Abstract
Multidrug-resistant bacterial infections mediated by metallo-β-lactamases (MβLs) have grown into an emergent health threat, and development of novel antimicrobials is an ideal strategy to combat the infections. Herein, a novel vancomycin derivative Vb was constructed by conjugation of triazolylthioacetamide and vancomycin molecules, characterized by reverse-phase high performance liquid chromatography (HPLC) and confirmed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). The biological assays revealed that Vb effectively inhibited S. aureus and methicillin-resistant S. aureus (MRSA), gradually increased the antimicrobial effect of β-lactam antibiotics (cefazolin, meropenem and penicillin G) and exhibited a dose-dependent synergistic antibacterial effect against eight resistant strains tested, which was confirmed by the time-kill curves determination. Most importantly, Vb increased the antimicrobial effect of meropenem against the clinical isolates EC08 and EC10 and E. coli producing ImiS and CcrA, resulting in a 4- and 8-fold reduction in MIC values, respectively, at a dose up to 32 μg/mL. This work offers a promising scaffold for the development of MβLs inhibitors, specifically antimicrobials for clinically drug-resistant isolates.
1. Introduction
Bacterial resistance has become a global problem threatening human life and health [1]. Multidrug-resistant bacterial infections are on the rise as many clinical antibiotics have been rendered ineffective [2]. Among them, resistant Gram-positive Enterococcus faecium and Staphylococcus aureus are listed as high priorities for new treatments in “ESKAPE pathogens” and in “the list of drug-resistant bacteria” released by the World Health Organization recently [3,4]. Gram-negative pathogens, such as P. aeruginosa and K. pneumoniae, have presented an enormous clinical challenge due to the dearth of effective antibiotics against these bacteria [5]. In addition, superbugs mediated by metallo-β-lactamases (MβLs) are almost resistant to clinically used antibiotics, including penicillins, cephalosporins and carbapenems [6,7].
Vancomycin (Van), a clinically glycopeptide antibiotic, is known as the “antibiotic last resort” for the treatment of Gram-positive bacterial infections, especially for methicillin-resistant S. aureus (MRSA) [8,9]. However, over the past decades, the increased clinical use of vancomycin has led to the emergence of vancomycin-resistant bacteria, including vancomycin-resistant S. aureus (VRSA) and E. faecium (VRE), which has become a new challenge for antibacterial therapy [10,11,12]. Vancomycin specifically binds to the D-Ala-D-Ala terminal of the cell-wall pentapeptide precursor to inhibit the cell wall biosynthesis of Gram-positive bacteria [13]. Bacteria acquired resistance to vancomycin by mutating the pathogen peptidoglycan sequence from D-Ala-D-Ala to D-Ala-D-Lac, resulting in an overall 1000-fold decrease in the binding affinity to vancomycin [14,15].
At present, VRE has become one of the most common acquired pathogens in hospitals and the treatment options for these drug-resistant infections are severely limited, which has created an urgent need for new clinical agents with activity against resistant pathogens [16]. Thus, various modification strategies of novel vancomycin derivatives have been developed to combat vancomycin resistance, such as lipophilic modification of vancomycin, enhancing the binding affinity of the drug for bacterial ligands, pyrophosphate-targeting designs of cell wall phospholipids and the modification of the vancomycin main structure by total synthesis [17,18,19,20,21]. Recently, our group creatively modified the photosensitizer porphyrin onto the vancomycin molecule; the photosensitizer porphyrin–vancomycin molecule was targeted and enriched on drug-resistant bacteria cells and then the Gram-positive bacteria were inactivated at a specific wavelength by photodynamic therapy [22]. Furthermore, it has been reported that the lipophilic and cationic motifs on vancomycin were modified in combination to enhance the ability of vancomycin to penetrate the bacterial membrane, including vancomycin derivatives carrying C-terminal lipophilic quaternary ammonium moieties and carrying the lysine-rich lipopeptides [23,24]. It was found that vancomycin derivatives with hydrophobic substituents in the disaccharide moiety have significant antibacterial activity against drug-resistant strains including MRSA and VRE [25]. Recently, Venkateswarlu et al. developed a dipicolyl–vancomycin (Dipi-van) conjugate as an inhibitor for the NDM-1 enzyme, which has the ability to penetrate the outer membrane of Gram-negative pathogens (GNPs) and reinstate the activity of carbapenem [26].
We have previously reported the triazolylthioacetamides with different substitutional groups, which exhibited good inhibitory activity against the bacterial resistance target MβLs and restored the antibacterial activity of antibiotics against P. aeruginosa and ImiS-producing E. coli [27,28]. In this work, we constructed a novel vancomycin derivative Vb (Scheme 1) by modifying the molecule with a hydrophobic triazolylthioacetamide that has low steric hindrance and a carboxyl group (see Supplementary Materials). Vb was characterized by reverse-phase HPLC and confirmed by MALDI-TOF MS. The antibacterial activity of Vb and its antibacterial activity synergizing with β-lactam antibiotics against resistant Gram-negative bacteria that produce MβLs and Gram-positive bacteria were evaluated (see Supplementary Materials).
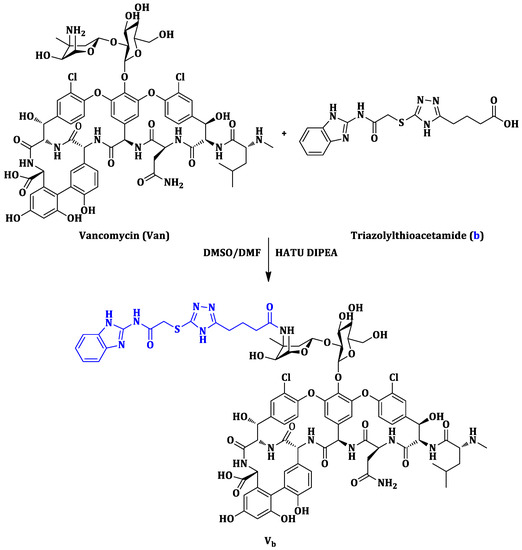
Scheme 1.
Synthetic route of the vancomycin derivative Vb.
2. Results and Discussion
The synthetic pathway of the vancomycin derivative Vb is shown in Scheme 1. Triazolylthioacetamide (b) was synthesized with previously reported methods [27], characterized by 1H and 13C NMR, and further confirmed by MS (see Supplementary Materials). The synthesis of the vancomycin derivative Vb was adapted from literature procedure [13,22]. Briefly, vancomycin hydrochloride and the synthesized triazolylthioacetamide b were dissolved in 2 mL dry cosolvent (DMF/DMSO = 1/1) at 0 °C. Then a solution of HATU in DMF was added dropwise, followed by diisopropylethylamine (DIPEA). The reaction mixture was allowed to stir for 20 h at room temperature. The resulting crude product was loaded onto a Sephadex G-25 column to offer the purified Vb as a white powder with a total yield of 18%.
The obtained Vb was analyzed by reverse-phase HPLC using a C18 column (4.6 × 250 mm) and a UV detector (280 nm), and the column was eluted with a gradient of 5–70% acetonitrile containing 0.1% TFA in 30 min at a flow rate of 1 mL/min. The liquid product Vb was first treated with a 0.22 μm filter membrane. The HPLC analysis result for Vb is shown in Figure 1, indicating that the purity of this compound was more than 95%.
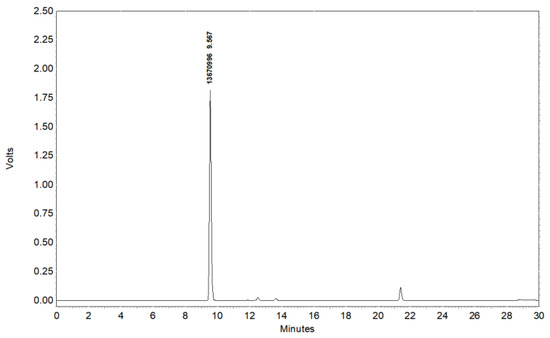
Figure 1.
HPLC analysis of vancomycin derivative Vb.
The purified Vb (white powder) was confirmed by MALDI-TOF MS. As shown in Figure 2, the peak at 1792.14 (m/z: calculated for [M+H]+ = 1792.63) corresponding to Vb was clearly observed, demonstrating that triazolylthioacetamide b was conjugated with the vancomycin successfully.
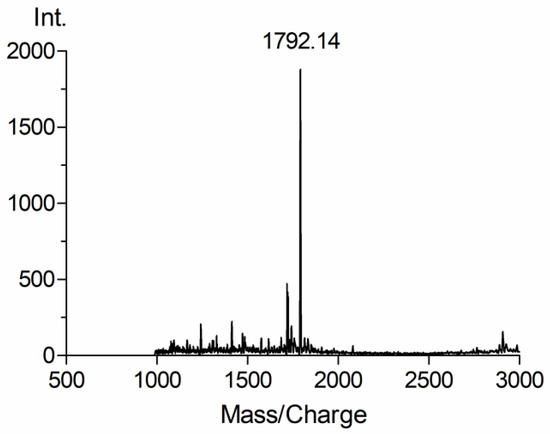
Figure 2.
MALDI-TOF mass spectrum of vancomycin derivative Vb.
The antibacterial activities of vancomycin and Vb were evaluated in vitro by determining the minimum inhibitory concentrations (MICs) according to the Clinical and Laboratory Standards Institute (CLSI) broth micro-dilution method [29]. The employed resistant Gram-positive pathogens were S. aureus, MRSA and VRE. Resistant Gram-negative bacteria were K. pneumoniae, the clinical isolates E. coli producing New Delhi metallo-β-lactamases (NDMs), including E. coli 08 (EC08), E. coli 10 (EC10) and E. coli BL21 (DE3) producing MβL ImiS or MβL CcrA. The collected MIC data are summarized in Table 1.

Table 1.
Antibacterial activities (MICs, μg/mL) of vancomycin and vancomycin derivative Vb against the resistant Gram-positive and Gram-negative strain at the diluted concentrations from 4 to 512 μg/mL.
The collected MIC data indicated that Vb had effective antibacterial activity against S. aureus and MRSA, which was similar to the parent vancomycin molecule. However, the low antimicrobial activities against Gram-negative bacteria and MβL-producing resistant strains were also observed. Next, we assessed the synergistic effects of Vb with three β-lactam antibiotics (cefazolin, meropenem and penicillin G) against the above eight resistant strains. The MIC values for Gram-positive S. aureus, MRSA, VRE and Gram-negative K. pneumoniae are listed in Table 2, and for four MβLs-producing bacteria are listed in Table 3.

Table 2.
Antibacterial activities (MICs, μg/mL) of vancomycin derivative Vb synergizing with β-lactam antibiotics against the resistant strains at a dose in the range of 1–32 μg/mL.

Table 3.
Antibacterial activities (MICs, μg/mL) of vancomycin derivative Vb synergizing with β-lactam antibiotics against resistant E. coli producing MβLs (NDMs, ImiS and CcrA) at a dose of 8, 16, and 32 μg/mL.
The MIC data indicated that Vb gradually increased the antimicrobial effect of all tested β-lactams with an increasing dose and exhibited a dose-dependent synergistic antibacterial effect against the eight resistant strains. The highest dose of Vb (4 μg/mL) resulted in a maximum 128-fold MIC decrease in the antibiotics against S. aureus and MRSA, respectively, and a dose of 16 μg/mL Vb resulted in a 128-fold MIC decrease in cefazolin against VRE. Vb also increased the antimicrobial effect of all tested β-lactams against K. pneumoniae, resulting in a 16–32-fold reduction in MICs. Importantly, as shown in Table 3, Vb increased the antimicrobial effect of meropenem against the clinical isolates E. coli 08 and E. coli 10, resulting in a 4-fold reduction in MIC value at a dose up to 32 μg/mL. Furthermore, Vb resulted in an 8-fold MIC decrease in the antibiotics against resistant E. coli producing ImiS and CcrA at a dose of 32 μg/mL.
The potent synergistic antibacterial activity of Vb was verified by time-kill curves (see Supplementary Materials) against S. aureus and K. pneumonia as shown in Figure 3. Figure 3A shows that the population of S. aureus decreased after treatment with Vb alone for 12 h, indicating that Vb has bactericidal activity against S. aureus. Furthermore, compared with cefazolin treatment alone, the synergistic therapy of cefazolin with Vb resulted in a significant reduction in the population of S. aureus.
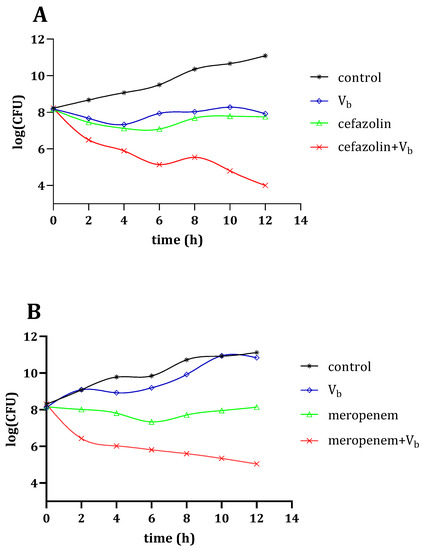
Figure 3.
Time-kill kinetic analysis of Vb-, antibiotics- and synergetic therapy-treated S. aureus (A) and K. pneumonia (B) for 12 h.
Furthermore, Figure 3B shows that the population of K. pneumonia at the exponential phase is significantly reduced upon exposure to the synergistic therapy of meropenem with Vb for 12 h. Indeed, the time-kill curves against S. aureus and K. pneumonia confirmed the synergistic antibacterial effect of Vb with β-lactam antibiotics.
3. Conclusions
A novel vancomycin derivative Vb was constructed by conjugation of the triazolylthioacetamide b and vancomycin molecule, characterized by reverse-phase HPLC and confirmed by MALDI-TOF MS. The biological assays showed that Vb had effective antibacterial activity against S. aureus and MRSA, but low antimicrobial activities against Gram-negative K. pneumoniae. Moreover, Vb gradually increased the antimicrobial effect of three β-lactam antibiotics tested (cefazolin, meropenem and penicillin G), exhibited a dose-dependent synergistic antibacterial effect against the eight resistant strains, and the synergistic effect of Vb and β-lactam antibiotics against S. aureus and K. pneumonia was confirmed by the time-kill curves determination. Most importantly, Vb increased the antimicrobial effect of meropenem against the clinical isolates EC08 and EC10 and E. coli producing ImiS and CcrA, resulting in a 4- and 8-fold reduction in MIC value at a dose up to 32 μg/mL. This work offers a promising scaffold for the development of MβLs inhibitors, specifically antimicrobials for clinically drug-resistant isolates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27227685/s1, Materials and instruments; Synthesis of vancomycin derivative Vb; Antibacterial activity assay in vitro; Time-kill kinetic analysis. All four Supplementary Materials references have been appeared in the maintext’s reference list: the first ref is in line with Ref. [27]; second ref is in line with Ref. [13]; third ref is in line with Ref. [22]; The last is in line with Ref. [29].
Author Contributions
Conceptualization, L.Z. and K.-W.Y.; methodology, L.Z. and K.-W.Y.; software, L.Z., J.X., Y.-L.Z. and K.-W.Y.; validation, L.Z. and K.-W.Y.; formal analysis, L.Z. and K.-W.Y.; investigation, Y.L., Y.J., L.-Y.K., Y.S. and Y.-X.W.; resources, L.Z. and K.-W.Y.; data curation, L.Z. and K.-W.Y.; writing—original draft preparation, Y.L.; writing—review and editing, L.Z. and K.-W.Y.; supervision, K.-W.Y.; project administration, K.-W.Y.; funding acquisition, K.-W.Y., J.X., L.Z. and Y.-L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the grant (22077100, to K.-W. Yang) from the National Natural Science Foundation of China and Innovation Capability Support Program of Shaanxi (No. 2022TD-63, to J. Xiao), and the grant from the Shaanxi Education Commission (17JS007, to L. Zhai). The authors also thank for the Youth Talent Promotion Program of Shaanxi University Association for Science and Technology (20200208, to Y.-L. Zhang).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Guan, D.; Chen, F.; Qiu, Y.; Jiang, B.; Gong, L.; Lan, L.; Huang, W. Sulfonium, an underestimated moiety for structural modification, alters antibacterial profile of vancomycin against multidrug-resistant bacteria. Angew. Chem. 2019, 131, 6750–6754. [Google Scholar] [CrossRef]
- Brogan, D.M.; Mossialos, E. A critical analysis of the review on antimicrobial resistance report and the infectious disease financing facility. Glob. Health 2016, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nature 2017, 543, 15. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Hansford, K.A.; Butler, M.S.; Jia, Z.G.; Mark, A.E.; Cooper, M.A. New developments in glycopeptide antibiotics. ACS Infect. Dis. 2018, 4, 715–735. [Google Scholar] [CrossRef] [PubMed]
- Antonoplis, A.; Zang, X.; Wegner, T.; Wender, P.A.; Cegelski, L. Vancomycin-Arginine Conjugate Inhibits Growth of Carbapenem-Resistant E. coli and Targets Cell-Wall Synthesis. ACS Chem. Biol. 2019, 14, 2065–2070. [Google Scholar] [CrossRef]
- King, D.T.; Strynadka, N.C. Targeting metallo-β-lactamase enzymes in antibiotic resistance. Future Med. Chem. 2013, 5, 1243–1263. [Google Scholar] [CrossRef]
- Bahr, G.; González, L.J.; Vila, A.J. Metallo-β-lactamases in the age of multidrug resistance: From structure and mechanism to evolution, dissemination, and inhibitor design. Chem. Rev. 2021, 121, 7957–8094. [Google Scholar] [CrossRef]
- Kahne, D.; Leimkuhler, C.; Lu, W.; Walsh, C. Glycopeptide and Lipoglycopeptide Antibiotics. Chem. Rev. 2005, 105, 425–448. [Google Scholar] [CrossRef]
- Hubbard, B.K.; Walsh, C.T. Vancomycin Assembly: Nature’s Way. Angew. Chem. Int. Ed. 2003, 42, 730–765. [Google Scholar] [CrossRef]
- Walsh, C.T.; Fisher, S.L.; Park, I.S.; Prahalad, M.; Wu, Z. Bacterial resistance to vancomycin: Five genes and one missing hydrogen bond tell the story. Cell Chem. Biol. 1996, 3, 21–28. [Google Scholar] [CrossRef]
- Pootoolal, J.; Neu, J.; Wright, G.D. Glycopeptide antibiotic resistance. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 381–408. [Google Scholar] [CrossRef]
- Mccomas, C.C.; Crowley, B.M.; Boger, D.L. Partitioning the Loss in Vancomycin Binding Affinity for d-Ala-d-Lac into Lost H-Bond and Repulsive Lone Pair Contributions. J. Am. Chem. Soc. 2003, 125, 9314–9315. [Google Scholar] [CrossRef]
- Yarlagadda, V.; Sarkar, P.; Samaddar, S.; Haldar, J. A Vancomycin Derivative with a Pyrophosphate-Binding Group: A Strategy to Combat Vancomycin-Resistant Bacteria. Angew. Chem. Int. Ed. 2016, 55, 7836–7840. [Google Scholar] [CrossRef]
- Li, L.; Xu, B. Multivalent vancomycins and related antibiotics against infectious diseases. Curr. Pharm. Des. 2005, 11, 3111–3124. [Google Scholar] [CrossRef]
- Fan, C.; Moews, P.C.; Walsh, C.T.; Knox, J.R. Vancomycin resistance: Structure of D-alanine:D-alanine ligase at 2.3 A resolution. Science 1994, 266, 439–443. [Google Scholar] [CrossRef]
- Taubes, G. The bacteria fight back. Science 2008, 321, 356–361. [Google Scholar] [CrossRef]
- Butler, M.S.; Hansford, K.A.; Blaskovich, M.A.T.; Halai, R.; Cooper, M.A. Glycopeptide antibiotics: Back to the future. J. Antibiot. 2014, 67, 631–644. [Google Scholar] [CrossRef]
- Yarlagadda, V.; Konai, M.M.; Manjunath, G.B.; Ghosh, C.; Haldar, J. Tackling vancomycin-resistant bacteria with ‘lipophilic–vancomycin–carbohydrate conjugates’. J. Antibiot. 2014, 68, 302–312. [Google Scholar] [CrossRef]
- Mu, Y.Q.; Nodwell, M.; Pace, J.L.; Shaw, J.P.; Judice, J.K. Vancomycin disulfide derivatives as antibacterial agents. Cheminform 2004, 14, 735–738. [Google Scholar]
- Okano, A.; Isley, N.A.; Boger, D.L. Peripheral modifications of [Ψ[CH2NH]Tpg4]vancomycin with added synergistic mechanisms of action provide durable and potent antibiotics. Proc. Natl. Acad. Sci. USA 2017, 114, E5052–E5061. [Google Scholar] [CrossRef]
- Okano, A.; Nakayama, A.; Schammel, A.W.; Boger, D.L. Total synthesis of [Ψ[C(=NH)NH]Tpg4]vancomycin and its (4-chlorobiphenyl)methyl derivative: Impact of peripheral modifications on vancomycin analogues redesigned for dual D-Ala-D-Ala and D-Ala-D-Lac binding. J. Am. Chem. Soc. 2014, 136, 13522–13525. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Yang, K.W. Porphyrin-vancomycin: A highly promising conjugate for the identification and photodynamic inactivation of antibiotic resistant Gram-positive pathogens. Dyes and Pigments 2015, 120, 228–238. [Google Scholar] [CrossRef]
- Yarlagadda, V.; Akkapeddi, P.; Manjunath, G.B.; Haldar, J. Membrane Active Vancomycin Analogues: A Strategy to Combat Bacterial Resistance. J. Med. Chem. 2014, 57, 4558–4568. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Hansford, K.A.; Gong, Y.; Butler, M.S.; Muldoon, C.; Huang, J.X.; Ramu, S.; Silva, A.B.; Cheng, M.; Kavanagh, A.M.; et al. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 2018, 9, 22. [Google Scholar] [CrossRef]
- Cooper, R.D.G.; Snyder, N.J.; Zweifel, M.J.; Staszak, M.A.; Wilkie, S.C.; Nicas, T.I.; Mullen, D.L.; Butler, T.F.; Roderguez, M.J.; Huff, B.E.; et al. Reductive Alkylation of Glycopeptide Antibiotics: Synthesis and Antibacterial Activity. J. Antibiot. 1996, 49, 575–581. [Google Scholar] [CrossRef]
- Yarlagadda, V.; Sarkar, P.; Samaddar, S.; Manjunath, G.B.; Mitra, S.D.; Paramanandham, K.; Shome, B.R.; Haldar, J. Vancomycin Analogue Restores Meropenem Activity against NDM-1 Gram-negative Pathogens. ACS Infect. Dis. 2018, 4, 1093–1101. [Google Scholar] [CrossRef]
- Yang, S.K.; Kang, J.S.; Oelschlaeger, P.; Yang, K.W. Azolylthioacetamide: A Highly Promising Scaffold for the Development of Metallo-β-lactamase Inhibitors. ACS Med. Chem. Lett. 2015, 6, 455–460. [Google Scholar] [CrossRef]
- Zhai, L.; Zhang, Y.L.; Kang, J.S.; Oelschlaeger, P.; Xiao, L.; Nie, S.S.; Yang, K.W. Triazolylthioacetamide: A Valid Scaffold for the Development of New Delhi Metallo-β-Lactmase-1 (NDM-1) Inhibitors. ACS Med. Chem. Lett. 2016, 7, 413–417. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI standard M07; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2018. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).