Abstract
Blumea lanceolaria (Roxb.) Druce, a flowering plant, is used for treating cancer and inflammatory diseases. In this study, we determined the chemical composition of the EOs extracted from the leaves (LBEO), stem (SBEO), and roots (RBEO) of B. lanceolaria and analyzed their anti-inflammation potential. Overall, 30 compounds representing 99.12%, 98.44%, and 96.89% of total EO constituents of the leaves, stem, and roots, respectively, were identified using GC-MS. ELISA, Western blotting, and qRT-PCR studies showed that LBEO, SBEO, and RBEO inhibited multiple steps in the inflammatory responses in the RAW 264.7 cell model, including NO production; TNF-α, IL-6, iNOS, and COX-2 transcription and translation; and phosphorylation of IκBα and p65 of the NF-κB pathway. In the carrageenan-induced paw edema model, all three EOs inhibited paw edema at both early and delayed phases. Molecular docking studies indicated that the main components of B. lanceolaria EOs (BEOs) targeted and inhibited major components of inflammation-related pathways, including the arachidonic acid metabolic pathway, NF-κB pathway, and MAPK pathway. We present the first study to characterize the chemical composition of BEOs and confirm their potent anti-inflammatory effects in in vitro, in vivo, and in silico analysis. These results can facilitate the development of effective anti-inflammatory drugs with limited side effects in the future.
1. Introduction
Inflammation is a protective response of the immune system to tissue damage or foreign stimuli, including human pathogens, allergens, toxic chemicals, or irradiation [1]. The vital functions of inflammation involve repair of damaged tissues, elimination of harmful stimuli, and maintenance of homeostasis and health [1]. However, uncontrolled inflammatory responses may lead to the initiation and progression of numerous diseases, including diabetes, Alzheimer’s disease, heart diseases, autoimmune disorders, and cancer [2,3,4]. The nonsteroidal anti-inflammatory drugs (NSAIDs) are current, widely used anti-inflammatory drugs, which inhibit the production of prostaglandins, key inflammatory mediators, by inhibiting the activity of cyclooxygenase isoenzymes (COX-1 and COX-2) [5]. As COX-1 and COX-2 are both involved in regulation of many important physiologic processes, selective or non-selective inhibition of COX-1 and COX-2 can adversely affect the stomach, kidneys, and liver [5,6,7]. Therefore, new potent anti-inflammatory drugs, especially those derived from plants with different drug targets or molecular mechanism of actions, as well as limited side effects, are urgently required to circumvent these problems [5,8].
Suitable anti-inflammatory models should be used to search for new potential anti-inflammatory drug candidates. Currently, several cell models are available for testing anti-inflammatory activities, such as RAW 264.7, THP-1, and endothelial cells [9,10]. However, the RAW 264.7 macrophages are the most commonly used cell model for assessing anti-inflammatory activities. Lipopolysaccharide (LPS) can bind to Toll-like receptor 4 (TLR4) and stimulate various inflammatory pathways in RAW 264.7 macrophages [10], including MAPK and NF-κB pathways [11,12]. The MAPK pathway—with key components such as ERK1/2, JNK, and p38—can be activated by TRAF6, which leads to the activation of AP-1 and the expression of pro-inflammatory factors [12]. The NF-κB pathway can be activated by the MyD88-dependent TLR pathway [10]. In unstimulated cells, key transcription factors of the NF-κB pathway (p50-p65) exist in a latent inactive form complexed to an inhibitor protein, IκBα. Under conditions of inflammation, IκBα is phosphorylated [13], and then degraded to induce the phosphorylation of NF-κB p65 (p-p65) [14]. The phosphorylated NF-κB p65 migrates to the nucleus and induces the expression of pro-inflammatory factors [14]. Therapeutic agents prevent inflammation by inhibiting inflammation-related signaling pathways, such as the MAPK pathway and NF-κB pathway, thereby inhibiting the production of pro-inflammatory factors, including inducible nitric oxide synthase (iNOS), cyclooxygenase 2 (COX-2), TNF-α, IL-6, NO, and PGE2.
Evaluation of bioactive compounds using animal models is important for discovering and developing new potential anti-inflammatory drugs. Different animal models are available for evaluating anti-inflammatory drugs, including chemical-induced paw edema, ear edema, vascular permeability, and pleurisy models [15,16]. Among these animal models, the carrageenan-induced edema model is widely used [17]. Inflammation induced by carrageenan is acute, local, non-immune, and reproducible in nature [15,18], which also triggers the activation of the NF-kB pathway via TLRs [19]. Involvement of multiple mechanisms of inflammation render this model suitable for preliminarily screening of anti-inflammatory drugs and assessing the anti-inflammatory activities of natural and synthetic compounds [18].
Blumea lanceolaria (Roxb.) Druce is a flowering plant belonging to the Asteraceae family, which is widely distributed across tropical and subtropical Asia, including India, China, Taiwan, Philippines, and Vietnam [20,21]. In folk medicine, B. lanceolaria is used to treat cancer and inflammation-associated diseases, such as fever, cough, asthma, chronic ulcers, wounds, and dysentery [20,21]. Moreover, this medicinal herb is edible and widely used in daily life for cooking fish, shrimp, pork, or snake meat [20,21]. Therefore, the chemical constituents of B. lanceolaria exhibit anti-inflammatory activities and are safe through oral absorption. Previous studies indicated that the methanol extracts of the leaves or roots of B. lanceolaria possess antioxidant and antimicrobial activities [22,23]. Using capillary gas chromatography (GC) and mass spectrometry (MS), Dung et al. (1991) found that the essential oil (EO) of B. lanceolaria contained mainly (95%) methyl thymol [24]. However, the chemical components of the EOs obtained from the leaves, stem, and roots of B. lanceolaria and their anti-inflammatory potential remain unclear.
This study aimed to analyze in detail the chemical composition of the EOs extracted from the leaves (LBEO), stem (SBEO), and roots (RBEO) of B. lanceolaria plants collected in Vietnam. The anti-inflammatory activities of these EOs were analyzed using in vitro and in vivo models. The mechanism underlying the molecular interactions and affinities with key proteins in three important inflammation-related pathways, namely the arachidonic acid (AA), NF-κB, and MAPK pathways, were analyzed using molecular docking analysis.
2. Results and Discussion
2.1. Chemical Composition of the BEOs
Table 1 shows the chemical components of LBEO, RBEO, and SBEO including the relative content (%), retention time (RT), and retention index (RI) of each constituent. In total, 30 compounds (Figure 1 and Table 1) were identified, representing 99.12%, 98.44%, and 95.62% of the total EO constituents of the leaves, stem, and roots, respectively. Among these, 5, 15, and 20 constituents were identified in the LBEO, SBEO, and RBEO, respectively, according to their mass spectra and relative RI (Table 1).

Table 1.
Chemical compositions of the BEOs from the leaves, stem, and roots analyzed using GC-MS/GC-flame ionization detection (FID).
Figure 1.
Chemical structure of the volatile compounds (1–30) identified from total essential oils of B. lanceolaria.
Among the five volatile constituents of LBEO, two major compounds were identified to be o-cymene (38.29%) and carvacrol methyl ether (58.28%), along with three other minor constituents, namely thymol methyl ether (0.24%), β-caryophyllene (0.26%), and hexadecanol (2.05%). The LBEO was found to be rich in monoterpene hydrocarbons (96.81%), followed by other alcoholic compounds (2.05%) and sesquiterpenes (0.26%) (Figure S1).
Among the 15 compounds of SBEO distilled from the B. lanceolaria stem (Figure S2), carvacrol methyl ether accounted for 89.40% of the SBEO, followed by o-cymene (3.35%) and α-pinene (2.28%). The remaining 12 compounds were in minority, with content <1%. Monoterpenes were most abundant (95.51%), followed by sesquiterpenes (2.42%) and other compounds (0.51%).
In the EO obtained from B. lanceolaria root, 25 components were structurally identified using GC/MS with the HP-5 MS column. Four major compounds were found, namely, α-pinene (33.36%), thymol isobutyrate (29.23%), thymol methyl ether (21.70%), and thymol (4.25%). The others were minor constituents, with content <1% (Figure S3). Similar to that observed in LBEO and SBEO, RBEO contained the highest amount of monoterpenes (61.94%), followed by thymol isobutyrate (29.23%) and sesquiterpenes (4.45%).
Comparison of the major chemical components of the LBEO, SBEO, and RBEO revealed considerable variability among some of the major components in the samples. The content of carvacrol methyl ether (10), contributing to 58.28% of the components in LBEO, increased to 89.40% in SBEO (highest among all EOs), but disappeared in RBEO (0%). Similarly, β-caryophyllene (16) content increased from 0.26% in LBEO to 0.82% in SBEO and then decreased to 0.53% in RBEO. The contents of monoterpene o-cymene (from 38.29% to 3.35% and 0.92%) and hexadecanol (from 2.05% to 0.26% and 0%) tended to decrease from LBEO to SBEO and RBEO. However, α-pinene (2) was not found in LBEO, although its content increased to 2.28% and 33.36% in SBEO and RBEO, respectively. The second highest increase in content was observed for thymol methyl (9) ether; its content increased from 0.24% in LBEO to 0.36% in SBEO and 21.70% in RBEO. The levels of α-copaene (from 0% to 0.37% and 0.65%), α-humulene (from 0% to 0.17% and 0.24%), and δ-cadinene (from 0% to 0.18% and 0.26%) also increased. Among the 30 compounds identified from the three EO samples, three compounds (10% of the total) were only present in the stem (SBEO), including linalool (0.12%), α-cadinol (0.27%), and hexadecanal (0.25%). Two compounds, carvacrol methyl ether and hexadecanol, were present in the stem and leaf, but not in the root (Table 1), while fifteen compounds were found to be present only in RBEO, including thymol isobutyrate, which accounted for the greatest proportion of the components (29.23%).
The EOs obtained from the leaves and stem of B. lanceolaria harvested in December 2020 differed considerably from that published previously. In our plant samples, 0.24% and 0.36% thymol methyl ether (9) were present in the leaves and stem of B. lanceolaria EOs, respectively. However, Dung et al. reported that thymol methyl ether contributed to 94.96% of the content of EOs obtained from the leaves and stem of B. lanceolaria harvested in June 1989 [24]. The content of methyl carvacrol (10) was highest in our samples, with 58.28% in the leaves and 89.40% in the stem, whereas only 0.02% was reported by Dung et al. Similarly, β-caryophyllene (16) content was also high in our samples, with 0.26% in LBEO and 0.82% in SBEO, compared to 0.04% reported previously. o-Cymene (3) content was the second highest in our leaf (38.29%) and stem (3.35%) samples, although it was not reported by Dung et al. (1991). Furthermore, according to Dung et al., the contents of limonene (4) and α-thujene (1) were 0.12% and 0.04%; whereas we could not detect them in our leaf and stem samples, and only 0.14% was detected in the root sample. These differences in results can be attributed to differences in geographic factor, time of collection (December 2020 versus June 1989), and technique used (15 compounds were found in our leaf and stem samples prepared using steam distillation compared to 11 compounds reported in literature) [24]. In addition, these differences may be the result of differences in analytical method, Dung et al. used a 25 m × 0.25 mm I.D.-fused silica OV-1 (0.25 pm) column and the nitrogen gas was carried at a flow rate of 1.2 mL/min. The oven was programmed after 5 min at 60 °C, at 5 °C/min to 220 °C, with a final hold time of 20 min. In our analyses, the HP-5 MS column with a dimension of 60 m × 0.25 mm and film thickness of 0.25 μm was used for separation. Running conditions were set as follows: injector temperature at 250 °C; initial temperature started from 60 °C then increased to 240 °C with increasing step of 4 °C/min; the carrier gas was helium with the flowrate of 1 mL/min; full scan modes under electron ionization with voltage: 70 eV, emission current: 40 mA; and mass range scan: 35–450 a.m.u.
2.2. Effect of the BEOs on Macrophage Viability
We evaluated the toxicity of BEOs in RAW 264.7 using the CCK-8 kit (Abcam, Cambridge, UK). The results indicate that at concentrations of 0–50 µg/mL, none of the three essential oil samples were toxic to the RAW 264.7 cells (Figure 2). However, 100 µg/mL LBEO, SBEO, and RBEO were cytotoxic, causing 34.89%, 22.12%, and 37.28% cell death, respectively. When the concentration of the BEOs was increased to 150 µg/mL, the cell survival rate decreased to <50%. Thus, 50 µg/mL BEO is relatively safe for cells, with a survival rate of more than 95%. The calculated IC50 values of LBEO, SBEO, and RBEO are 151.11, 335.60, and 123.05 µg/mL, respectively. Therefore, the optimal concentration range of 5–50 µg/mL, at which the EOs were not toxic but active, was selected for anti-inflammatory studies.
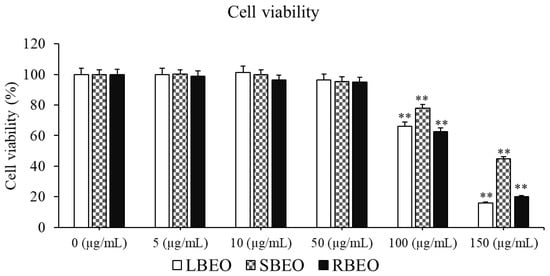
Figure 2.
Effect of BEOs on the viability of RAW 264.7 macrophages. Various concentrations of LBEO, SBEO, and RBEO (0, 5, 10, 50, 100, and 150 µg/mL) were added to macrophage cell cultures for 24 h at 37 °C, the cell viability was then assessed using the CCK-8 kit. Data from three independent experiments were used to calculate mean values and standard deviation. ** p < 0.001 vs. negative control (0 µg/mL EOs).
2.3. Inhibition of NO Production by BEOs
Inhibitory effects of BEOs on NO production in RAW 264.7 cells were assessed using the Griess reagent (Promega, Madison, WI, USA). As shown in Figure 3, all three EOs (BLEO, SBEO, and RBEO) remarkably inhibited NO production (p < 0.05) in a concentration-dependent manner. Among the EOs, LBEO showed the best NO inhibition ability at all the three tested concentrations (5, 10, and 50 µg /mL). Both 50 µg/mL LBEO and RBEO showed the highest NO inhibition ability (approximately 50%). Compared to LBEO and RBEO, 50 µg/mL SBEO showed the lowest NO inhibitory activity of 33.15 ± 1.01%. Thus, all three EOs markedly inhibited NO production in RAW 264.7 macrophages.
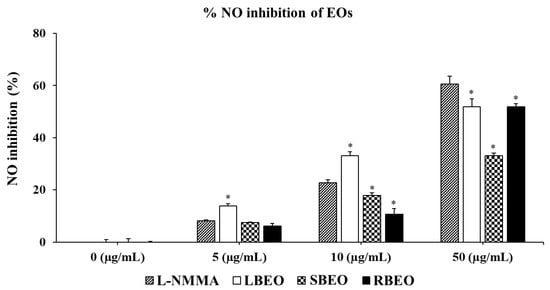
Figure 3.
Effects of LBEO, SBEO, and RBEO on NO production in RAW 264.7 macrophages. NO secretion in the macrophages treated with 5, 10, and 50 µg/mL BEOs was measured using the Griess reagent system. Data from three independent experiments were used to calculate mean values and standard deviation * p < 0.01 vs. positive control group (L-NMMA).
2.4. Effect of the BEOs on Transcription and Translation of TNF-α and IL-6
The ability to inhibit the production of TNF-α and IL-6 is an important indicator for evaluating the anti-inflammatory property of BEOs. ELISA results (Figure 4) show that the level of TNF-α increased to more than 2000 pg/mL when stimulated by LPS. TNF-α production decreased from 2000 pg/mL to 61, 91, and 44 pg/mL in cells treated with 50 µg/mL BLEO, SBEO, and RBEO, respectively. These results indicate that all three BEOs strongly inhibit TNF-α production. Similarly, in the presence of BEOs, the level of IL-6 decreased significantly compared to that in the control sample (LPS-treated macrophages). The amount of IL-6 decreased from 250 pg/mL to 94.67, 92.67, and 88 pg/mL when treated with increasing concentrations (up to 50 µg/mL) of LBEO, SBEO, and RBEO, respectively (Figure 4d–f).
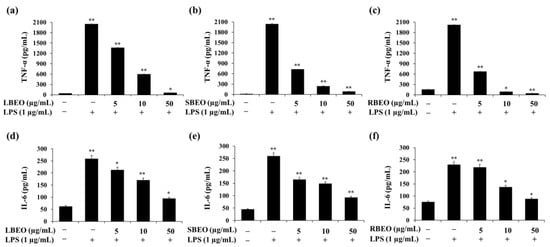
Figure 4.
Inhibitory effects of LBEO, SBEO, and RBEO (5, 10, and 50 µg/mL) on TNF-α and IL-6 production in RAW 264.7 macrophages. After 24 h of stimulation by LPS, the inhibition of TNF-α and IL-6 production by (a,d) LBEO, (b,e) SBEO, and (c,f) RBEO were quantified by ELISA. Cytokine concentrations were calculated using a standard curve. Data from three independent experiments were used to calculate mean values and standard deviation. * p < 0.05, ** p < 0.01 vs. the untreated group.
The expression of the TNF-α mRNA in LPS-treated cells (Figure 5a–c) was approximately 17.71-fold higher than that in cells not stimulated by LPS; this decreased to 5.76-, 8.09-, and 1.44-fold when the cells were incubated with 50 µg/mL of LBEO, SBEO, and RBEO, respectively. The IL-6 mRNA level increased by approximately 11.6-fold in the LPS-supplemented samples, which decreased to 5.73-, 4.75-, and 2.02-fold in the presence of 50 µg/mL of LBEO, SBEO, and RBEO, respectively (Figure 5d–f). These results suggest that LBEO, SBEO, and RBEO acted as anti-inflammatory agents in LPS-induced RAW 264.7 cells by suppressing the production of TNF-α and IL-6. Among the three EOs, RBEO inhibited the transcription and translation of the pro-inflammatory cytokines the most (Figure 4 and Figure 5).
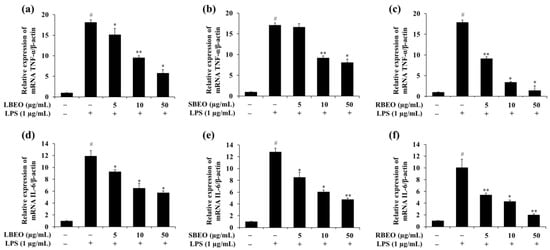
Figure 5.
Effect of BEOs on the TNF-α and IL-6 mRNA levels of macrophages. Effects of (a) BLEO, (b) SBEO, and (c) RBEO on the TNF-α mRNA expression and effects of (d) BLEO, (e) SBEO, and (f) RBEO on the IL-6 mRNA expression in RAW 264.7 cell model after LPS stimulation were analyzed by qRT-PCR. The β-actin mRNA level was used as the interior standard to normalize the data, and the results were compared with those for the untreated cells. Data from three independent experiments were used to calculate mean values and standard deviation; # p < 0.05 vs. the LPS-untreated cells group, * p < 0.05, and ** p < 0.01 vs. the LPS-treated group.
2.5. Effect of the BEOs on the Transcription and Translation of iNOS and COX-2
COX-2 and iNOS are important pro-inflammatory proteins responsible for the synthesis of NO and PGE2. Western blotting and qRT-PCRwere performed to evaluate whether the EOs inhibited expression of iNOS and COX-2 at both transcription (mRNA) and translation (protein) levels in the RAW 264.7 cell model. Corresponding to the results of NO production mentioned previously (Figure 3), the expression levels of iNOS at transcription level (Figure 6j–l) and translation level (Figure 6a–c,g–i) were significantly inhibited in the presence of BEOs. The inhibition of the iNOS expression (both mRNA and protein levels) was most effective in the RBEO sample (Figure 6c,i,l) where the mRNA and protein levels decreased to the levels observed in LPS-untreated cells.
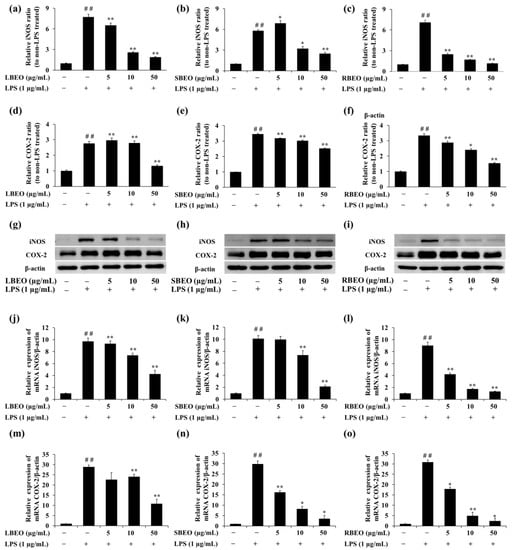
Figure 6.
Inhibitory effects of BEOs on iNOS and COX-2 expression in RAW 264.7 macrophages. The iNOS and COX-2 protein levels in (a,d,g) LBEO-, (b,e,h) SBEO-, and (c,f,i) RBEO-pre-treated cells were assessed using Western blotting after 24 h LPS induction. The iNOS and COX-2 mRNA expression levels in (j,m) LBEO-, (k,n) SBEO-, and (l,o) RBEO-pre-treated RAW 264.7 cells were quantified using qRT-PCR after 24 h LPS induction. The β-actin mRNA level was used to normalize the data. The results were compared to those in the untreated cells. Results are shown as mean ± SD (n = 3). ## p < 0.01 vs. the LPS-untreated cells; * p < 0.05, ** p < 0.01 vs. the LPS alone-treated cells.
COX-2 is an important enzyme that controls the production of PGE2 in the inflammatory response [25,26]. Figure 6 shows the effect of BEOs on COX-2 expression at both transcription and translation levels. The COX-2 protein expression increased significantly in response to LPS stimulation. However, in the presence of BEOs (5, 10, or 50 μg/mL), the COX-2 protein expression decreased in a concentration-dependent manner (Figure 6d–f). Moreover, COX-2 mRNA levels (Figure 6m–o) also decreased in the presence of BLEO, SBEO, and RBEO. Thus, BEOs can inhibit COX-2 expression at mRNA and protein levels.
2.6. Inhibitory Effect of BEOs on the NF-κB Pathway in RAW 264.7 Macrophages
The NF-κB pathway regulates many genes involved in the inflammatory response [15,27,28]. To further investigate the inhibitory targets of EOs, we analyzed whether BEOs can affect the phosphorylation of two key proteins (IkBα and p65) in NF-κB pathway by monitoring the levels of IκBα, phosphorylated IκBα (p-IκBα), p65, and phosphorylated p65 (p-p65) in RAW 264.7 macrophages using Western blotting. In the presence of increasing concentrations of LBEO, SBEO, and RBEO (Figure 7a–f), the level of phosphorylated IκBα in LPS-stimulated macrophages decreased gradually to that observed in the unstimulated stage. The levels of p-p65 gradually decreased to the level in the unstimulated state when the EO concentration reached 50 µg/mL (Figure 7a–c,g–i). These results suggest that BEOs inhibited the NF-κB pathway by inhibiting p65 and IκBα phosphorylation.
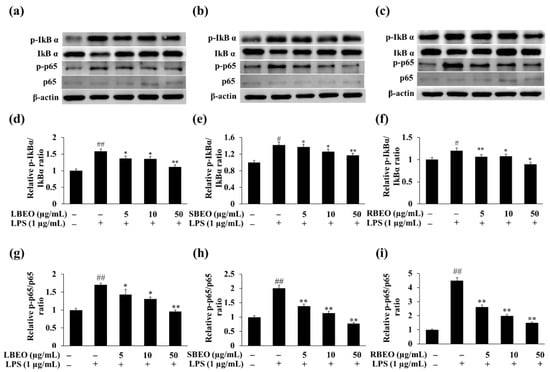
Figure 7.
Effects of LBEO, SBEO, and RBEO on protein levels of IκBα, p-IκBα, p65, and p-p65 in RAW 264.7 macrophages. Western blot analysis of non-phosphorylated and phosphorylated forms of IκBα and p65 in LPS-induced macrophages treated with (a) LBEO, (b) SBEO, and (c) RBEO. Relative expression level of p-IκBα and p-p65 in (d,g) LBEO-, (e,h) SBEO-, and (f,i) RBEO-treated group compared to non-stimulated group. The results are expressed relative to those for the LPS-untreated cells as mean ± SD (n = 3). # p < 0.05, and ## p < 0.01 vs. the LPS-untreated cells; * p < 0.05, and ** p < 0.01 relative to the LPS alone-treated cells.
2.7. In Vivo Anti-Inflammatory Assay
To evaluate the anti-inflammatory effect of BEOs at the in vivo level, we used Swiss mice as an animal model for the carrageenan-induced paw edema test. As shown in Figure 8a, all the EOs (LBEO, SBEO, and RBEO) inhibited paw edema in the Swiss mice model compared to that observed after treatment with indomethacin, a common nonsteroidal anti-inflammatory drug. Among the four samples (LBEO, SBEO, RBEO, and indomethacin), LBEO showed the maximum inhibition of paw edema throughout the trial period of 1–6 h after carrageenan-induced inflammation, with percentage inhibition of paw edema being 35.66 ± 8.29%, 43.11 ± 5.12%, 67.22 ± 3.31%, 70.74 ± 1.40%, and 55.45 ± 6.61% after 1, 2, 3, 4, and 6 h, respectively. The edema inhibition abilities of SBEO and RBEO were similar to that of indomethacin from 2 to 6 h after carrageenan induction. Compared to the other time points, at about 3 h after carrageenan induction, all three essential oils showed the highest inhibition (p < 0.001) of paw edema in mice; LBEO showed 67.22 ± 3.31% inhibition, while, LBEO, SBEO, and the positive control (indomethacin) showed 48.03 ± 5.05%, 47.01 ± 2.87%, and 46.38 ± 6.59% inhibition, respectively. The percentage increase in paw edema in Figure 8b and the images of the mouse paw edema (Figure 8c) at 3 h also indicated that the inhibitory effects of LBEO, SBEO, and RBEO on paw edema were similar or even better than that of indomethacin.
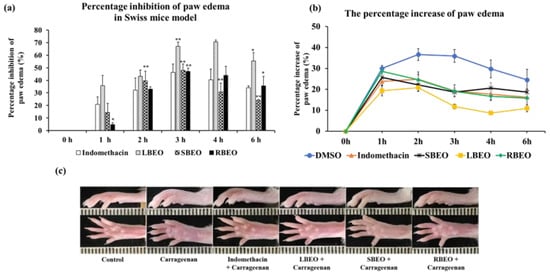
Figure 8.
Inhibition of paw edema in mice by LBEO, SBEO, and RBEO. (a) The percentage inhibition of paw edema in mice over time was monitored after treatment with LBEO, SBEO, RBEO, and the positive control (indomethacin). Data are shown as mean ± SD (n = 6 mice/group), * (p < 0.01), and ** (p < 0.001) vs. indomethacin group. (b) The percentage increase in paw edema. (c) Image of hind paws of mice at 3 h after carrageenan injection.
Carrageenan-induced paw edema model is a well-known in vivo model for studying acute inflammation. In this model, carrageenan induces edema in two-phase responses, namely, the early and delayed phases. The early phase (0−2 h) involves the production of histamines, serotonin, and bradykinin, while the delayed phase (3−6 h) involved the release of NO and pro-inflammatory cytokines, such as IL-6 and TNF-α. Our in vivo results show that all three EOs inhibited paw edema at both phases. This agrees with our in vitro results showing that LBEO, SBEO, and RBEO can inhibit multiple steps in the inflammatory responses of the RAW 264.7 cell model, including (1) NO production; (2) TNF-α, IL-6, iNOS, and COX-2 expression at both mRNA and protein levels; and (3) IκBα and p65 phosphorylation in the NF-κB pathway.
2.8. Molecular Docking of 12 Main Compounds with Anti-Inflammatory Protein Targets
We showed that BEOs possessed anti-inflammatory activities and also revealed some of the important targets of these activities using Western blotting, ELISA, and RT-PCR. However, the underlying molecular mechanisms and interaction affinities of the EO components with inflammatory protein targets remain unknown. Hence, to further investigate the potential interaction affinity of BEO compounds with inflammatory protein targets, we selected 12 main compounds (Table 2) present in BEOs, which were then docked with inflammatory protein targets [28,29,30] of the AA, NF-κB, and MAPK pathways. The results of molecular docking are summarized in Table 2.

Table 2.
Docking energy of the 12 main compounds with target proteins.
2.8.1. Validation of the Molecular Docking Method
To obtain reliable results in molecular docking analysis, the molecular docking method should be validated. Toward this, reference ligands were obtained from native co-crystal structures, including meloxicam for COX-1 (PDB ID: 4O1Z), celecoxib for COX-2 (PDB ID: 6COX), 30Z for 5-LOX (PDB ID: 6N2W), RS7 for ALOX15 (PDB ID: 2P0M), AT2 for iNOS (PDB ID: 3E7G), SB0 for p38MAPK (PDB ID: 4FA2), ligand PDB ID:519 for JNK (PDB ID: 4Y5H), ANP for ERK5 (PDB ID: 4IC7), AQ4 (erlotinib) for EGFR (PDB ID: 1M17), and 03Q for ERBB2 (PDB ID: 3PP0). All the reference ligands docked back well to the target protein structures (Figure 9). The re-docked conformations overlaid well with their crystal conformations, with the RMSD values ranging from 0.049 Å (COX-1) to 1566 Å (5-LOX). These results indicate that our docking method provided reliable results and can be used to further analyze the interaction of other potential ligands with the target protein structures.
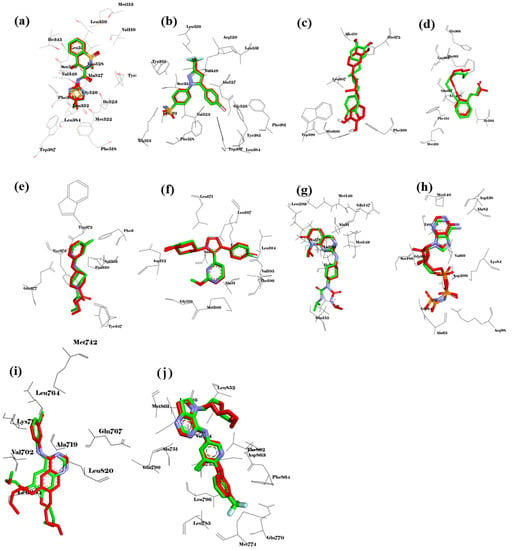
Figure 9.
Validation of the molecular docking method. Overlay of redocked conformation (green carbons) with co-crystalized conformation (red carbons) of (a) meloxicam to COX-1 (4O1Z), (b) celecoxib to COX-2 (6COX), (c) 30Z to 5-LOX (6N2W), (d) RS7 to ALOX15 (2P0M), (e) AT2 to iNOS (3E7G), (f) SB0 to p38MAPK (4FA2), (g) PDB ID:519 to JNK (4Y5H), (h) ANP to ERK5 (4IC7), (i) AQ4 to EGFR (1M17), and (j) 03Q to ERBB2 (3PP0).
2.8.2. AA Metabolic Pathway
The AA metabolic pathway is associated with the development and elimination of inflammation [15,28,31,32]. Key proteins associated with inflammatory responses of the AA pathway, including COX-1, COX-2, 5-LOX, and ALOX15, were selected to evaluate their interaction with 12 selected compounds present in the BEOs. The docking results (Table 2) show that all 12 compounds (except compound 30) showed good binding affinity for COX-1, COX-2, and ALOX15, with binding values below −6.0 kcal/mol. The lower the calculated binding values, the better the binding ability. Thus, compounds 14, 16, 19, 20, 23, and 27 showed extremely strong binding affinity for COX-1, COX-2, and ALOX15, with binding values lower than −7.0 kcal/mol. All the compounds showed weaker binding with 5-LOX, among which, the binding affinities of compounds 14, 16, 19, 20, 23, and 27 ranged from −6.1 to 6.4 kcal/mol. These results suggest that the main compounds of BEOs can target and interact with multiple targets of the AA pathway. In particular, compound 16 (β-caryophyllene), present in all three types of EOs (leaf, stem, and root), showed considerably high binding affinity for COX-1 (−9.0 kcal/mol), COX-2 (−8.1 kcal/mol), and ALOX15 (−8.2 kcal/mol). This partially explained why all three types of BEOs showed good anti-inflammatory effects, although their compound compositions may differ. Compound 27 (present in SBEO and RBEO) also showed very strong binding with COX-1 (−7.0 kcal/mol), COX-2 (−7.8 kcal/mol), 5-LOX (−6.4 kcal/mol), and especially ALONX15 (−9.1 kcal/mol). The binding locations of 16 and 27 with COX-1, COX-2, and ALOX15 (Figure 10) are similar to the binding locations of the reference co-crystal ligands (Figure 9), indicating the reliability of the docking method. These results show that the bioactive compounds of the BEOs can target key components of the AA pathway, thereby inhibiting inflammatory responses.
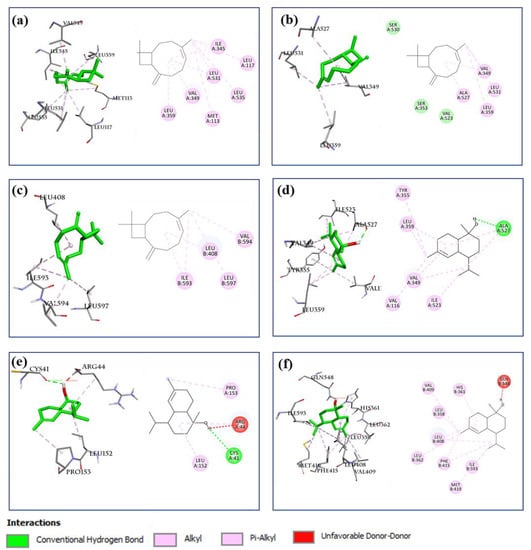
Figure 10.
Docking analysis of compounds 16 and 27 with COX-1, COX-2, and ALOX15. Interaction analysis of compound 16 (left: 3D, right: 2D) with (a) COX-1 (PDB ID: 4O1Z), (b) COX-2 (PDB ID: 6COX), and (c) ALOX15 (PDB ID: 2P0M). Interaction analysis of compound 27 (left: 3D, right: 2D) with (d) COX-1(PDB ID: 4O1Z), (e) COX-2(PDB ID: 6COX), and (f) ALOX15 (PDB ID: 2P0M).
2.8.3. NF-κB Signaling Pathway
The NF-κB pathway is also associated with inflammation [15,27,28]. Therefore, important components of the NF-κB signaling pathway (TLR, NF-κB p50p65, TNF-α, and iNOS) were selected for studying their interactions with components of BEOs using molecular docking analysis. No co-crystal structure of inhibitor/NF-κB complex is reported so far. Therefore, the re-dock strategy cannot be used to validate docking methods for NF-κB. However, a docking method that was confirmed to be effective for NF-κB proteins in previous studies [33,34] can be used, in which the binding region of NF-κB with DNA fragment was used as the binding site for docking. The grid box was set at 50 × 50 × 50 Å with a 0.375 Å space to cover the DNA binding pocket of the most prevalent form of NF-κB, p50-p60 (PDB ID: 1VKX), and all other docking parameters were set to default values. The docking results (Table 2) show that compounds 10, 11, and 16 have good binding affinity for the NF-κB p50-p65 homodimer complex, with the calculated binding values of −6.2, −6.1, and −6.7 kcal/mol, respectively. Compounds 20, 23, and 27 showed strong binding affinities for NF-κB p50-p65, with the calculated binding values of −7.0, −7.6, and −7.1 kcal/mol, respectively. Interestingly, these compounds interacted with the cavity near the cysteine 359 of p50 (Figure 11a–c). This result agrees with that of a previous study showing that the binding region of the p50 inhibitor (andrographolide) is near the cysteine 359 of p50 [35]. This suggests that many compounds of BEOs can interact and inhibit NF-κB p50.
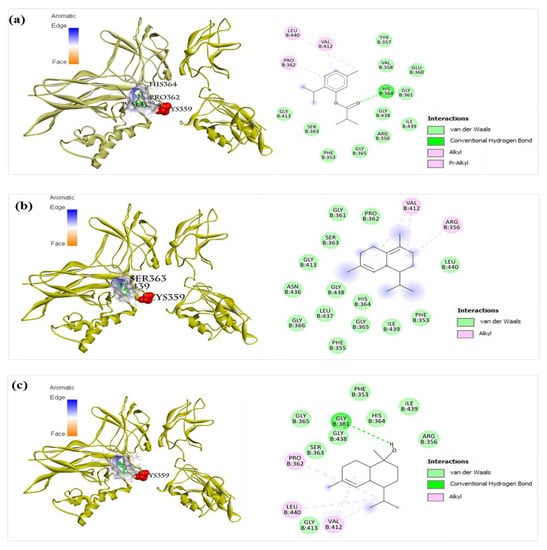
Figure 11.
Docking analysis of compound 20, 23, and 27 with NF-κB p50-p65. (a) Compound 20 (left: 3D, right: 2D) showed pi-alkyl interaction with Val412, alkyl interaction with Pro362 and Leu440, and hydrogen bonding with His364 of NF-κB (PDB ID: 1VKX). (b) Compound 23 (left: 3D, right: 2D) showed alkyl interaction with Val412 and Arg356, and van der Waals interactions with Phe353, Phe355, Gly361, Pro362, Ser363, His364, Gly366, Gly413, Gly365, Asn436, Leu437, Gly438, Ile439, and Leu440, of NF-κB (PDB ID: 1VKX). (c) Compound 27 (left: 3D, right: 2D) showed alkyl interaction with Pro362, Val412, and Leu440, and hydrogen bonding with Gly361 residues of p50 (PDB ID: 1VKX).
Another important component of the NF-κB pathway is iNOS. Compounds 10, 11, 14, 16, 19, 20, 23, and 27 showed good interaction with iNOS, with binding affinity values lower than -6.0 kcal/mol. Compounds 14, 16, 19, 20, 23, and 27 also showed good interaction with TNF-α, with binding affinity values lower than -6.0 kcal/mol. However, TLR did not appear to be the inhibitory target of BEO components when all the calculated binding values ranged from -3.1 to -5.2 kcal/mol (Table 2). These results suggest that NF-κB, TNF-α, and iNOS are the main targets of the bioactive compounds in BEOs.
2.8.4. MAPK Signaling Pathway
The MAPK signaling pathway is one of the key pathways for inducing inflammatory responses [15,27,28]. In this study, important components of the MAPK pathway mediating inflammatory responses, including JNK, p38 MAPK, ERK5, EGFR [36], and ERBB2 [37], were selected for the docking study with the twelve main components of BEOs. The docking results (Table 2) suggest that almost all compounds (except for compound 30) docked well with five target proteins of the MAPK pathway, with calculated binding affinities lower than −6.0 kcal/mol. Compounds 14, 16, 19, 23, and 27 showed strong binding affinities for all the five target proteins (p38MAPK, JNK, ERK5, EGFR, and ERBB2), as their calculated binding affinities were lower than -7.0 kcal/mol. Compounds 16 and 23 showed strong binding ability with p38 MAPK, with the binding affinity values of −8.2 and −8.5 kcal/mol, respectively, which are better than that of a reference compound (SB0) (−7.8 kcal/mol) for p38 MAPK. The binding sites of compounds 16 and 23 with p38 MAPK (Figure 12) are similar to that of SB0 (Figure 9), indicating the reliability of our docking method. Compound 16 showed very high binding with p38MAPK, JNK, ERK5, and ERBB2, with calculated binding affinity values of −8.2, −8.0, −7.2, and −7.7 kcal/mol, respectively. These results suggest that many bioactive compounds of BEOs can interact and inhibit multiple important targets of the MAPK pathway.
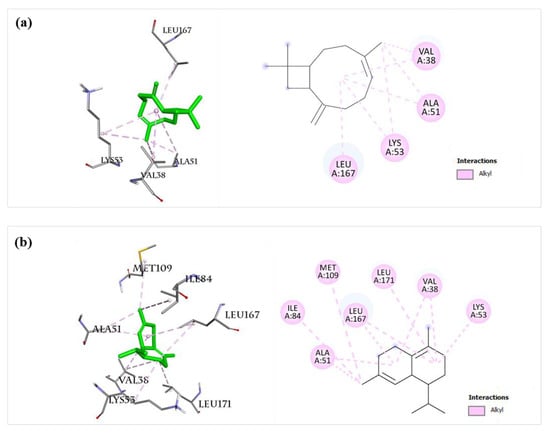
Figure 12.
Docking analysis of compounds 16 and 23 with p38MAPK. (a) Compound 16 (left: 3D, right: 2D) showed alkyl interaction with Val38, Ala51, Lys53, and Leu167 residues of p38MAPK (PDB ID: 4FA2). (b) Compound 23 (left: 3D, right: 2D) showed alkyl interaction with Val38, Ala51, Lys53, Ile84, Met109, Leu167, and Leu171 residues of p38MAPK (PDB ID: 4FA2).
2.8.5. Inflammatory Interleukins (IL-6 and IL-23R)
Inflammatory interleukins also play essential roles in inflammatory responses [29,38]. In this study, IL-6 and IL-23 receptors (IL-23R) were selected for molecular docking studies. Almost all the 12 selected compounds showed low binding affinity for IL-6 and IL-23R (ranging from −3.6 to 5.9). Only compound 27 showed good binding affinity for IL-6 and IL-23R, with binding values of −6.2, and −6.1 kcal/mol, respectively. Compound 16 showed good binding affinity for IL-6, with a binding affinity value of −6.1 kcal/mol. Overall, the binding abilities of the selected compounds of BEOs with inflammatory cytokines (IL-6, IL-23R) were not as high as those of other inflammatory targets, such as COX-1, COX-2, ALONX15, NF-κB, p38MAPK, JNK, and ERBB2. These results suggest that IL-6 and IL-23R are not the key anti-inflammatory targets of BEOs.
Although the 12 main compounds of BEOs have good docking scores with important anti-inflammatory protein targets, the anti-inflammatory effects of BEOs may also be due to interactions with other inflammatory protein targets. The in silico anti-inflammatory effects of the 18 other compounds of low abundance in the BEOs (especially RBEO) have to be analyzed using molecular docking in the future.
3. Materials and Methods
3.1. Experimental Animals and Ethics Statement
Swiss mice (female, 8–12-week-old, 20–25 g) from the National Institute of Hygiene and Epidemiology, Hanoi, Vietnam were maintained at the Animal Facility, Faculty of Biology, HUS, Vietnam National University, Vietnam, using standard conditions with commercial food and water ad libitum. Ethical approval for animal studies was obtained from the Ethic Committee of the Dinh Tien Hoang Institute of Medicine (certificate number: IRB-A-2102).
3.2. Plant Material
The whole plant, containing leaves, stem, and roots of B. lanceolaria were collected from Hanoi city (21°2′46.46′′ N, 105°24′56.59′′ E), Vietnam, in December 2020. The plant samples were identified and deposited at the Center of Life Science, Faculty of Biology, University of Science, Vietnam National University, Hanoi, Vietnam, under specimen vouchers (No. 20201201, No. 20201202, and No. 20201203).
3.3. Preparation of EOs
The plant materials were separated into three parts: leaves, stem, and roots. These materials were then sliced into small pieces before being steam-distilled to obtain three essential oils, namely, LBEO (0.1677%), SBEO (0.0111%), and RBEO (0.0177%).
3.4. GC/MS Analyses
The GC-MS/GC-flame ionization detection (FID) comprising of an HP7890A model GC and HP5975C MS detector (Agilent Technologies, Santa Clara, CA, USA) was used for analyzing the chemical constituents of LBEO, SBEO, and RBEO. A HP-5 MS column with a dimension of 60 m × 0.25 mm and film thickness of 0.25 μm was used for separation. The running condition was set as follows: injector temperature at 250 °C; initial temperature started at 60 °C then increased to 240 °C with an increasing step of 4 °C/min; the carrier gas was used of helium and the flowrate was set as 1 mL/min; the split ratio was 100:1; full scan modes under electron ionization with voltage: 70 eV, emission current: 40 mA; mass range scan: 35–450 a.m.u. Chemical constituents in each essential oil were identified by analysis of RI and MS values in comparing with standard compounds in the NIST database and literature [39].
3.5. Chemicals and Reagents
Escherichia coli LPS, and λ-carrageenan were obtained from Sigma (St. Louis, MO, USA). Primary antibodies (Anti-iNOS, anti-COX2, anti-IκBα, anti-p-IκBα, anti-β-actin antibodies) and secondary antibody (goat anti-rabbit IgG (H + L), HRP) were purchased from Invitrogen (Rockford, IL, USA). Anti-p65 and anti-phospho- p65 were obtained from Bio-Rad (Oxford, UK).
3.6. Cell Culture
The RAW 264.7 cells were obtained from the Animal Cell Culture Lab, Faculty of Biology, University of Science, Vietnam National University. The cells were grown in DMEM medium at 37 °C with 10% FBS, 50 µg/mL streptomycin/penicillin, and 5% CO2.
3.7. Cell Viability Assay
The toxicity of the EO samples toward RAW 246.7 macrophages was determined using CCK-8 (ab228554, Abcam, Cambridge, UK). The macrophages were maintained at 37 °C for 24 h in a 96-well plate (2 × 104 cells in 100 µL culture per well) before being treated with various concentrations of EOs from B. lanceolaria (BEOs) for another 24 h. Each sample was incubated with 10 μL of CCK-8 solution for 2 h at 37 °C before being measured at the OD of 450 nm.
3.8. Measuring NO Production
RAW 264.7 macrophages (2 × 105 cells/well) were cultured in FBS-free DMEM for 3 h before being incubated with EOs (0, 5, 10, and 50 µg/mL) for 2 h. The cells were then incubated with LPS (1 μg/mL) for 24 h to stimulate NO production. NG-methyl-L-arginine acetate (L-NMMA) was used as the positive control. Cell culture medium (100 μL) was mixed with 100 μL Griess reagent (Promega, Madison, WI, USA) at 25 °C for 10 min and the absorbance at 540 nm was then recorded using the SpectraMax Plus384 microplate reader (Molecular Devices, California, USA).
The inhibition of NO production was determined using the formula:
3.9. ELISA
The macrophages (2 × 105 cells/well) were maintained for 24 h at 37 °C before being treated to 5, 10, and 50 µg/mL BEOs for 2 h. The cells were then stimulated by LPS (1 µg/mL) for 24 h. The supernatant was used to measure the concentrations of TNF-α and IL-6 produced using ELISA kits (Invitrogen, Vienna, Austria).
3.10. Western Blotting
The effects of BEOs on the expression of important proteins involved in the inflammatory response in RAW 264.7 macrophages were assessed by Western blotting. The cells (4 × 105 cells/well in 6-wells plate) were cultured for 24 h before incubating with BEOs for 2 h. The cells were then stimulated by LPS (1 µg/mL) for 24 h before being harvested and resuspended in radioimmunoprecipitation assay lysis buffer (Thermo Scientific, Rockford, IL, USA) to obtain the total protein. Total protein samples (20 µg each) were analyzed using SDS-PAGE before being transferred to a PVDF membrane. The membrane was blocked with T-TBS (Tris-buffered saline, 0.1% Tween 20) containing 2% bovine serum albumin (Biobasic, Markham, Ontario, Canada) for 1 h. The membrane was then incubated with primary antibodies (1:1000 dilutions) for detecting β-actin, iNOS, COX-2, IκBα, p-IκBα, NF-κB p65, and p-NF-κBp65 for 16 h at 4 °C. The membranes were then washed thrice with T-TBS before being incubated with HRP-conjugated goat anti-rabbit IgG for 1 h at 25 °C. The membranes were washed thrice and then incubated with Clarity MaxTM Western ECL substrate (Bio-Rad, Milan, Italy) and the signals were detected using a ChemmiDocTM imaging system (Bio-Rad, Hercules, CA, USA). Protein quantities from Western blot results were calculated by Image Lab software Version 6.1 (Bio-Rad, Hercules, CA, USA).
3.11. qRT-PCR
To determine the effect of BEOs on the mRNA production of IL-6, TNF-α, iNOS, and COX-2, the RAW 264.7 macrophages (4 × 105 cells/well) were cultured for 24 h before being treated with LBEO, SBEO, and RBEO (5, 10, and 50 µg/mL) for 2 h. Inflammatory responses were stimulated by LPS (1 µg/mL) for 16 h. Total RNA (5 µg/mL) purified from each treated sample using the TRIzol™ reagent (Invitrogen, Rockford, Illinois, USA) was converted to cDNA using M-MLV reverse transcriptase (Thermo Fisher Scientific). The qPCR reaction (10 µL in total volume) contained 2 µL cDNA templates, 5 µL GoTaq® qPCR master mix (Promega, Madison, WI, USA), 0.25 µL forward/reverse primers (Table 3), and 2.5 µL H2O. All reactions were run with 40 cycles; each cycle included DNA denaturation step at 95 °C for 30 s, primer annealing step at 60 °C for 30 s, and DNA strand extension step at 72 °C for 30 s. Each reaction was run in triplicate. The relative mRNA levels were calculated using the 2-∆∆Ct method [40]; β-actin mRNA level was used as an internal standard for normalizing expression data.

Table 3.
Primers for qRT-PCR.
3.12. Anti-Inflammation Assay Using Carrageenan-Induced Edema Model
The effects of BEOs on acute inflammation in an animal model were evaluated using the carrageenan-induced paw edema model [44]. Swiss mice (21–24 g) were stabilized under laboratory conditions for at least 7 days prior to testing. Five groups of mice (n = 6 mice/group) were prepared, including: (1) negative control group (dimethyl sulfoxide or DMSO, 10 mL/kg), (2) positive control group (indomethacin, 10 mg/kg), (3) LBEO (50 mg/kg), (4) SBEO (50 mg/kg), and (5) RBEO (50 mg/kg). Mice were orally administered with indomethacin, or BEOs, and DMSO (the vehicle control). After 60 min, 0.025 mL of 2% carrageenan suspension (Sigma-Aldrich) was injected under the soles of the right hind paws to induce inflammation. The changes in the right hind paw thickness of the mice at 0, 1, 2, 3, 4, and 6 h after carrageenan injection was measured using a micrometer (Mitutoyo, Japan). The changes in the hind paw thickness in the BEO-treated groups were compared to those in the control group (DMSO) at the same time to evaluate the anti-inflammatory effect of the tested samples. The BEOs or the reference drug (indomethacin) were considered to show an acute anti-inflammatory effect if the extent of reduction in paw edema was statistically significant compared to that in the negative control group (DMSO). The percentage inhibition of inflammation was calculated using the following formulae:
where
I (%): Percentage inhibition at t h.
∆CC: Percentage increase in the thickness of the hind paw in the negative control group at t h compared to 0 h.
∆Ct: Percentage increase in the thickness of the hind paw in the positive control group or treated groups at t h compared to 0 h.
3.13. Molecular Docking
Molecular docking was used to explore the potential anti-inflammatory targets of B. lanceolaria EOs. Twelve main components of the B. lanceolaria EO samples from the leaves, stem, and roots (Table 2 and Figure 1) were docked with the key anti-inflammatory protein targets [15,28,29,30,31], including COX-1 (PDB ID: 4O1Z) [45], COX-2 (PDB ID: 6COX) [46], 5-LOX (PDB ID: 6N2W) [47], ALOX15 (PDB ID: [48]), TLR (PDB ID: 2Z7X) [49], NF-κB (PDB ID: 1VKX) [50], TNF-α (PDB ID: 2E7A) [51], iNOS (PDB ID: 3E7G) [52], p38MAPK (PDB ID: 4FA2) [53], JNK (PDB ID: 4Y5H) [54], ERK5 (PDB ID: 4IC7) [55], EGFR (PDB ID: 1M17) [56], ERBB2 (PDB ID: 3PP0) [57], IL-6 (PDB ID: 1P9M) [58], and IL-23R (PDB ID: 3DUH) [59]. All the PDB files were downloaded from the RCSB PDB database “https://www.rcsb.org/ (accessed on 8 May 2022)” and processed using the Discovery Studio Visualizer 2021 [60] and AutoDockTools v1.5.7 [61] to remove water and co-crystalized ligands. Polar hydrogen atoms and Kollman charges were added to the protein structures before converting to the pdbqt file format. The SDF files of the twelve main EO compounds and reference compounds were downloaded from PubChem [62] and converted to pdb format using PyMol 2.4.0 [63]. The structure files were then processed using AutoDock tools v1.5.7 [61] to add Gasteiger charges and define rotatable bonds. The grid box of 20 × 20 × 20 Å (8000 Å3) with a 0.375 Å space was generated at the ligand binding site of each protein. For molecular docking, Autodock Vina 1.1.2 [64] with the Lamarckian genetic algorithm was used. The default docking protocol with a rigid protein, flexible ligand, and exhaustiveness value of 8 was used. The lowest binding energy conformation of each ligand was selected for the interaction analysis using PyMol 2.4.0 [63] and the Discovery Studio Visualizer 2021 [60].
3.14. Statistical Analysis
The mean values were statistically compared using one-way analysis of variance (ANOVA) with Tukey’s test. The differences were considered significant for p < 0.05. The statistical tests were applied using OriginPro, version 8.5.1 (OriginLab Corp, Northampton, MA, USA).
4. Conclusions
In this study, the chemical components and their concentrations in the EOs from the leaf, stem, and root samples of B. lanceolaria, collected from Vietnam, were successfully elucidated. GC-MS/GC-FID analysis identified 30 compounds in the BEOs, among which, LBEO, SBEO, and RBEO contained 5, 15, and 20 compounds, respectively. Despite the variability among some of the major components in all three types of EOs, all of them showed remarkable anti-inflammatory effects in in vitro and in vivo models of inflammation. LBEO, SBEO, and RBEO inhibited multiple steps in the inflammatory responses of the RAW 264.7 cell model, including (1) NO production, (2) TNF-α, IL-6, iNOS, and COX-2 expression at both mRNA and protein levels, and (3) IκBα and p65 phosphorylation in the NF-κB pathway. In the carrageenan-induced paw edema model, all three EOs inhibited paw edema at both early and delayed phases. These in vivo results are in agreement with the in vitro results, as the inhibition of paw edema at both phases is also associated with the inhibition of key inflammatory components, including COX-1, iNOS production, nitric oxide (NO), and pro-inflammatory cytokines (TNF-α and IL-6). Furthermore, our molecular docking simulation suggested that the chemical components of B. lanceolaria EOs target and bind very strongly with many protein components of all three important signaling pathways related to inflammation, including the AA metabolic pathway (COX-1, COX-2, and ALOX15), NF-κB pathway (NF-κB, TNF-α, and iNOS), and MAPK pathway (p38MAPK, JNK, ERK5, EGFR, and ERBB2).
Overall, our results show, for the first time, the detailed chemical composition of BEOs and confirmed their potent anti-inflammatory effects using an in vitro cell model (RAW 264.7), in vivo animal model (carrageenan-induced mouse model of edema) and in silico model (molecular docking of key inflammatory components). These results demonstrate the promising anti-inflammatory potential of BEOs, which can be further utilized to develop effective anti-inflammatory drugs with limited side effects in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27227839/s1, Figure S1. GC-MS chromatogram of the essential oils of B. lanceolaria leaf (LBEO); Figure S2. GC-MS chromatogram of the essential oils of B. lanceolaria stem (SBEO); Figure S3. GC-MS chromatogram of the essential oils of B. lanceolaria root (RBEO).
Author Contributions
Conceptualization, T.T.H.D., T.U.N., P.-H.N. and V.S.N.; methodology, T.T.H.D., T.U.N., T.T.H.N., T.Y.H., T.L.H.P., P.-H.N. and V.S.N.; validation, T.U.N., P.-H.N. and V.S.N.; formal analysis, T.T.H.D., T.U.N., T.T.H.N., T.Y.H., T.S.L., T.H.V.N., Q.H.N., P.-H.N. and V.S.N.; investigation and data curation, T.T.H.D., T.U.N., T.T.H.N., T.Y.H., T.S.L., T.L.H.P., T.H.V.N., Q.H.N., P.-H.N. and V.S.N.; funding acquisition, V.S.N. The manuscript was written and edited by all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106-NN.02-2016.58.
Institutional Review Board Statement
The ethical approval was obtained from Ethic Committee at Dinh Tien Hoang Institute of Medicine with certificate number IRB-A-2102 for using animals in this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples are available from the authors.
References
- Medzhitov, R. Inflammation 2010: New Adventures of an Old Flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Scrivo, R.; Vasile, M. Inflammation as “common soil” of the multifactorial diseases. Autoimmun. Rev. 2011, 10, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Gu, Y.; Wang, C.; Zhang, J.; Zhang, J.; Wang, G.; Wang, F. A Systematic Review of the Anti-Inflammatory and Immunomodulatory Properties of 16 Essential Oils of Herbs. Evid. Based Complement. Alternat. Med. 2020, 2020, 8878927. [Google Scholar] [CrossRef] [PubMed]
- Arulselvan, P.; Fard, M.T.; Tan, W.S.; Gothai, S.; Fakurazi, S.; Norhaizan, M.E.; Kumar, S.S. Role of Antioxidants and Natural Products in Inflammation. Oxid. Med. Cell. Longev. 2016, 2016, 5276130. [Google Scholar] [CrossRef] [PubMed]
- McCarberg, B.; Gibofsky, A. Need to Develop New Nonsteroidal Anti-Inflammatory Drug Formulations. Clin. Ther. 2012, 34, 1954–1963. [Google Scholar] [CrossRef]
- Davies, N.M.; Reynolds, J.K.; Undeberg, M.R.; Gates, B.J.; Ohgami, Y.; Vega-Villa, K.R. Minimizing risks of NSAIDs: Cardiovascular, gastrointestinal and renal. Expert Rev. Neurother. 2006, 6, 1643–1655. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, L.G.; Jick, H. Risk of upper gastrointestinal bleeding and perforation associated with Individual non-steroidal anti-inflammatory drugs. Lancet 1994, 343, 769–772. [Google Scholar] [CrossRef]
- Mahesh, G.; Kumar, K.A.; Reddanna, P. Overview on the Discovery and Development of Anti-Inflammatory Drugs: Should the Focus Be on Synthesis or Degradation of PGE2? J. Inflamm. Res. 2021, 14, 253–263. [Google Scholar] [CrossRef]
- Kim, K.-N.; Ko, S.C.; Ye, B.R.; Kim, M.S.; Kim, J.; Ko, E.Y.; Cho, S.-H.; Kim, D.; Heo, S.-J.; Jung, W.-K. 5-Bromo-2-hydroxy-4-methyl-benzaldehyde inhibited LPS-induced production of pro-inflammatory mediators through the inactivation of ERK, p38, and NF-κB pathways in RAW 264.7 macrophages. Chem. -Biol. Interact. 2016, 258, 108–114. [Google Scholar] [CrossRef]
- Saleh, H.A.; Yousef, M.H.; Abdelnaser, A. The anti-inflammatory properties of phytochemicals and their effects on epigenetic mechanisms involved in TLR4/NF-κB-mediated inflammation. Front. Immunol. 2021, 12, 606069. [Google Scholar] [CrossRef]
- Fujihara, M.; Muroi, M.; Tanamoto, K.-I.; Suzuki, T.; Azuma, H.; Ikeda, H. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: Roles of the receptor complex. Pharmacol. Ther. 2003, 100, 171–194. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy—From molecular mechanisms to therapeutic benefits. Biochim. Biophys. Acta 2005, 1754, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Patil, K.R.; Mahajan, U.B.; Unger, B.S.; Goyal, S.N.; Belemkar, S.; Surana, S.J.; Ojha, S.; Patil, C.R. Animal Models of Inflammation for Screening of Anti-inflammatory Drugs: Implications for the Discovery and Development of Phytopharmaceuticals. Int. J. Mol. Sci. 2019, 20, 4367. [Google Scholar] [CrossRef] [PubMed]
- Hongzhi, D.; Xiaoying, H.; Yujie, G.; Le, C.; Yuhuan, M.; Dahui, L.; Luqi, H. Classic mechanisms and experimental models for the anti-inflammatory effect of traditional Chinese medicine. Anim. Model. Exp. Med. 2022, 5, 108–119. [Google Scholar] [CrossRef]
- Mahajan, A.; Hardyal, S.; Tung, B. Effect of concurrent use of cimetidine and anti-inflammatory agents in experimental models of peptic ulcer and inflammation. Indian J. Pharmacol. 1984, 16, 132. [Google Scholar]
- Morris, C.J. Carrageenan-induced paw edema in the rat and mouse. Methods Mol. Biol. 2003, 225, 115–121. [Google Scholar]
- Tsuji, R.; Hoshino, K.; Noro, Y.; Tsuji, N.M.; Kurokawa, T.; Masuda, T.; Akira, S.; Nowak, B. Suppression of allergic reaction by λ-carrageenan: Toll-like receptor 4/MyD88-dependent and-independent modulation of immunity. Clin. Exp. Allergy 2003, 33, 249–258. [Google Scholar] [CrossRef]
- Loi, D.T. Những Cây Thuốc và vị Thuốc Việt Nam [Book in Vietnamese]; Medica Publishing House: Hanoi, Vietnam, 1999; pp. 689–690. [Google Scholar]
- Joshi, R.K. GC-MS Analysis of Volatile Organic Constituents of Traditionally Used Medicinal Plants from the Western Ghatsof India: Blumea lanceolaria (Roxb.) Druce., Heliotropium indicum L. and Triumfetta rhomboidea Jacq. J. Mex. Chem. Soc. 2020, 64, 74–82. [Google Scholar] [CrossRef]
- Lalmuanthanga, C.; Roy, D.C.; Roy, R.K.; Sarma, Y.; Borah, P.; Tamuli, S.; Hmarthansanga, L.; Inaotombi, L.; Ali, D.M.A. Antioxidant activity of methanolic extract of Blumea lanceolaria. Int. J. Chem. Stud. 2019, 7, 3546–3548. [Google Scholar]
- Vineet, K.M.; Ajit, K.P.; Nachimuthu, S.K.; Bhim, P.S.; Mishra, V.K.; Passari, A.; Vanlalhmangaihi, K.; Kumar, N.S.; Singh, B.P. Antimicrobial and antioxidant activities of Blumea lanceolaria (Roxb.). J. Med. Plants Res. 2015, 9, 84–90. [Google Scholar] [CrossRef][Green Version]
- Nguyêñ Xuân Duñg, D.T.L.; Dô Tât, H.ù.n.g.; Piet, A. Leclercq Chemical Composition of the Oil of Blumea lanceolaria (Roxb.) Druce from Vietnam. J. Essent. Oil Res. 1991, 3, 285–286. [Google Scholar] [CrossRef]
- FitzGerald, G.A.; Patrono, C. The Coxibs, Selective Inhibitors of Cyclooxygenase-2. N. Engl. J. Med. 2001, 345, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Kobayashi, K. Macrophages in inflammation. Curr. Drug Targets-Inflamm. Allergy 2005, 4, 281–286. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef]
- Bai, J.; Zhang, Y.; Tang, C.; Hou, Y.; Ai, X.; Chen, X.; Zhang, Y.; Wang, X.; Meng, X. Gallic acid: Pharmacological activities and molecular mechanisms involved in inflammation-related diseases. Biomed. Pharmacother. 2020, 133, 110985. [Google Scholar] [CrossRef]
- Li, G.; Fang, Y.; Ma, Y.; Dawa, Y.; Wang, Q.; Gan, J.; Dang, J. Screening and Isolation of Potential Anti-Inflammatory Compounds from Saxifraga atrata via Affinity Ultrafiltration-HPLC and Multi-Target Molecular Docking Analyses. Nutrients 2022, 14, 2405. [Google Scholar] [CrossRef]
- Alamzeb, M.; Setzer, W.N.; Ali, S.; Khan, B.; Rashid, M.-U.; Salman, S.M.; Omer, M.; Ali, J. Spectral, Anti-Inflammatory, Anti-Pyretic, Leishmanicidal, and Molecular Docking Studies, Against Selected Protein Targets, of a New Bisbenzylisoquinoline Alkaloid. Front. Chem. 2021, 9, 711190. [Google Scholar] [CrossRef]
- Wang, B.; Wu, L.; Chen, J.; Dong, L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Signal Transduct. Target. Ther. 2021, 6, 94. [Google Scholar] [CrossRef]
- Wang, T.; Fu, X.; Chen, Q.; Patra, J.K.; Wang, D.; Wang, Z.; Gai, Z. Arachidonic Acid Metabolism and Kidney Inflammation. Int. J. Mol. Sci. 2019, 20, 3683. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Jaiswal, V.; Akhtar, S.; Jayashree, B.S.; Dhar, K.L. Isolation of isoflavones from Iris kashmiriana Baker as potential anti proliferative agents targeting NF-kappaB. Phytochemistry 2017, 136, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Piccagli, L.; Thangavel, S.; Alam, A. Virtual screening against nuclear factor kappaB (NF-kappaB) of a focus library: Identification of bioactive furocoumarin derivatives inhibiting NF-kappaB dependent biological functions involved in cystic fibrosis. Bioorg. Med. Chem. 2010, 18, 8341–8349. [Google Scholar] [CrossRef]
- Nguyen, V.S.; Loh, X.Y.; Wijaya, H.; Wang, J.; Lin, Q.; Lam, Y.; Wong, W.-S.F.; Mok, Y.K. Specificity and inhibitory mechanism of andrographolide and its analogues as antiasthma agents on NF-kappaB p50. J. Nat. Prod. 2015, 78, 208–217. [Google Scholar] [CrossRef]
- Elkamhawy, A.; Hassan, A.H.; Paik, S.; Lee, Y.S.; Lee, H.-H.; Shin, J.-S.; Lee, K.-T.; Roh, E.J. EGFR inhibitors from cancer to inflammation: Discovery of 4-fluoro-N-(4-(3-(trifluoromethyl)phenoxy)pyrimidin-5-yl)benzamide as a novel anti-inflammatory EGFR inhibitor. Bioorganic Chem. 2019, 86, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Yang, Y.; Tan, Y.; Lu, C.; Pan, Y.; Chen, L.; Lu, G. ERBB2-induced inflammation in lung carcinogenesis. Mol. Biol. Rep. 2012, 39, 7911–7917. [Google Scholar] [CrossRef] [PubMed]
- Gabay, C. Interleukin-6 and chronic inflammation. Arthritis Res. Ther. 2006, 8 (Suppl. S2), S3. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/mass Spectrometry, 4th ed.; Allured Publishing Corporation: Carol Stream, IL, USA, 2007. [Google Scholar]
- Gepdiremen, A.; Mshvildadze, V.; Süleyman, H.; Elias, R. Acute and chronic antiinflammatory effects of Hedera colchica in rats. J. Ethnopharmacol. 2004, 94, 191–195. [Google Scholar] [CrossRef]
- Lee, W.-S.; Shin, J.S.; Jang, D.S.; Lee, K.T. Cnidilide, an alkylphthalide isolated from the roots of Cnidium officinale, suppresses LPS-induced NO, PGE2, IL-1β, IL-6 and TNF-α production by AP-1 and NF-κB inactivation in RAW 264.7 macrophages. Int. Immunopharmacol. 2016, 40, 146–155. [Google Scholar] [CrossRef]
- Li, C.; Wang, M.-H. Anti-inflammatory effect of the water fraction from hawthorn fruit on LPS-stimulated RAW 264.7 cells. Nutr. Res. Pr. 2011, 5, 101–106. [Google Scholar] [CrossRef]
- Liao, J.; Xie, X.; Wang, W.; Gao, Y.; Cai, Y.; Peng, J.; Li, T.; Yi, Q.; He, C.; Wang, L. Anti-inflammatory activity of essential oil from leaves of Blumea balsamifera (L.) DC through inhibiting TLR4/NF-kB signaling pathways and NLRP3 inflammasome activation in LPS-induced RAW264. 7 macrophage cells. J. Essent. Oil Bear. Plants 2021, 24, 160–176. [Google Scholar] [CrossRef]
- Winter, C.A.; Risley, E.A.; Nuss, G.W. Carrageenin-induced edema in hind paw of the rat as an assay for antiinflammatory drugs. Proc. Soc. Exp. Biol. Med. 1962, 111, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hermanson, D.J.; Banerjee, S.; Ghebreselasie, K.; Clayton, G.M.; Garavito, R.M.; Marnett, L.J. Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network. J. Biol. Chem. 2014, 289, 6799–6808. [Google Scholar] [CrossRef]
- Kurumbail, R.G.; Stevens, A.M.; Gierse, J.K.; McDonald, J.J.; Stegeman, R.A.; Pak, J.Y.; Gildehaus, D.; Iyashiro, J.M.; Penning, T.D.; Seibert, K.; et al. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature 1996, 384, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.C.; Gerstmeier, J.; Schexnaydre, E.E.; Börner, F.; Garscha, U.; Neau, D.B.; Werz, O.; Newcomer, M.E. Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products. Nat. Chem. Biol. 2020, 16, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chon, J.K.; Kim, S.; Shin, W. Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins 2008, 70, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.-G.; Lee, H.; Lee, J.-O. Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.E.; Huang, D.B.; Chen, Y.Q.; Ghosh, G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 1998, 391, 410–413. [Google Scholar] [CrossRef]
- Shibata, H.; Yoshioka, Y.; Ohkawa, A.; Minowa, K.; Mukai, Y.; Abe, Y.; Taniai, M.; Nomura, T.; Kayamuro, H.; Nabeshi, H.; et al. Creation and X-ray structure analysis of the tumor necrosis factor receptor-1-selective mutant of a tumor necrosis factor-alpha antagonist. J. Biol. Chem. 2008, 283, 998–1007. [Google Scholar] [CrossRef]
- Garcin, E.D.; Arvai, A.S.; Rosenfeld, R.J.; Kroeger, M.D.; Crane, B.R.; Andersson, G.; Andrews, G.; Hamley, P.; Mallinder, P.R.; Nicholls, D.J.; et al. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat. Chem. Biol. 2008, 4, 700–707. [Google Scholar] [CrossRef]
- Watterson, D.M.; Grum-Tokars, V.L.; Roy, S.M.; Schavocky, J.P.; Bradaric, B.; Bachstetter, A.; Xing, B.; Dimayuga, E.; Saeed, F.; Zhang, H.; et al. Development of Novel In Vivo Chemical Probes to Address CNS Protein Kinase Involvement in Synaptic Dysfunction. PLoS ONE 2013, 8, e66226. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Park, C.M.; Iqbal, S.; Hernandez, P.; Park, H.; LoGrasso, P.V.; Feng, Y. Pyridopyrimidinone Derivatives as Potent and Selective c-Jun N-Terminal Kinase (JNK) Inhibitors. ACS Med. Chem. Lett. 2015, 6, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Glatz, G.; Gógl, G.; Alexa, A.; Reményi, A. Structural Mechanism for the Specific Assembly and Activation of the Extracellular Signal Regulated Kinase 5 (ERK5) Module. J. Biol. Chem. 2013, 288, 8596–8609. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the Epidermal Growth Factor Receptor Kinase Domain Alone and in Complex with a 4-Anilinoquinazoline Inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef]
- Aertgeerts, K.; Skene, R.; Yano, J.; Sang, B.-C.; Zou, H.; Snell, G.; Jennings, A.; Iwamoto, K.; Habuka, N.; Hirokawa, A.; et al. Structural Analysis of the Mechanism of Inhibition and Allosteric Activation of the Kinase Domain of HER2 Protein. J. Biol. Chem. 2011, 286, 18756–18765. [Google Scholar] [CrossRef] [PubMed]
- Boulanger, M.J.; Chow, D.C.; Brevnova, E.E.; Garcia, K.C. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science 2003, 300, 2101–2104. [Google Scholar] [CrossRef]
- Lupardus, P.J.; Garcia, K.C. The structure of interleukin-23 reveals the molecular basis of p40 subunit sharing with interleukin-12. J. Mol. Biol. 2008, 382, 931–941. [Google Scholar] [CrossRef]
- BIOVIA, D.S. Discovery Studio Visualizer; v21.1.0.20298; Dassault Systèmes: San Diego, CA, USA, 2021; Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 12 May 2022)v21.1.0.20298.
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Bryant, S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009, 37, W623–W633. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System; Version 2.4.0; Schrodinger, L.L.C.: New York, NY, USA, 2020.
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).