Folding Dynamics of 3,4,3-LI(1,2-HOPO) in Its Free and Bound State with U4+ Implicated by MD Simulations
Abstract
1. Introduction
2. Results and Discussion
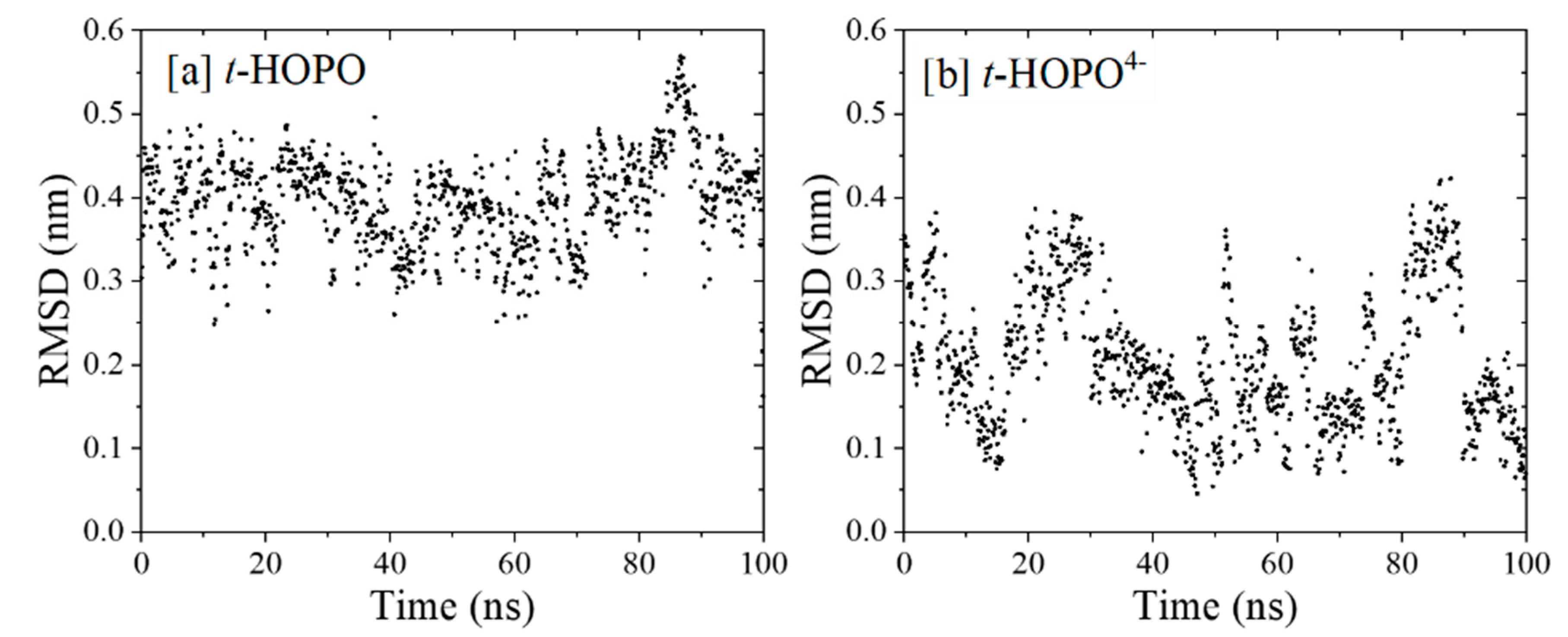
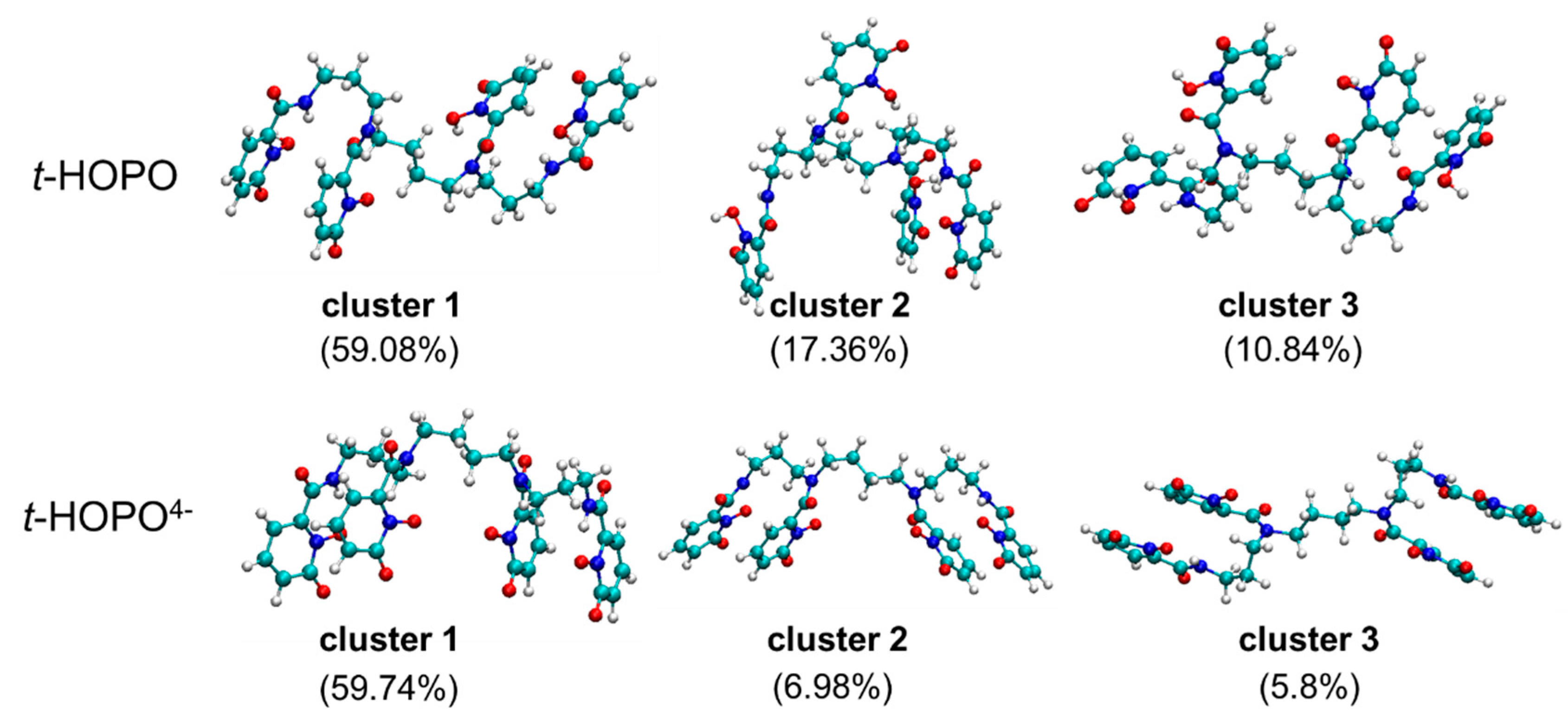
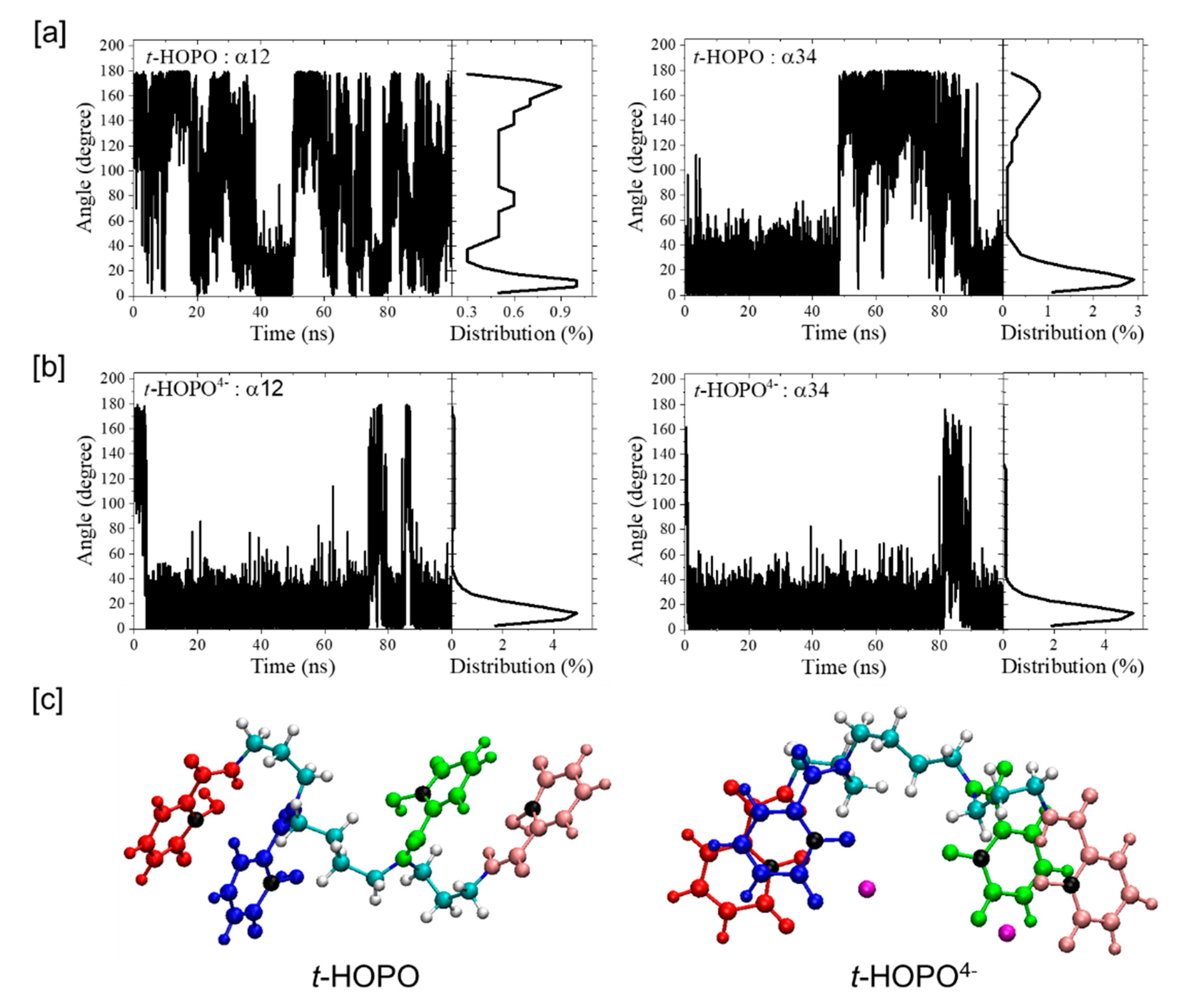
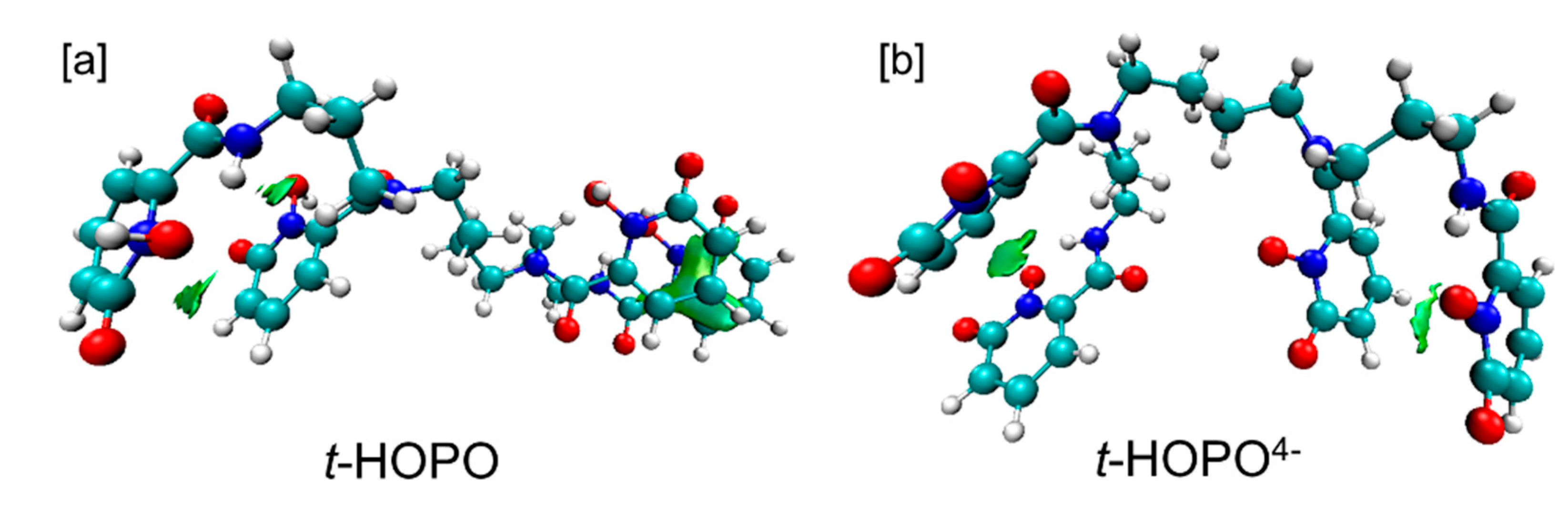
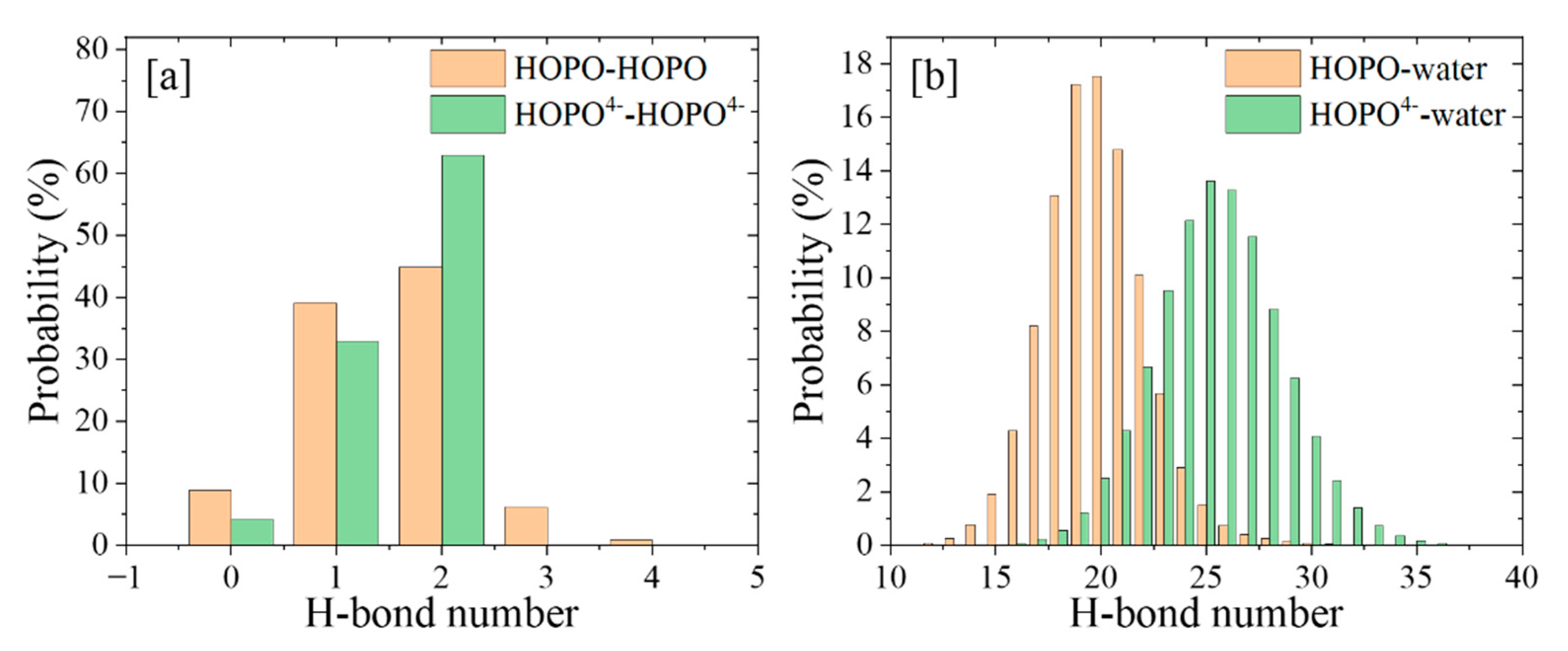
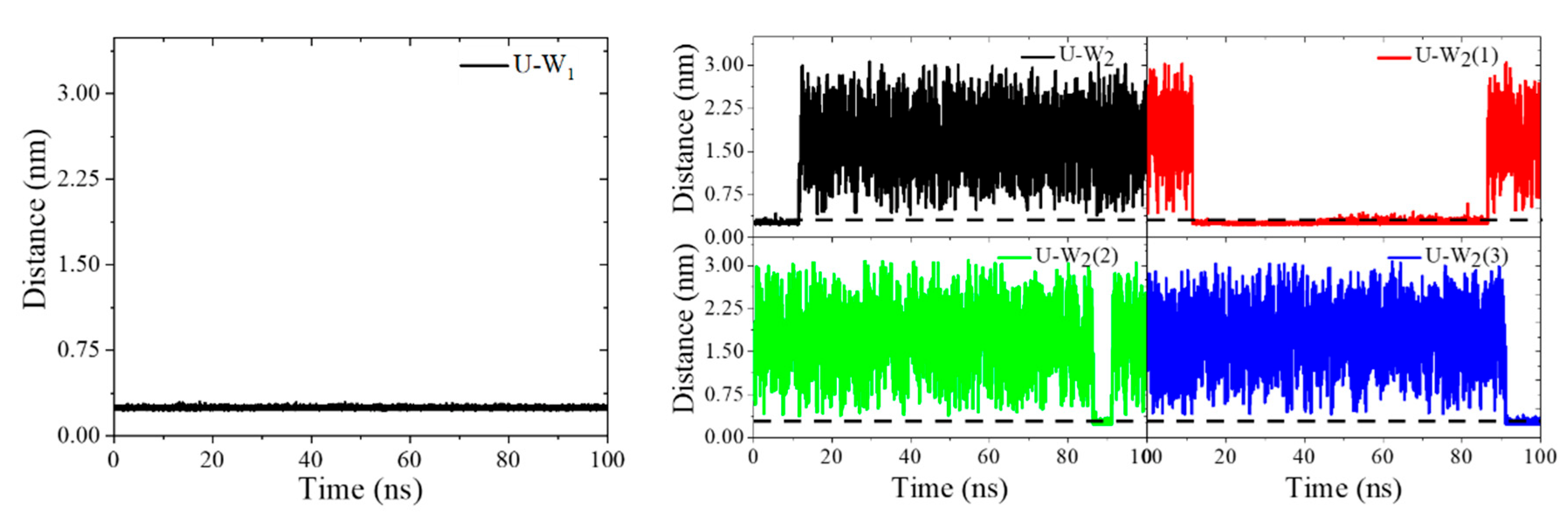
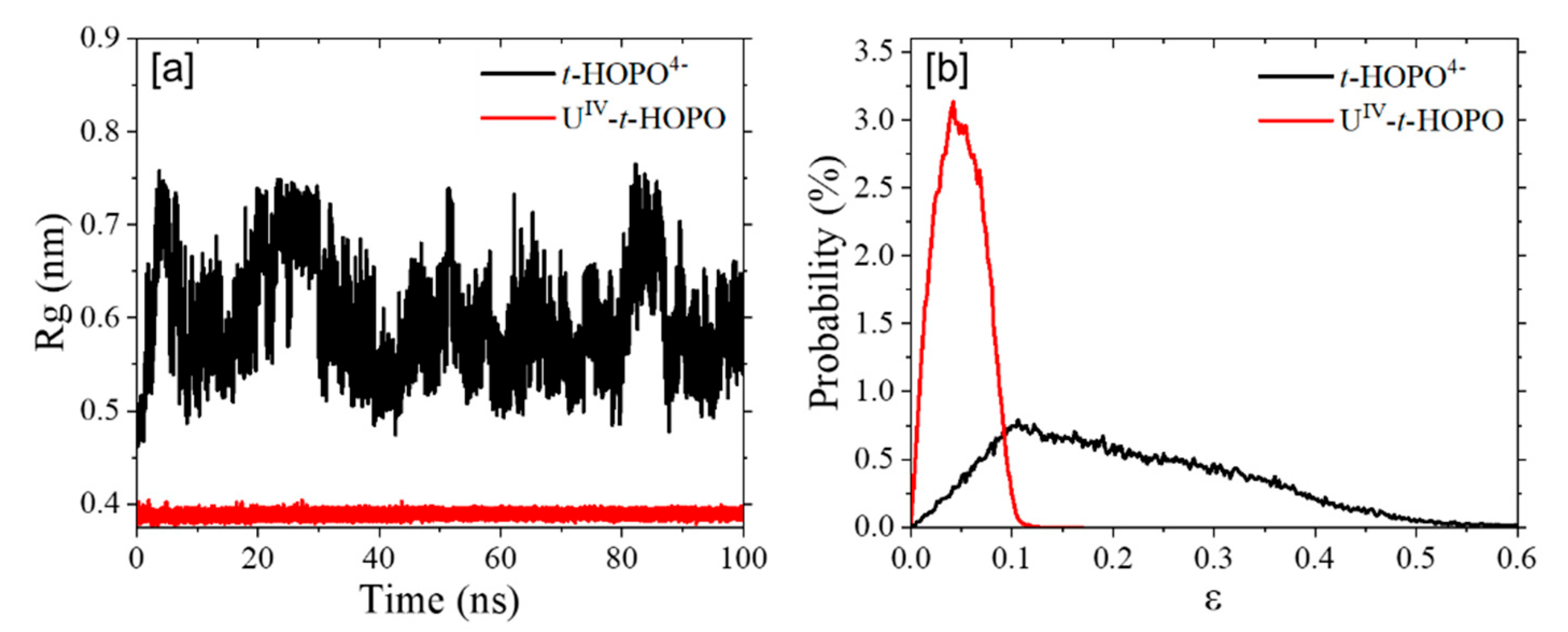
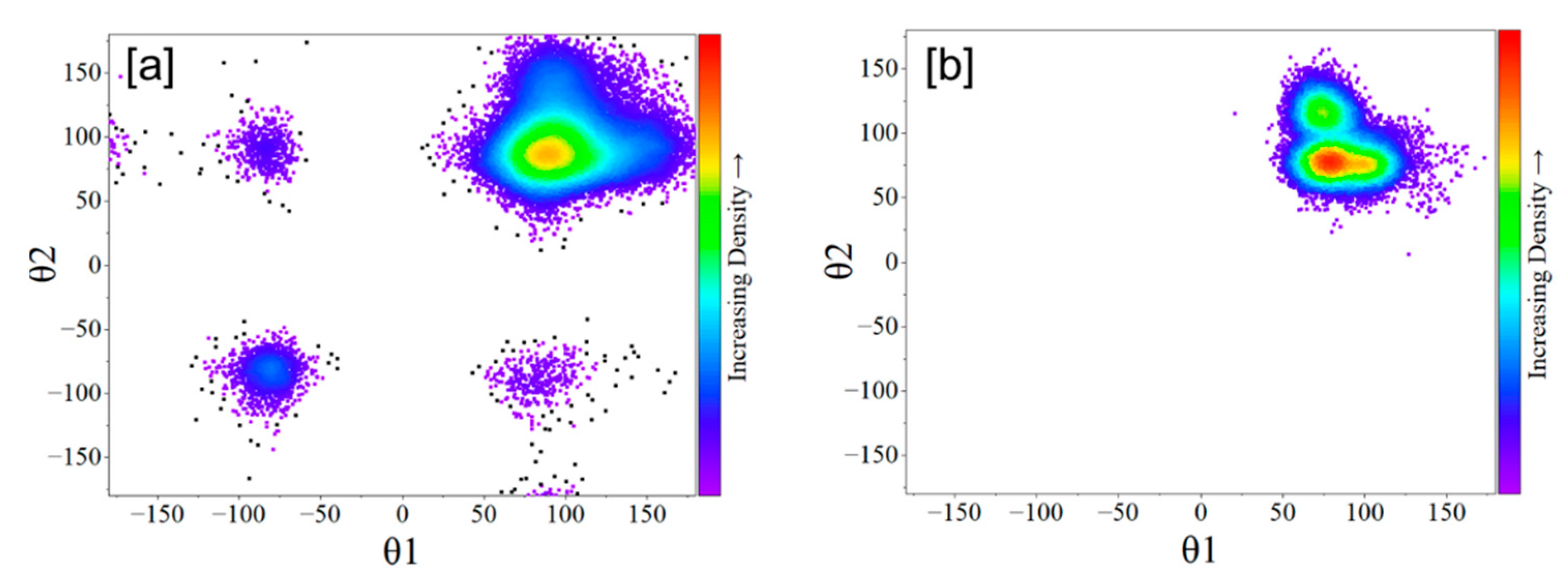
2.1. HOPO in Its Neutral and Deprotonated States
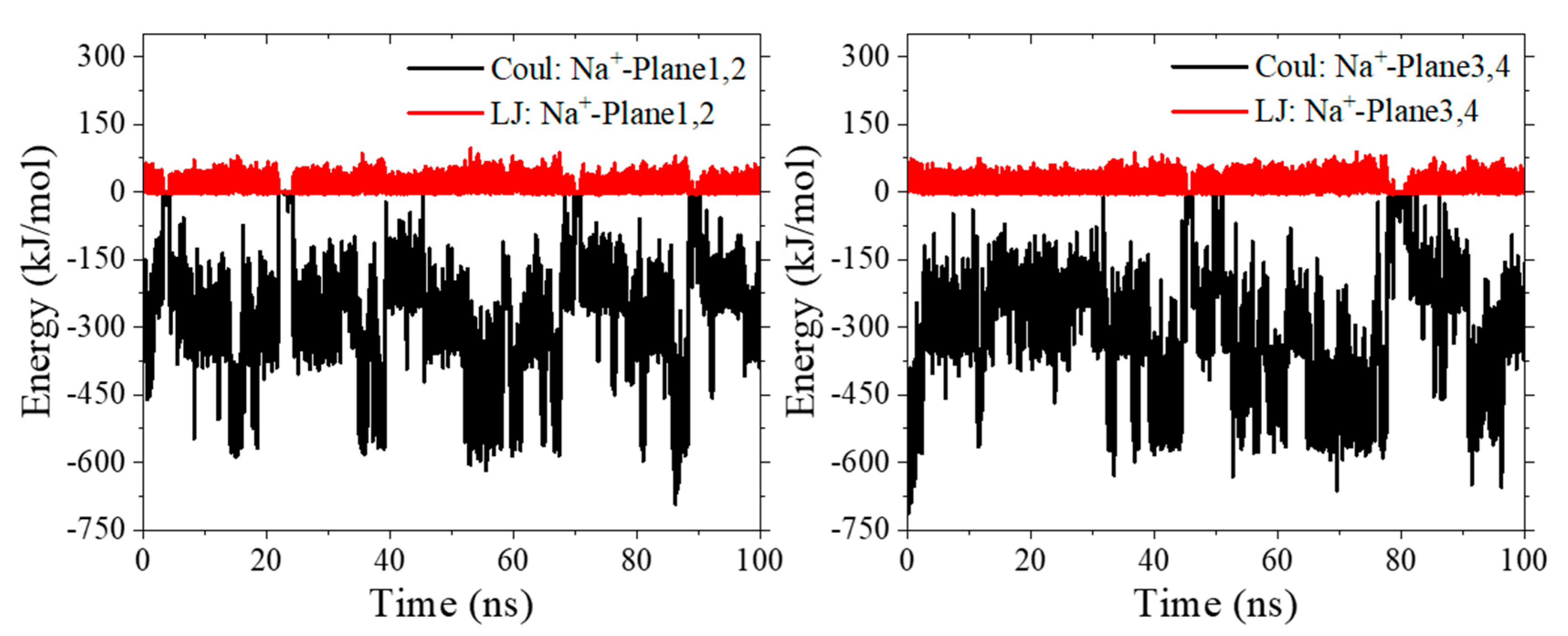
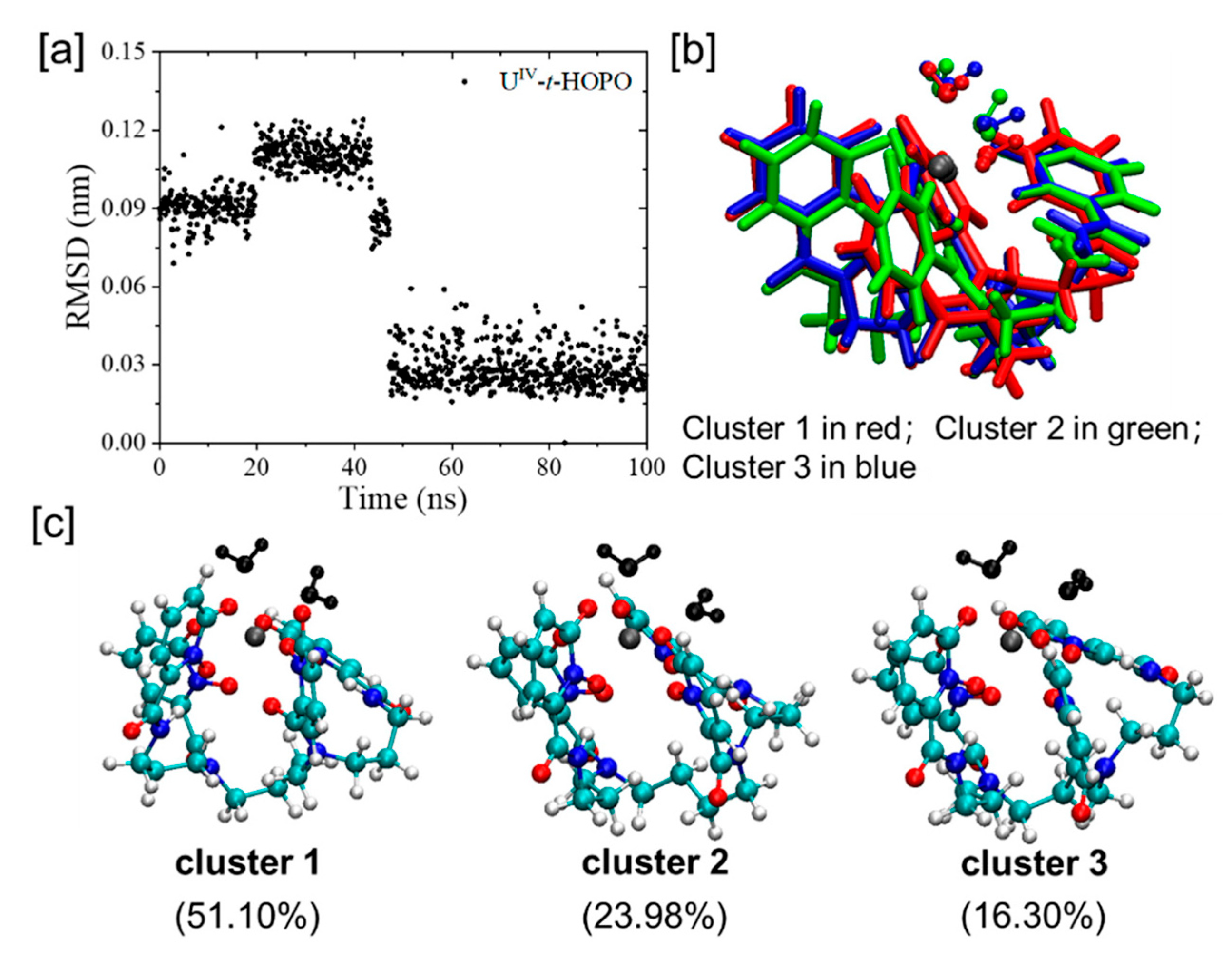
2.2. UⅣ-t-HOPO Complexes
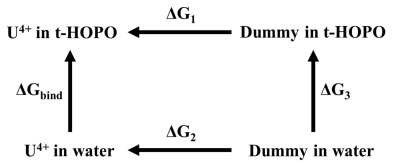 |
3. Computational Detail Methodology
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jee, S.S.W. The Health Effects of Plutonium and Radium: Proceedings of the Symposium, Sun Valley, Idaho, USA, 6–9 October 1975; JW Press: Martin, TN, USA, 1976. [Google Scholar]
- Nénot, J.-C.; Stather, J.W. The Toxicity of Plutonium, Americium and Curium: A Report Prepared under Contract for the Commission of the European Communities within Its Research and Development Programme on Plutonium Recycling in Light Water Reactors; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Sigel, H. Metal Ions in Biological Systems: Volume 42: Metal Complexes in Tumor Diagnosis and as Anticancer Agents; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Meunier, B.; Robert, A.; Crisponi, G.; Nurchi, V.; Lachowicz, J.I.; Nairz, M.; Weiss, G.; Pai, A.B.; Ward, R.J.; Crichton, R.R. Essential Metals in Medicine: Therapeutic Use and Toxicity of Metal Ions in the Clinic; Walter de Gruyter GmbH & Co KG: Berlin, Germany, 2019; Volume 19. [Google Scholar]
- Barnham, K.J.; Bush, A.I. Biological metals and metal-targeting compounds in major neurodegenerative diseases. Chem. Soc. Rev. 2014, 43, 6727–6749. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.G.; Crowley, J.; Foreman, H. Tracer studies: The metabolism of the fission products. In Medical and Health Physics Divisions Quarterly Report; University of California Radiation Laboratory: Berkeley, CA, USA, 1950; pp. 8–22. [Google Scholar]
- Hamilton, J.G.; Scott, K.G. Effect of calcium salt of versene upon metabolism of plutonium in the rat. Proc. Soc. Exp. Biol. Med. 1953, 83, 301–305. [Google Scholar] [CrossRef] [PubMed]
- SillÉN, L.G.; Martell, A.E. Stability Constants of Metal-Ion Complexes; Chemical Society: New York, NY, USA, 1964. [Google Scholar]
- Dagirmanjian, R.; Maynard, E.; Hodge, H. The effects of calcium disodium ethylenediamine tetraacetate on uranium poisoning in rats. J. Pharmacol. Exp. Ther. 1956, 117, 20–28. [Google Scholar] [PubMed]
- Smith, V.H. Removal of internally deposited plutonium. Nature 1958, 181, 1792–1793. [Google Scholar] [CrossRef] [PubMed]
- Volf, V. Treatment of Incorporated Transuranium Elements; Technical Reports Series, No. 184; IAEA: Vienna, Austria, 1978. [Google Scholar]
- Guilmette, R.A.; Lindhorst, P.S.; Hanlon, L.L. Interaction of Pu and Am with bone mineral in vitro. Radiat. Prot. Dosim. 1998, 79, 453–458. [Google Scholar] [CrossRef]
- Stradling, G.N. Decorporation of actinides: A review of recent research. J. Alloy. Compd. 1998, 271, 72–77. [Google Scholar] [CrossRef]
- Gorden, A.E.V.; Xu, J.; Raymond, K.N.; Durbin, P. Rational design of sequestering agents for plutonium and other actinides. Chem. Rev. 2003, 103, 4207–4282. [Google Scholar] [CrossRef]
- Wang, X.; Shi, C.; Guan, J.; Chen, Y.; Xu, Y.; Diwu, J.; Wang, S. The development of molecular and nano actinide decorporation agents. Chinese Chem. Lett. 2022, 33, 3395–3404. [Google Scholar] [CrossRef]
- Cilibrizzi, A.; Abbate, V.; Chen, Y.-L.; Ma, Y.; Zhou, T.; Hider, R.C. Hydroxypyridinone Journey into Metal Chelation. Chem. Rev. 2018, 118, 7657–7701. [Google Scholar] [CrossRef]
- Choi, T.A.; Furimsky, A.M.; Swezey, R.; Bunin, D.I.; Byrge, P.; Iyer, L.V.; Chang, P.Y.; Abergel, R.J. In Vitro Metabolism and Stability of the Actinide Chelating Agent 3,4,3-LI (1,2-HOPO). J. Pharm. Sci. 2015, 104, 1832–1838. [Google Scholar] [CrossRef]
- Sturzbecher-Hoehne, M.; Deblonde, G.J.P.; Abergel, R.J. Solution thermodynamic evaluation of hydroxypyridinonate chelators 3,4,3-LI(1,2-HOPO) and 5-LIO(Me-3,2-HOPO) for UO2(VI) and Th(IV) decorporation. Radiochim. Acta 2013, 101, 359–366. [Google Scholar] [CrossRef]
- Abergel, R.J.; Durbin, P.W.; Kullgren, B.; Ebbe, S.N.; Xu, J.; Chang, P.Y.; Bunin, D.I.; Blakely, E.A.; Bjornstad, K.A.; Rosen, C.J. Biomimetic actinide chelators: An update on the preclinical development of the orally active hydroxypyridonate decorporation agents 3,4,3-LI (1,2-HOPO) and 5-LIO (Me-3,2-HOPO). Health Phys. 2010, 99, 401–407. [Google Scholar] [CrossRef]
- Sturzbecher-Hoehne, M.; Choi, T.A.; Abergel, R.J. Hydroxypyridinonate complex stability of group (IV) metals and tetravalent f-block elements: The key to the next generation of chelating agents for radiopharmaceuticals. Inorg. Chem. 2015, 54, 3462–3468. [Google Scholar] [CrossRef]
- An, D.D.; Kullgren, B.; Jarvis, E.E.; Abergel, R.J. From early prophylaxis to delayed treatment: Establishing the plutonium decorporation activity window of hydroxypyridinonate chelating agents. Chem. Biol. Interact. 2017, 267, 80–88. [Google Scholar] [CrossRef]
- An, D.D.; Villalobos, J.A.; Morales-Rivera, J.A.; Rosen, C.J.; Bjornstad, K.A.; Gauny, S.S.; Choi, T.A.; Sturzbecher-Hoehne, M.; Abergel, R.J. 238Pu elimination profiles after delayed treatment with 3,4,3LI (1,2HOPO) in female and male Swiss-Webster mice. Int. J. Radiat. Biol. 2014, 90, 1055–1061. [Google Scholar] [CrossRef]
- Sturzbecher-Hoehne, M.; Kullgren, B.; Jarvis, E.E.; An, D.D.; Abergel, R.J. Highly Luminescent and Stable Hydroxypyridinonate Complexes: A Step Towards New Curium Decontamination Strategies. Chem. Eur. J. 2014, 20, 9962–9968. [Google Scholar] [CrossRef]
- Deblonde, G.J.P.; Sturzbecher-Hoehne, M.; Rupert, P.B.; An, D.D.; Illy, M.-C.; Ralston, C.Y.; Brabec, J.; de Jong, W.A.; Strong, R.K.; Abergel, R.J. Chelation and stabilization of berkelium in oxidation state+IV. Nat. Chem. 2017, 9, 843–849. [Google Scholar] [CrossRef]
- Sturzbecher-Hoehne, M.; Leung, C.N.P.; D’Aléo, A.; Kullgren, B.; Prigent, A.-L.; Shuh, D.K.; Raymond, K.N.; Abergel, R.J. 3,4,3-LI (1,2-HOPO): In vitro formation of highly stable lanthanide complexes translates into efficacious in vivo europium decorporation. Daltion T. 2011, 40, 8340–8346. [Google Scholar] [CrossRef]
- Kelley, M.P.; Deblonde, G.J.P.; Su, J.; Booth, C.H.; Abergel, R.J.; Batista, E.R.; Yang, P. Bond covalency and oxidation state of actinide ions complexed with therapeutic chelating agent 3,4,3-LI (1,2-HOPO). Inorg. Chem. 2018, 57, 5352–5363. [Google Scholar] [CrossRef]
- Carter, K.P.; Smith, K.F.; Tratnjek, T.; Deblonde, G.J.P.; Moreau, L.M.; Rees, J.A.; Booth, C.H.; Abergel, R.J. Controlling the Reduction of Chelated Uranyl to Stable Tetravalent Uranium Coordination Complexes in Aqueous Solution. Inorg. Chem. 2021, 60, 973–981. [Google Scholar] [CrossRef]
- Younes, A.; Creff, G.; Beccia, M.R.; Moisy, P.; Roques, J.; Aupiais, J.; Hennig, C.; Solari, P.L.; Den Auwer, C.; Vidaud, C. Is hydroxypyridonate 3,4,3-LI(1,2-HOPO) a good competitor of fetuin for uranyl metabolism? Metallomics 2019, 11, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham, T.E.; DeBolt, S.; Ferguson, D.; Seibel, G.; Kollman, P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General AMBER Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Grigera, J.R.; Straatsma, T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987, 91, 6269–6271. [Google Scholar] [CrossRef]
- Li, P.; Song, L.F.; Merz, K.M. Parameterization of Highly Charged Metal Ions Using the 12-6-4 LJ-Type Nonbonded Model in Explicit Water. J. Phys. Chem. B 2015, 119, 883–895. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Edit. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Lefebvre, C.; Rubez, G.; Khartabil, H.; Boisson, J.-C.; Contreras-García, J.; Hénon, E. Accurately extracting the signature of intermolecular interactions present in the NCI plot of the reduced density gradient versus electron density. Phys. Chem. Chem. Phys. 2017, 19, 17928–17936. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Zwanzig, R.W. High-Temperature Equation of State by a Perturbation Method. I. Nonpolar Gases. J. Chem. Phys. 1954, 22, 1420–1426. [Google Scholar] [CrossRef]
- Bennett, C.H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 1976, 22, 245–268. [Google Scholar] [CrossRef]


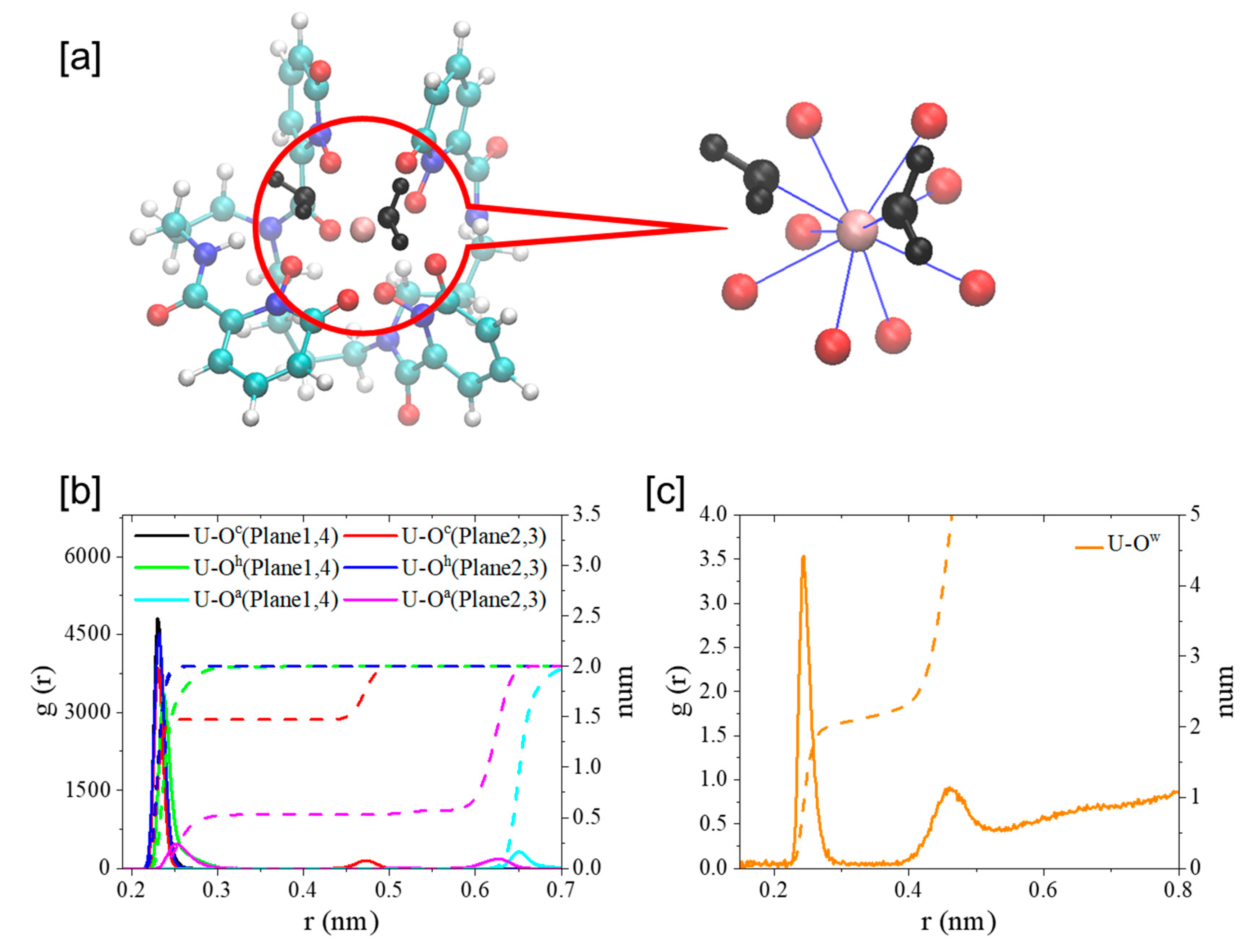
| Charge | dM-O (nm) | C.N. | |
|---|---|---|---|
| Oh (Plane1, 4) | −0.667321 | 0.238 | 2 |
| Oc (Plane1, 4) | −0.745996 | 0.230 | 2 |
| Oh (Plane2, 3) | −0.723067 | 0.232 | 2 |
| Oc (Plane2, 3) | −0.783067 | 0.230 | 1 |
| Oa (Plane2, 3) | −0.652427 | 0.252 | 1 |
| Ow | −0.847600 | 0.244 | 2 |
| Thermodynamic Cycle | Decomposed Interaction Energies | |||
|---|---|---|---|---|
| UIV-t-HOPO | U-t-HOPO (U4+ Bound with t-HOPO) | U-Water (U4+ in Water) | ||
| ΔG1 | −1330.14 | Eele | −750.50 | −141.90 |
| ΔG2 | −1248.84 | ELJ | 70.97 | 13.51 |
| ΔG | −81.30 | Eele + ELJ | −679.52 | −128.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Liu, Z.; Song, Y.-F.; Wang, D. Folding Dynamics of 3,4,3-LI(1,2-HOPO) in Its Free and Bound State with U4+ Implicated by MD Simulations. Molecules 2022, 27, 8151. https://doi.org/10.3390/molecules27238151
Wang Q, Liu Z, Song Y-F, Wang D. Folding Dynamics of 3,4,3-LI(1,2-HOPO) in Its Free and Bound State with U4+ Implicated by MD Simulations. Molecules. 2022; 27(23):8151. https://doi.org/10.3390/molecules27238151
Chicago/Turabian StyleWang, Qin, Ziyi Liu, Yu-Fei Song, and Dongqi Wang. 2022. "Folding Dynamics of 3,4,3-LI(1,2-HOPO) in Its Free and Bound State with U4+ Implicated by MD Simulations" Molecules 27, no. 23: 8151. https://doi.org/10.3390/molecules27238151
APA StyleWang, Q., Liu, Z., Song, Y.-F., & Wang, D. (2022). Folding Dynamics of 3,4,3-LI(1,2-HOPO) in Its Free and Bound State with U4+ Implicated by MD Simulations. Molecules, 27(23), 8151. https://doi.org/10.3390/molecules27238151







