Antitumor Activity and Mechanism of Action of the Antimicrobial Peptide AMP-17 on Human Leukemia K562 Cells
Abstract
1. Introduction
2. Results
2.1. AMP-17 Exhibits Antiproliferative Activity on K562 Cells
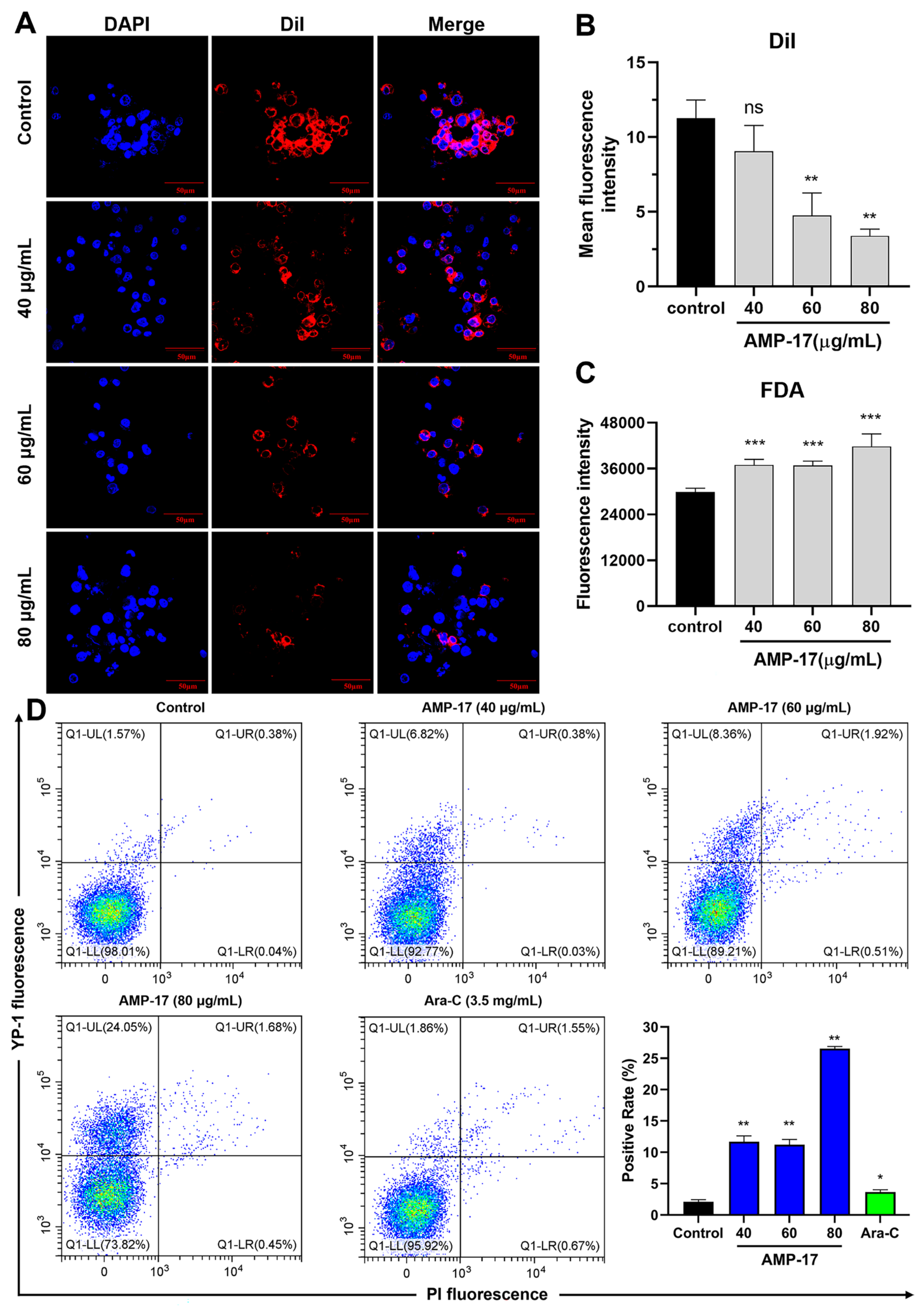
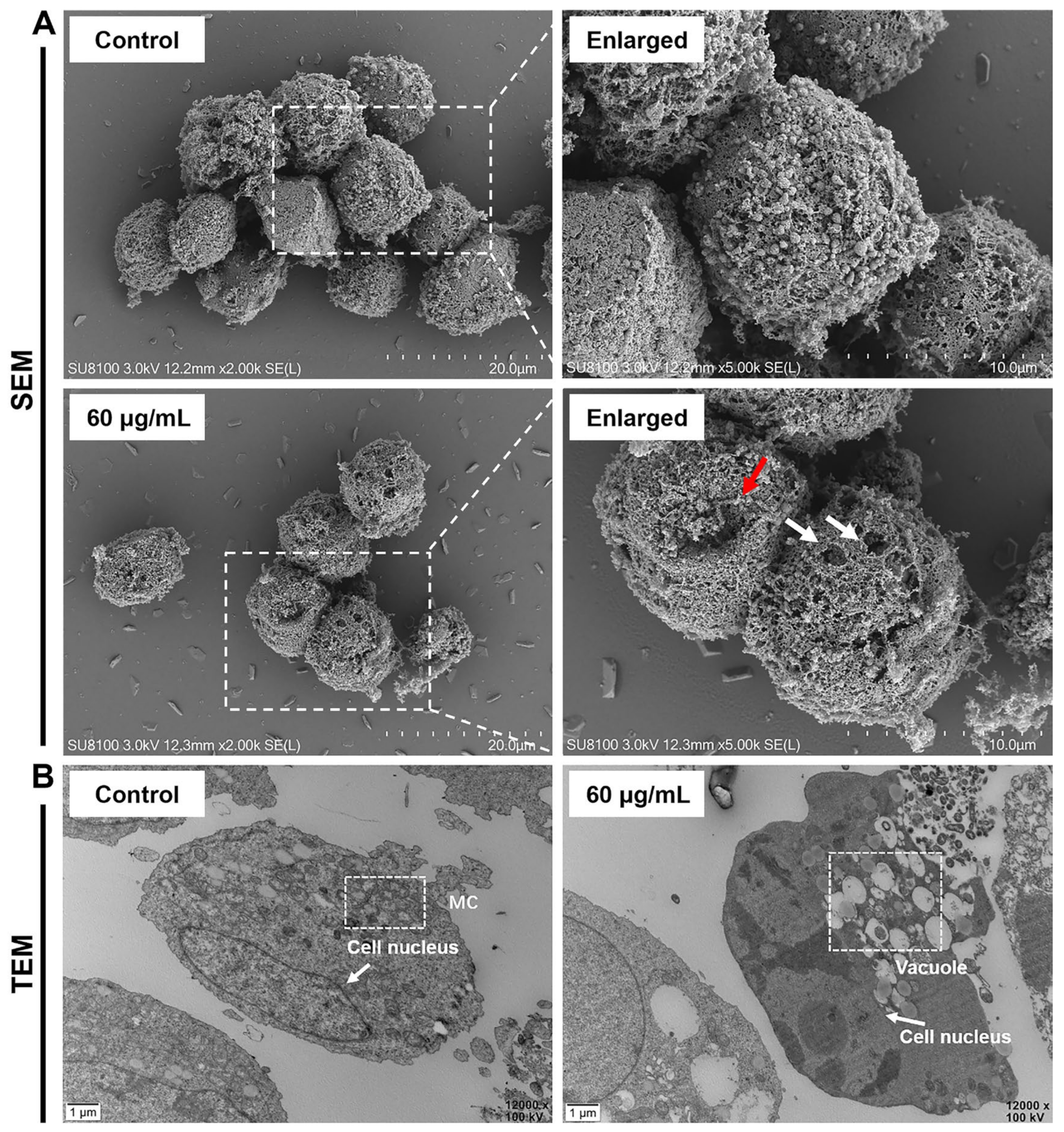
2.2. AMP-17 Induces Membrane Disruption of K562 Cells
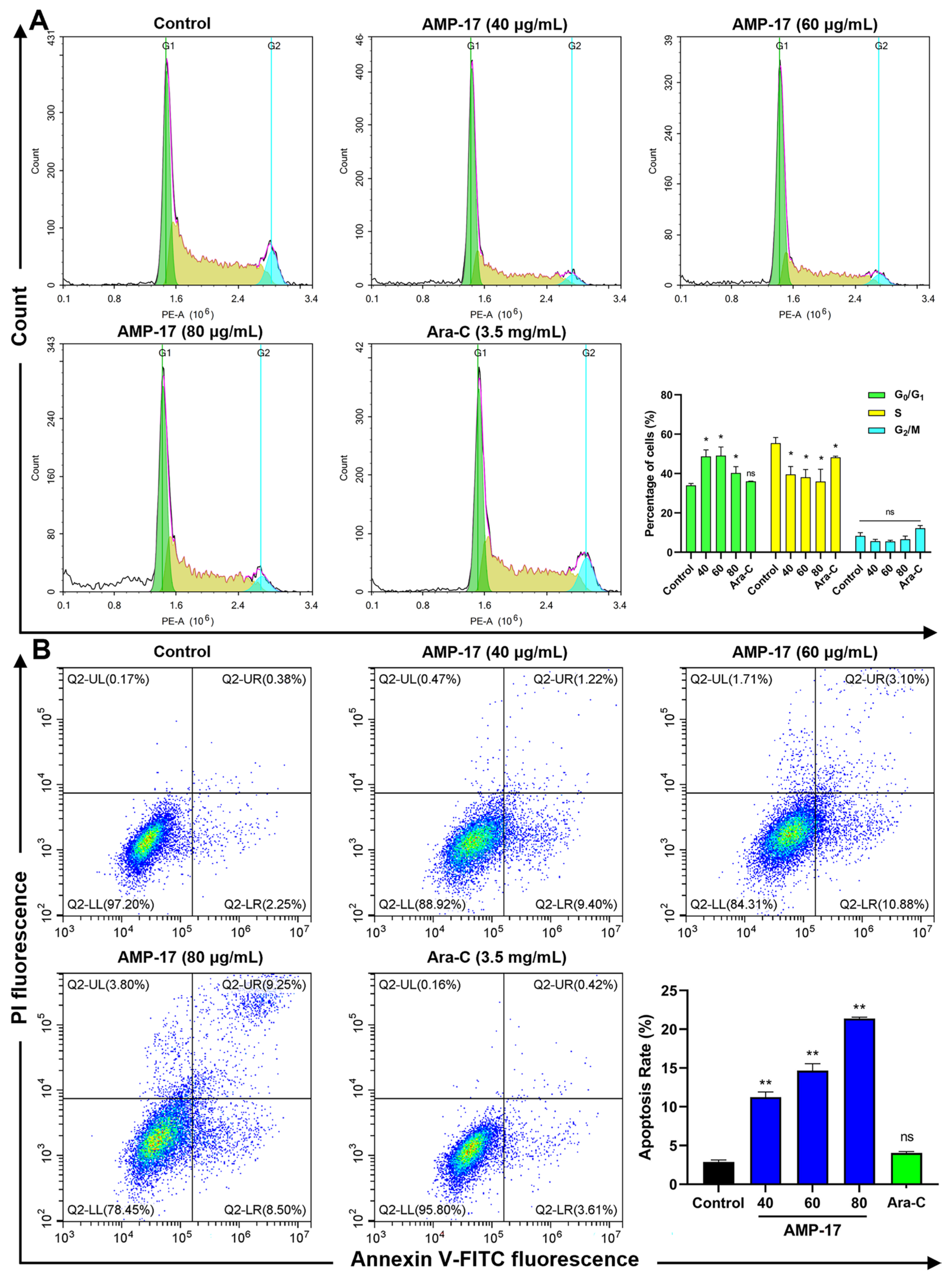
2.3. AMP-17 Induces Apoptosis in K562 Cells
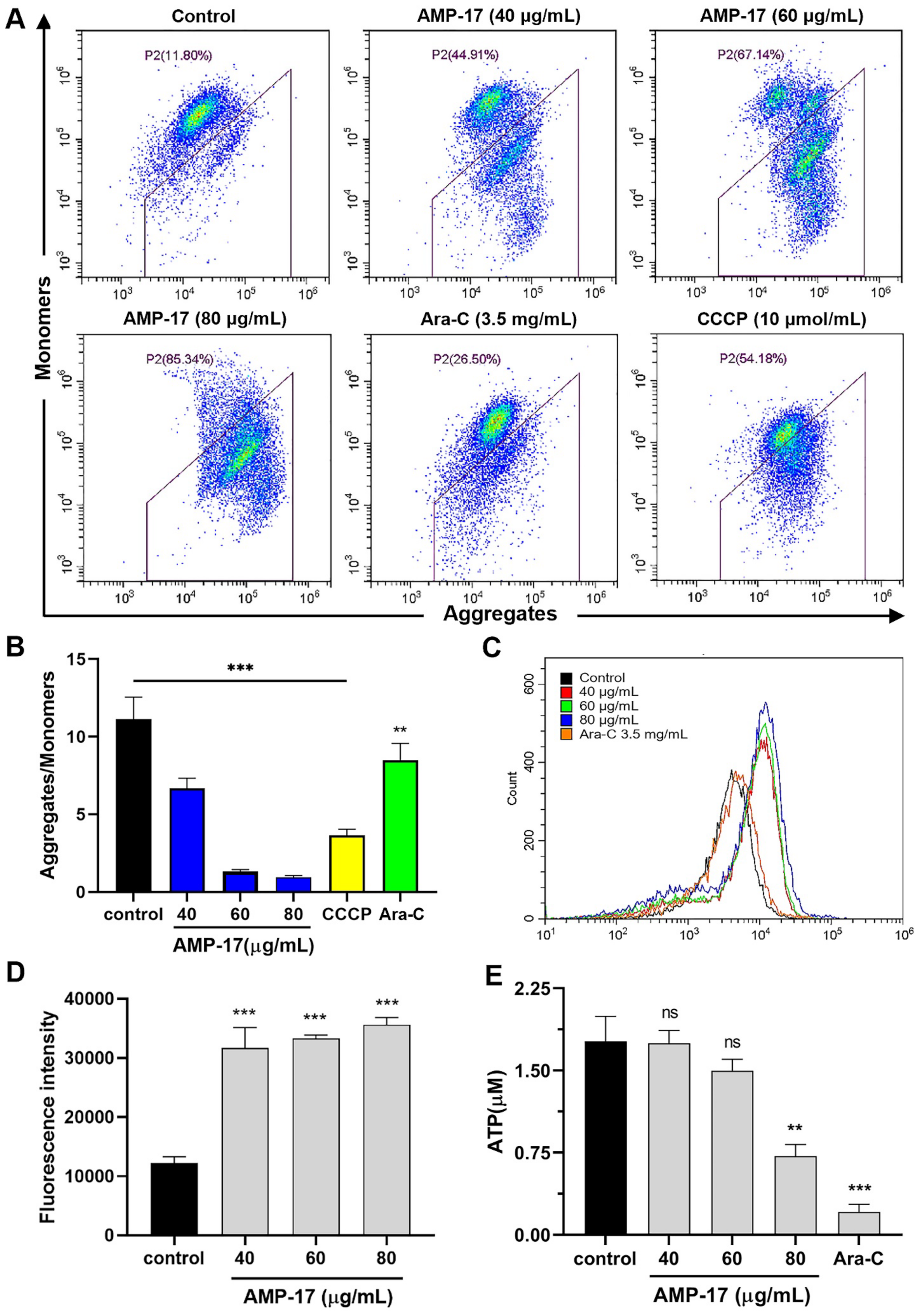
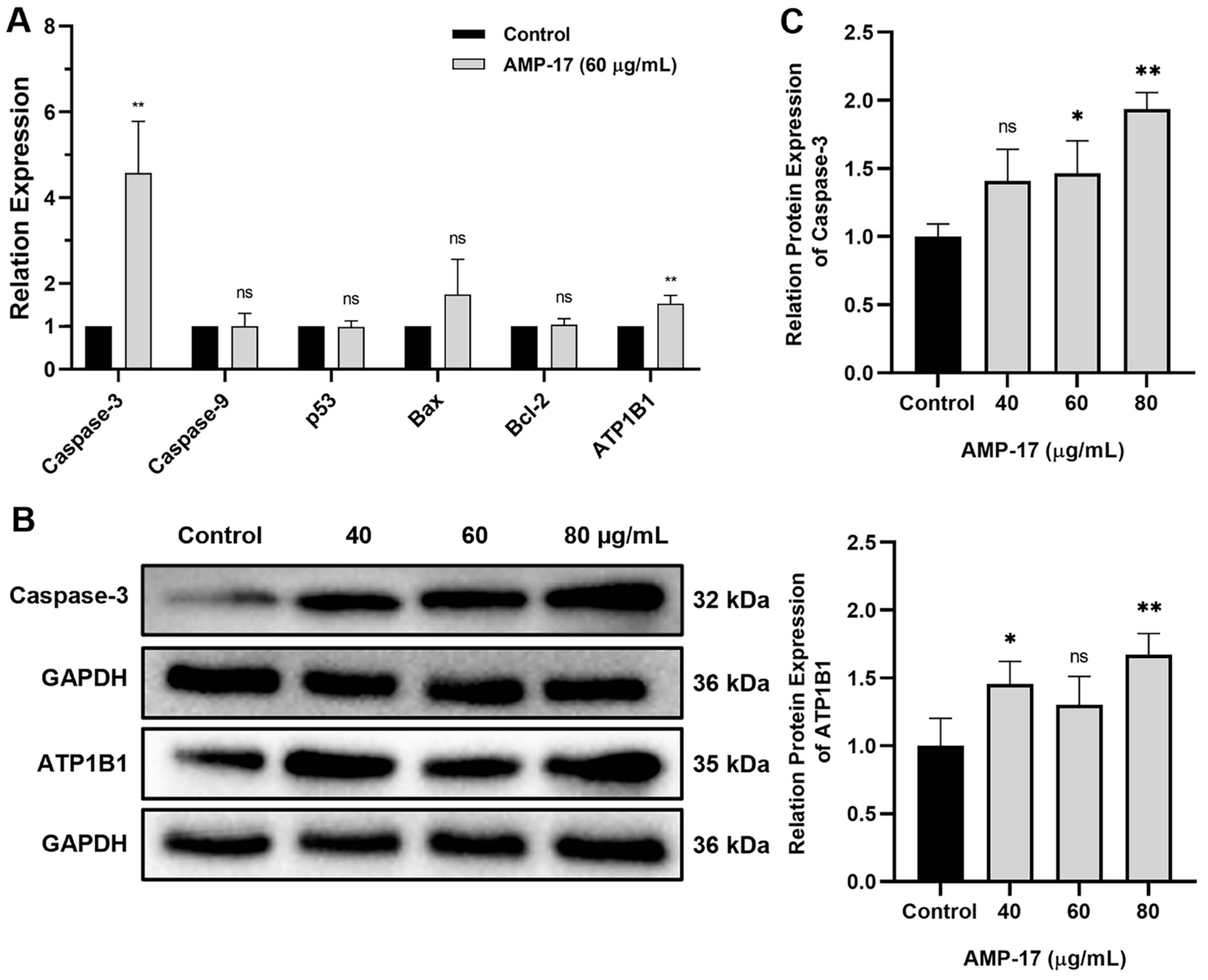
2.4. AMP-17-Induced Apoptosis Is Caspase-3 and ATP1B1 Dependent
3. Discussion
4. Material and Methods
4.1. Antimicrobial Peptide
4.2. Cell Culture
4.3. Cell Proliferation and Viability Assays
4.4. Trypan Blue Staining
4.5. Cell Cycle Analysis
4.6. Apoptosis Analysis
4.7. Membrane Permeability Assay
4.8. Confocal Laser Scanning Microscope
4.9. Scanning Electron Microscope
4.10. Transmission Electron Microscope
4.11. ROS Assay
4.12. Detection of Intracellular Ca2+ Concentration
4.13. Mitochondrial Membrane Potential Assay
4.14. Detection of ATP Content
4.15. Real-Time PCR Analysis
4.16. Western Blotting
4.17. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer. J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Babushok, D.V.; Bessler, M.; Olson, T.S. Genetic predisposition to myelodysplastic syndrome and acute myeloid leukemia in children and young adults. Leuk. Lymphoma 2016, 57, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Roboz, G.J. Current treatment of acute myeloid leukemia. Curr. Opin. Oncol. 2012, 24, 711–719. [Google Scholar] [CrossRef]
- Pisa, R.; Kapoor, T.M. Chemical strategies to overcome resistance against targeted anticancer therapeutics. Nat. Chem. Biol. 2020, 16, 817–825. [Google Scholar] [CrossRef]
- Bassan, R.; Hoelzer, D. Modern therapy of acute lymphoblastic leukemia. J. Clin. Oncol. 2011, 29, 532–543. [Google Scholar] [CrossRef]
- Gopal, S.; Miller, K.B.; Jaffe, I.Z. Molecular mechanisms for vascular complications of targeted cancer therapies. Clin. Sci. 2016, 130, 1763–1779. [Google Scholar] [CrossRef]
- Walz, C.; Sattler, M. Novel targeted therapies to overcome imatinib mesylate resistance in chronic myeloid leukemia (CML). Crit. Rev. Oncol. Hematol. 2006, 57, 145–164. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial peptides as anticancer agents: Functional properties and biological activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Zandsalimi, F.; Talaei, S.; Noormohammad Ahari, M.; Aghamiri, S.; Raee, P.; Roshanzamiri, S.; Yarian, F.; Bandehpour, M.; Zohrab Zadeh, Z. Antimicrobial peptides: A promising strategy for lung cancer drug discovery? Expert Opin. Drug Discov. 2020, 15, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Aghamiri, S.; Zandsalimi, F.; Raee, P.; Abdollahifar, M.A.; Tan, S.C.; Low, T.Y.; Najafi, S.; Ashrafizadeh, M.; Zarrabi, A.; Ghanbarian, H.; et al. Antimicrobial peptides as potential therapeutics for breast cancer. Pharmacol. Res. 2021, 171, 105777. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dong, C.; Li, X.; Han, W.; Su, X. Anticancer potential of bioactive peptides from animal sources (Review). Oncol. Rep. 2017, 38, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Fan, D.; Kong, L.; Yang, Q.; Zhu, Y.; Zhang, S.; Su, G.; Li, Y. Antimicrobial peptide Brevinin-1RL1 from frog skin secretion induces apoptosis and necrosis of tumor cells. Molecules 2021, 26, 2059. [Google Scholar] [CrossRef]
- Li, G.; Huang, Y.; Feng, Q.; Chen, Y. Tryptophan as a probe to study the anticancer mechanism of action and specificity of α-helical anticancer peptides. Molecules 2014, 19, 12224–12241. [Google Scholar] [CrossRef]
- Xuan, H.L.; Duc, T.D.; Thuy, A.M.; Chau, P.M.; Tung, T.T. Chemical approaches in the development of natural nontoxic peptide Polybia-MP1 as a potential dual antimicrobial and antitumor agent. Amino Acids 2021, 53, 843–852. [Google Scholar] [CrossRef]
- Han, Y.F.; Cui, Z.B.; Li, Y.H.; Hsu, W.H.; Lee, B.H. In Vitro and in Vivo anticancer activity of Pardaxin against proliferation and growth of oral squamous cell carcinoma. Mar. Drugs 2016, 14, 2. [Google Scholar] [CrossRef]
- Huang, T.C.; Lee, J.F.; Chen, J.Y. Pardaxin, an antimicrobial peptide, triggers caspase-dependent and ROS-mediated apoptosis in HT-1080 cells. Mar. Drugs 2011, 9, 1995–2009. [Google Scholar] [CrossRef]
- Li, C.; Dong, Y.; Wang, L.; Xu, G.; Yang, Q.; Tang, X.; Qiao, Y.; Cong, Z. Ginsenoside metabolite compound K induces apoptosis and autophagy in non-small cell lung cancer cells via AMPK-mTOR and JNK pathways. Biochem. Cell Biol. 2019, 97, 406–414. [Google Scholar] [CrossRef]
- Ren, S.X.; Shen, J.; Cheng, A.S.; Lu, L.; Chan, R.L.; Li, Z.J.; Wang, X.J.; Wong, C.C.; Zhang, L.; Ng, S.S.; et al. Correction: FK-16 Derived from the Anticancer Peptide LL-37 Induces Caspase-Independent Apoptosis and Autophagic Cell Death in Colon Cancer Cells. PLoS ONE 2015, 10, e0131750. [Google Scholar] [CrossRef]
- Guo, G.; Tao, R.; Li, Y.; Ma, H.; Xiu, J.; Fu, P.; Wu, J. Identification and characterization of a novel antimicrobial protein from the housefly Musca domestica. Biochem. Biophys. Res. Commun. 2017, 490, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.B.; Guo, G.; Zhao, X.Y.; Su, P.P.; Fu, P.; Peng, J.; Xiu, J.F.; Li, B.Y. Antifungal Activity and Physicochemical Properties of a Novel Antimicrobial Protein AMP-17 from Musca domestica. Pol. J. Microbiol. 2019, 68, 383–390. [Google Scholar] [CrossRef]
- Ma, H.; Zhao, X.; Yang, L.; Su, P.; Fu, P.; Peng, J.; Yang, N.; Guo, G. Antimicrobial Peptide AMP-17 Affects Candida albicans by Disrupting Its Cell Wall and Cell Membrane Integrity. Infect. Drug Resist. 2020, 13, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Blatt, S.; Dawood, M.; Klauck, S.M.; Fleischer, E.; Kämmerer, P.W.; Efferth, T. Novel artemisinin derivative FO8643 with anti-angiogenic activity inhibits growth and migration of cancer cells via VEGFR2 signaling. Eur. J. Pharmacol. 2022, 930, 175158. [Google Scholar] [CrossRef]
- Liu, K.; Ren, T.; Huang, Y.; Sun, K.; Bao, X.; Wang, S.; Zheng, B.; Guo, W. Apatinib promotes autophagy and apoptosis through VEGFR2/STAT3/BCL-2 signaling in osteosarcoma. Cell Death Dis. 2017, 8, e3015. [Google Scholar] [CrossRef]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Mani, S.; Swargiary, G.; Singh, K.K. Natural Agents Targeting Mitochondria in Cancer. Int. J. Mol. Sci. 2020, 21, 6992. [Google Scholar] [CrossRef]
- Delierneux, C.; Kouba, S.; Shanmughapriya, S.; Potier-Cartereau, M.; Trebak, M.; Hempel, N. Mitochondrial Calcium Regulation of Redox Signaling in Cancer. Cells 2020, 9, 432. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Li, B.; Tang, H.; Cheng, Z.; Zhang, Y.; Xiang, H. The Current Situation and Future Trend of Leukemia Mortality by Sex and Area in China. Front Public Health. 2020, 8, 598215. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Qiu, Q.; Yang, B.; Wang, X.; Huang, W.; Qian, H. Design, synthesis and biological evaluation of novel peptides with anti-cancer and drug resistance-reversing activities. Eur. J. Med. Chem. 2015, 89, 540–548. [Google Scholar] [CrossRef]
- Bakare, O.O.; Gokul, A.; Wu, R.; Niekerk, L.A.; Klein, A.; Keyster, M. Biomedical Relevance of Novel Anticancer Peptides in the Sensitive Treatment of Cancer. Biomolecules 2021, 11, 1120. [Google Scholar] [CrossRef]
- Leite, M.L.; da Cunha, N.B.; Costa, F.F. Antimicrobial peptides, nanotechnology, and natural metabolites as novel approaches for cancer treatment. Pharmacol. Ther. 2018, 183, 160–176. [Google Scholar] [CrossRef]
- Baxter, A.A.; Lay, F.T.; Poon, I.K.H.; Kvansakul, M.; Hulett, M.D. Tumor cell membrane-targeting cationic antimicrobial peptides: Novel insights into mechanisms of action and therapeutic prospects. Cell. Mol. Life Sci. 2017, 74, 3809–3825. [Google Scholar] [CrossRef] [PubMed]
- Felicio, M.R.; Silva, O.N.; Goncalves, S.; Santos, N.C.; Franco, O.L. Peptides with dual antimicrobial and anticancer activities. Front. Chem. 2017, 5, 5. [Google Scholar] [CrossRef]
- Connor, J.; Bucana, C.; Fidler, I.J.; Schroit, A.J. Differentiation-Dependent Expression of Phosphatidylserine in Mammalian Plasma-Membranes—Quantitative Assessment of Outer-Leaflet Lipid by Prothrombinase Complex-Formation. Proc. Natl. Acad. Sci. USA 1989, 86, 3184–3188. [Google Scholar] [CrossRef]
- Orrenius, S.; Gogvadze, V.; Zhivotovsky, B. Calcium and mitochondria in the regulation of cell death. Biochem. Biophys. Res. Commun. 2015, 460, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Zhivotovsky, B.; Orrenius, S. Calcium and cell death mechanisms: A perspective from the cell death community. Cell Calcium 2011, 50, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Costantini, P.; Jacotot, E.; Decaudin, D.; Kroemer, G. Mitochondrion as a novel target of anticancer chemotherapy. J. Natl. Cancer Inst. 2000, 92, 1042–1053. [Google Scholar] [CrossRef]
- Mignotte, B.; Vayssiere, J.L. Mitochondria and apoptosis. Eur. J. Biochem. 1998, 252, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ceremuga, M.; Stela, M.; Janik, E.; Gorniak, L.; Synowiec, E.; Sliwinski, T.; Sitarek, P.; Saluk-Bijak, J.; Bijak, M. Melittin—A natural peptide from bee venom which induces apoptosis in human leukaemia cells. Biomolecules 2020, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhu, M.; Xu, Z.; Li, W.; Dong, X.; Chen, Y.; Lin, B.; Li, M. Ropivacaine promotes apoptosis of hepatocellular carcinoma cells through damaging mitochondria and activating caspase-3 activity. Biol. Res. 2019, 52, 36. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Li, X.; Feng, S.; Cheng, W.; Tang, B.; Shi, Y.L.; Hua, Z.C. Plasma membrane depolarization and Na,K-ATPase impairment induced by mitochondrial toxins augment leukemia cell apoptosis via a novel mitochondrial amplification mechanism. Biochem. Pharmacol. 2009, 78, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Litan, A.; Li, Z.; Graves, B.; Lindsey, S.; Barwe, S.P.; Langhans, S.A. Na,K-ATPase β1-subunit is a target of sonic hedgehog signaling and enhances medulloblastoma tumorigenicity. Mol. Cancer 2015, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Rotoli, D.; Cejas, M.M.; Maeso, M.D.; Pérez-Rodríguez, N.D.; Morales, M.; Ávila, J.; Mobasheri, A.; Martín-Vasallo, P. The Na, K-ATPase β-Subunit Isoforms Expression in Glioblastoma Multiforme: Moonlighting Roles. Int. J. Mol. Sci. 2017, 18, 2369. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Marfell, B.J.; Christensen, M.E.; Waterhouse, N.J. Measuring cell death by trypan blue uptake and light microscopy. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Liang, X.; Wang, R.; Dou, W.; Zhao, L.; Zhou, L.; Zhu, J.; Wang, K.; Yan, J. Arminin 1a-C, a novel antimicrobial peptide from ancient metazoan Hydra, shows potent antileukemia activity against drug-sensitive and drug-resistant leukemia cells. Drug Des. Devel. Ther. 2018, 12, 3691–3703. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequences | |
|---|---|---|
| Caspase-3 | FORWARD | 5′-GCAGCAAACCTAGGGAAAC-3′ |
| REVERSE | 5′-TGTCGGCATACTGTTTCAGCA-3′ | |
| Caspase-9 | FORWARD | 5′-TGTGGTGGTCATCCTCTCTCA-3′ |
| REVERSE | 5′-GTCACTGGGGGTAGGCAAACT-3′ | |
| Bax | FORWARD | 5′ -CATCTCCTTGCTCGTAGTCTAGAGC-3′ |
| REVERSE | 5′ -CATTGTGATGGACTCCGGAGACGG-3′ | |
| Bcl-2 | FORWARD | 5′-TTTGAGTTCGGTGGGGTCAG-3′ |
| REVERSE | 5′-TGACTTCACTTGTGGCCCAG-3′ | |
| P53 | FORWARD | 5′-GGGTTAGTTTACAATCAGCCACATT-3′ |
| REVERSE | 5′-GGCCTTGAAGTTAGAGAAAATTCA-3′ | |
| ATP1B1 | FORWARD | 5′-GCCAGGATTAACACAGATTCCTCAG-3′ |
| REVERSE | 5′-CCTTATCTTCATCTCGCTTGCC-3′ | |
| GAPDH | FORWARD | 5′-ACACCCACTCCTCCACCTTT-3′ |
| REVERSE | 5′-TAGCCAAATTCGTTGTCATACC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Z.; Yang, L.; Huang, M.; Sun, C.; Chen, M.; Zhao, W.; Peng, J.; Guo, G. Antitumor Activity and Mechanism of Action of the Antimicrobial Peptide AMP-17 on Human Leukemia K562 Cells. Molecules 2022, 27, 8109. https://doi.org/10.3390/molecules27228109
Tian Z, Yang L, Huang M, Sun C, Chen M, Zhao W, Peng J, Guo G. Antitumor Activity and Mechanism of Action of the Antimicrobial Peptide AMP-17 on Human Leukemia K562 Cells. Molecules. 2022; 27(22):8109. https://doi.org/10.3390/molecules27228109
Chicago/Turabian StyleTian, Zhuqing, Longbing Yang, Mingjiao Huang, Chaoqin Sun, Mingming Chen, Wenjing Zhao, Jian Peng, and Guo Guo. 2022. "Antitumor Activity and Mechanism of Action of the Antimicrobial Peptide AMP-17 on Human Leukemia K562 Cells" Molecules 27, no. 22: 8109. https://doi.org/10.3390/molecules27228109
APA StyleTian, Z., Yang, L., Huang, M., Sun, C., Chen, M., Zhao, W., Peng, J., & Guo, G. (2022). Antitumor Activity and Mechanism of Action of the Antimicrobial Peptide AMP-17 on Human Leukemia K562 Cells. Molecules, 27(22), 8109. https://doi.org/10.3390/molecules27228109




