnant Women: In Silico Spontaneous Abortion Risk Evaluation and Control. Environ. Int. 2022, 158, 106911. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Li, X.A.; Zhao, Y.Y.; Pu, Q.K.; Li, Y.; Gu, W.W. Efficient and Synergistic Degradation of Fluoroquinolones by Bacteria and Microalgae: Design of Environmentally Friendly Substitutes, Risk Regulation and Mechanism Analysis. J. Hazard. Mater. 2022, 437, 129384. [Google Scholar] [CrossRef]
- Pu, Q.K.; Han, Z.Z.; Li, X.A.; Li, Q.; Li, Y. Designing and Screening of Fluoroquinolone Substitutes Using Combined in Silico Approaches: Biological Metabolism–bioconcentration Bilateral Selection and Their Mechanism Analyses. Green Chem. 2022, 249, 3778–3793. [Google Scholar] [CrossRef]


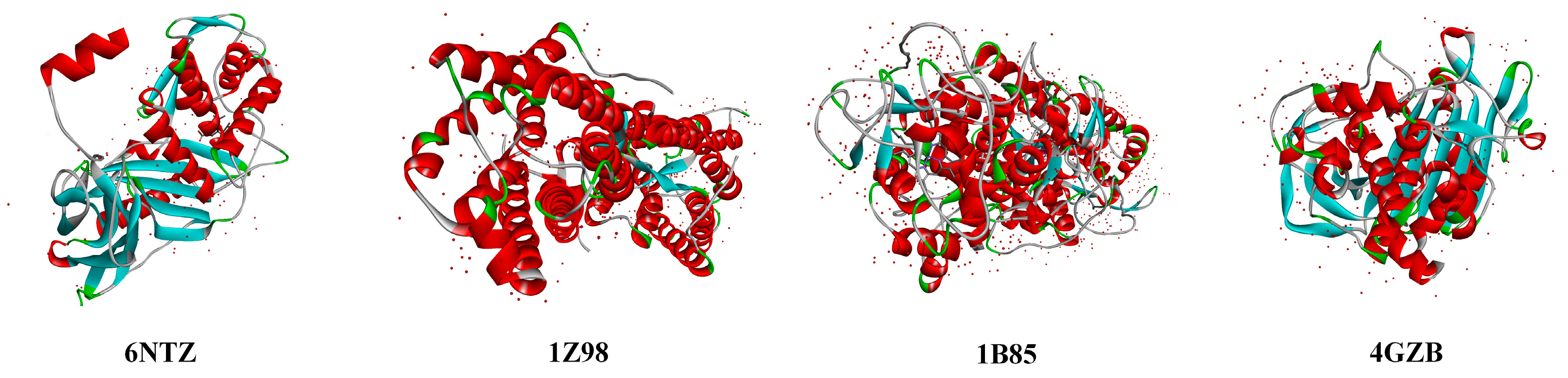

| No. | Compounds | 6NTZ (Å) | Change Rate (%) | 1Z98 (Å) | Change Rate (%) | 1B85 (Å) | Change Rate (%) | 4GZB (Å) | Change Rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| - | PNC | 70.00 | - | 95.11 | - | 67.49 | - | 69.72 | - |
| PNC-3 | PNC-18-N=NH2-17-NH-NH2 | 108.74 | 55.34 | 113.58 | 19.42 | 78.00 | 15.57 | 91.17 | 30.77 |
| PNC-14 | PNC-18-N=NH2-7-NH-NH2 | 111.76 | 15.75 | 113.56 | 8.85 | 82.69 | 22.51 | 111.76 | 15.75 |
| PNC-20 | PNC-18-N=NH2-7-SO | Failed | - | Failed | - | Failed | - | Failed | - |
| PNC-21 | PNC-18-N=NH2-7-SO2 | Failed | - | Failed | - | Failed | - | Failed | - |
| PNC-23 | PNC-18-NH-NH2 | 105.78 | 9.57 | 103.20 | −1.08 | 78.85 | 16.82 | 105.78 | 9.57 |
| PNC-28 | PNC-18-NO2-17-N=NH2 | 100.72 | 4.33 | 106.67 | 2.25 | 75.46 | 11.79 | 100.72 | 4.33 |
| PNC-34 | PNC-18-NO2-17-N=NH2-7-OCHO | 114.13 | 18.21 | 106.30 | 1.89 | 84.05 | 24.53 | 114.13 | 18.21 |
| PNC-36 | PNC-18-NO2-17-N=NH2-7-OOH | 114.56 | 18.66 | 103.20 | −1.08 | 81.57 | 20.86 | 114.56 | 18.66 |
| PNC-43 | PNC-18-NO2-17-N3-7-NH-NH2 | Failed | - | Failed | - | Failed | - | Failed | - |
| PNC-47 | PNC-18-NO2-17-N3-7-OOH | Failed | - | Failed | - | Failed | - | Failed | - |
| PNC-53 | PNC-18-NO2-17-NH-NH2-7-N3 | Failed | Failed | - | Failed | - | Failed | - | |
| PNC-66 | PNC-18-NO2-17-NO-7-NO | 111.66 | 15.65 | 104.66 | 0.32 | 89.07 | 31.97 | 111.66 | 15.65 |
| PNC-68 | PNC-18-NO2-17-NO-7-OCHO | 112.12 | 16.13 | 107.94 | 3.46 | 89.85 | 33.12 | 112.12 | 16.13 |
| PNC-69 | PNC-18-NO2-17-NO-7-ONO | 109.93 | 13.86 | 107.25 | 2.80 | 88.48 | 31.10 | 109.93 | 13.86 |
| PNC-71 | PNC-18-NO2-17-NO-7-SH | 101.27 | 4.89 | 110.64 | 6.05 | 83.90 | 24.31 | 101.27 | 4.89 |
| PNC-75 | PNC-18-NO2-17-NO2-7-N=NH2 | 109.58 | 13.50 | 110.73 | 6.14 | 83.67 | 23.97 | 109.58 | 13.50 |
| PNC-77 | PNC-18-NO2-17-NO2-7-NO2 | 109.72 | 13.64 | 111.22 | 6.61 | 82.36 | 22.02 | 109.72 | 13.64 |
| PNC-79 | PNC-18-NO2-17-NO2-7-ONO | 117.76 | 21.97 | 122.57 | 17.49 | 86.80 | 28.60 | 117.76 | 21.97 |
| PNC-81 | PNC-18-NO2-17-NO2-7-SH | 102.31 | 5.97 | 107.86 | 3.38 | 79.00 | 17.05 | 102.31 | 5.97 |
| PNC-86 | PNC-18-NO2-17-ONO | 104.62 | 8.36 | 112.15 | 7.50 | 81.55 | 20.83 | 104.62 | 8.36 |
| PNC-98 | PNC-18-ONO-17-NO2 | 107.56 | 11.40 | 107.11 | 2.66 | 74.38 | 10.21 | 107.56 | 11.40 |
| PNC-108 | PNC-18-SO2-17-N=NH2-7-N3 | Failed | Failed | - | Failed | - | Failed | - | |
| PNC-118 | PNC-18-SO2-17-N3 | Failed | Failed | - | Failed | - | Failed | - | |
| PNC-168 | PNC-18-SO2-17-SH-7-N3 | Failed | Failed | - | Failed | - | Failed | - | |
| PNC-196 | PNC-7-OOH | 78.49 | 12.13 | 112.88 | 18.68 | 78.22 | 15.89 | 92.43 | 32.06 |
| Name | Mutation Zone | Key Residues and Adjacent Amino Acid Sequences | LibDock Score (Å) | Change Rate (%) |
|---|---|---|---|---|
| PBP | - | ASLTKMM | 87.32 | - |
| PBP-1 | 72–78 | HFYFGDA | 93.27 | −6.82 |
| PBP-2 | 72–78 | QCIDETT | 90.76 | −3.94 |
| PBP-3 | 72–78 | YIWCQDY | 95.91 | −9.84 |
| PBP-4 | 72–78 | KPFPVYW | 88.03 | −0.81 |
| PBP-5 | 72–78 | VRYSVRW | 108.51 | −24.27 |
| PBP | - | YSIYKE | 72.51 | - |
| PBP-6 | 209–214 | VRPDKF | 102.90 | −41.91 |
| PBP-7 | 209–214 | PWGNSP | 108.49 | −49.61 |
| PBP-8 | 209–214 | QCSQTT | 113.24 | −56.16 |
| PBP-9 | 209–214 | MLISTN | 104.63 | −44.29 |
| PBP-10 | 209–214 | DDPDND | 106.83 | −47.32 |
| PBP | - | ASLTKMM, YSIYKE | 72.51 | - |
| PBP-11 | 72–78, 209–214 | RHRFCHR, WISQRE | 114.16 | −57.44 |
| PBP-12 | 72–78, 209–214 | REGTLEQ, FEYVRF | Failed | - |
| PBP-13 | 72–78, 209–214 | WRYSFQW, WESMDS | 117.49 | −62.02 |
| PBP-14 | 72–78, 209–214 | HVNWSPS, GANEVG | 58.11 | 19.86 |
| PBP-15 | 72–78, 209–214 | CALRDQY, PDGRGW | 71.79 | 1.00 |
| PBP-16 | 72–78, 209–214 | YFHMQNP, CVSPCN | 93.22 | −28.56 |
| PBP-17 | 72–78, 209–214 | FMYHCQV, FDVLLP | 108.13 | −49.12 |
| PBP-18 | 72–78, 209–214 | YHEGNAD, KRVRPF | 118.59 | −63.54 |
| PBP-19 | 72–78, 209–214 | ALVHGNC, HTKRYG | 85.59 | −18.04 |
| PBP-20 | 72–78, 209–214 | FNELHGF, GVNYCC | Failed | - |
| PBP-21 | 72–78, 209–214 | NKVFWAD, QWMRPS | Failed | - |
| PBP-22 | 72–78, 209–214 | YPTSRWV, NRTMVE | Failed | - |
| PBP-23 | 72–78, 209–214 | KVAEFLT, DDIHKR | 92.27 | −27.24 |
| PBP-24 | 72–78, 209–214 | EQFHREL, SIARGH | 92.07 | −26.97 |
| PBP-25 | 72–78, 209–214 | CWCWDLQ, MHAKMI | 117.05 | −61.41 |
| PBP-26 | 72–78, 209–214 | IHNYKHY, WHYFDI | 106.36 | −46.68 |
| PBP-27 | 72–78, 209–214 | WIHNIHF, SCDARW | 110.36 | −52.20 |
| PBP-28 | 72–78, 209–214 | FFTYLPD, HWGTNE | 115.45 | −59.21 |
| PBP-29 | 72–78, 209–214 | VMEEGNF, GDNDTC | 78.39 | −8.10 |
| PBP-30 | 72–78, 209–214 | FVSNSSK, AVGQRC | 114.91 | −58.47 |
| PBP-31 | 72–78, 209–214 | TWCRNST, EIVRNH | Failed | - |
| PBP-32 | 72–78, 209–214 | ILATADF, NWCFIH | 118.67 | −63.65 |
| PBP-33 | 72–78, 209–214 | RTNANWG, QALIRD | 85.10 | −17.35 |
| PBP-34 | 72–78, 209–214 | PKDTEHF, WCKNRT | 84.56 | −16.61 |
| PBP-35 | 72–78, 209–214 | AVEHVE, EKPHCT | 95.27 | −31.39 |
| No. | Compounds | LibDock Score (Å) | Change Rate (%) |
|---|---|---|---|
| - | PNC | 58.11 | - |
| PNC-3 | PNC-18-N=NH2-17-NH-NH2 | 63.36 | 9.04 |
| PNC-14 | PNC-18-N=NH2-7-NH-NH2 | 70.87 | 21.96 |
| PNC-28 | PNC-18-NO2-17-N=NH2 | Failed | - |
| PNC-34 | PNC-18-NO2-17-N=NH2-7-OCHO | 57.22 | −1.53 |
| PNC-66 | PNC-18-NO2-17-NO-7-NO | 31.14 | −46.41 |
| PNC-68 | PNC-18-NO2-17-NO-7-OCHO | 67.95 | 16.93 |
| PNC-69 | PNC-18-NO2-17-NO-7-ONO | Failed | - |
| PNC-71 | PNC-18-NO2-17-NO-7-SH | 71.06 | 22.29 |
| PNC-75 | PNC-18-NO2-17-NO2-7-N=NH2 | 62.13 | 6.92 |
| PNC-77 | PNC-18-NO2-17-NO2-7-NO2 | Failed | - |
| PNC-79 | PNC-18-NO2-17-NO2-7-ONO | Failed | - |
| PNC-81 | PNC-18-NO2-17-NO2-7-SH | 72.62 | 24.97 |
| PNC-86 | PNC-18-NO2-17-ONO | Failed | - |
| PNC-98 | PNC-18-ONO-17-NO2 | 43.55 | −25.06 |
| PNC-196 | PNC-7-OOH | 73.04 | 25.70 |
| PNC-PBP | |||
| Hydrophobicity | Hydrophilic | ||
| Non-polar amino acids | Polar amino acids | Polar positively charged amino acids (Basic amino acids) | Polar negatively charged amino acids (Acidic amino acids) |
| ALA86 ALA151 LEU161 | GLN85 | - | - |
| PNC-196-PBP | |||
| Hydrophobicity | Hydrophilic | ||
| Non-polar amino acids | Polar amino acids | Polar positively charged amino acids (Basic amino acids) | Polar negatively charged amino acids (Acidic amino acids) |
| ALA86 ALA89 ALA151 LEU161 | - | LYS91 | - |
| PNC-PBP resistant mutation | |||
| Hydrophobicity | Hydrophilic | ||
| Non-polar amino acids | Polar amino acids | Polar positively charged amino acids (Basic amino acids) | Polar negatively charged amino acids (Acidic amino acids) |
| - | TYR54 ASN55 | ARG203 LYS323 | ASP324 |
| PNC-196-PBP resistant mutation | |||
| Hydrophobicity | Hydrophilic | ||
| Non-polar amino acids | Polar amino acids | Polar positively charged amino acids (Basic amino acids) | Polar negatively charged amino acids (Acidic amino acids) |
| - | TYR54 ASN55 | ARG203 ARG373 | ASP324 |
| PNC-PBP | ||||
| Bond type | Bonding site | Active bond length | Average bond length | LibDock Score (Å) |
| Amide-Pi Stacked | GLN85-C1~C6 | 4.12 | 3.83 | 72.51 |
| Pi-Sigma | ALA86-C1~C6 | 3.72 | ||
| Carbon | ALA151-H33 | 2.25 | ||
| Pi-Alkyl | LEU161-C1~C6 | 5.23 | ||
| PNC-196-PBP | ||||
| Key bonds type | Bonding site | Key bonds length | Average bonds length | LibDock Score (Å) |
| Pi- Sigma | LEU161-C3~C8 | 3.02 | 3.96 | 76.14 |
| Pi-Alkyl | LYS91-C3~C8 | 5.11 | ||
| Pi-Alkyl | ALA151-C3~C8 | 4.66 | ||
| Alkyl | LYS91-C22 | 4.99 | ||
| Pi-Sigma | ALA86-C3~C8 | 3.42 | ||
| Carbon | ALA89-H35 | 2.57 | ||
| PNC-PBP resistant mutation | ||||
| Key bonds type | Bonding site | Key bonds length | Average bonds length | LibDock Score (Å) |
| Conventional | ASP324-H31 | 2.38 | 3.38 | 58.11 |
| Conventional | LYS323-O5 | 3.38 | ||
| Conventional | ASN55-S16 | 2.95 | ||
| Sulfur-X | ASN55-S16 | 3.31 | ||
| Carbon | ASN55-H33 | 2.95 | ||
| Carbon | ASN55-H32 | 2.96 | ||
| Pi-Alkyl | TYR54-H36 | 4.34 | ||
| Pi-Alkyl | TYR54-C20 | 4.56 | ||
| Alkyl | ARG203-C20 | 3.58 | ||
| PNC-196-PBP resistant mutation | ||||
| Key bonds type | Bonding site | Key bonds length | Average bonds length | LibDock Score (Å) |
| Pi-Alkyl | TYR54-C21 | 4.88 | 3.40 | 73.04 |
| Pi-Alkyl | TYR54-C22 | 4.54 | ||
| Alkyl | ARG203-C21 | 3.88 | ||
| Alkyl | ARG203-C22 | 4.20 | ||
| Donor-Donor | ARG373-H26 | 1.75 | ||
| Conventional | ASN55-S18 | 3.26 | ||
| Conventional | ASP324-H33 | 2.67 | ||
| Carbon | ASN55-H34 | 2.82 | ||
| Carbon | ASN55-H35 | 2.64 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Li, Z.; Ren, Z.; Li, Y. Multi-Dimensional Elimination of β-Lactams in the Rural Wetland: Molecule Design and Screening for More Antibacterial and Degradable Substitutes. Molecules 2022, 27, 8434. https://doi.org/10.3390/molecules27238434
Sun S, Li Z, Ren Z, Li Y. Multi-Dimensional Elimination of β-Lactams in the Rural Wetland: Molecule Design and Screening for More Antibacterial and Degradable Substitutes. Molecules. 2022; 27(23):8434. https://doi.org/10.3390/molecules27238434
Chicago/Turabian StyleSun, Shuhai, Zhuang Li, Zhixing Ren, and Yu Li. 2022. "Multi-Dimensional Elimination of β-Lactams in the Rural Wetland: Molecule Design and Screening for More Antibacterial and Degradable Substitutes" Molecules 27, no. 23: 8434. https://doi.org/10.3390/molecules27238434
APA StyleSun, S., Li, Z., Ren, Z., & Li, Y. (2022). Multi-Dimensional Elimination of β-Lactams in the Rural Wetland: Molecule Design and Screening for More Antibacterial and Degradable Substitutes. Molecules, 27(23), 8434. https://doi.org/10.3390/molecules27238434







