Abstract
To better guide microbial risk management and control, growth kinetic models of Salmonella with the coexistence of two other dominant background bacteria in pork were constructed. Sterilized pork cutlets were inoculated with a cocktail of Salmonella Derby (S. Derby), Pseudomonas aeruginosa (P. aeruginosa), and Escherichia coli (E. coli), and incubated at various temperatures (4–37 °C). The predictive growth models were developed based on the observed growth data. By comparing R2 of primary models, Baranyi models were preferred to fit the growth curves of S. Derby and P. aeruginosa, while the Huang model was preferred for E. coli (all R2 ≥ 0.997). The secondary Ratkowsky square root model can well describe the relationship between temperature and μmax (all R2 ≥ 0.97) or Lag (all R2 ≥ 0.98). Growth models were validated by the actual test values, with Bf and Af close to 1, and MSE around 0.001. The time for S. Derby to reach a pathogenic dose (105 CFU/g) at each temperature in pork was predicted accordingly and found to be earlier than the time when the pork began to be judged nearly fresh according to the sensory indicators. Therefore, the predictive microbiology model can be applied to more accurately predict the shelf life of pork to secure its quality and safety.
1. Introduction
Fresh pork is currently the most important meat product consumed by Chinese residents, which is rich in nutrients, but also a good medium for microbial growth during production and transportation. The continuous growth of microorganisms at a suitable temperature will eventually lead to meat spoilage or even foodborne disease [1]. Salmonella, as the main foodborne pathogen in pork [2,3], may cause infection due to a short cooking time and cross-contamination during treatment in the kitchen. Therefore, it is very important to control microbial contamination and continuous reproduction in pork to ensure the quality and safety of products.
The behavior of microorganisms is an important factor affecting the quality and safety of meat, and temperature is the most important factor that allows for microorganisms to continue to grow and even secrete harmful substances [1]. Traditional microbial detection with certain lag results is time-consuming and labor-intensive, and it cannot predict subsequent bacterial contamination [4]. On the other hand, predictive microbiology is computer-based, describing the growth, survival, and death of microorganisms under specific environmental conditions, and establishing mathematical models to provide a basis for risk assessment [5,6,7,8]. Using it can quickly predict the changes in pathogenic or spoilage bacteria in food according to the primary contaminated quantity without the traditional microbial detection, which realizes intelligent monitoring from raw materials to production to contribute to microbial risk management in food [9,10]. Therefore, predictive microbiology studies on pork are important.
In regard to the predictive microbiological model in pork, previous domestic studies mostly established predictive models for single spoilage bacteria or pathogenic bacteria [11,12,13]. However, in pork, there are often several types of dominant bacteria that contaminate a certain quantity and continue to proliferate at the same time. The specific spoilage bacteria, Pseudomonas, are the dominant background bacteria in fresh pork [14], and Escherichia coli (E. coli) is also a type of major background bacterium [15,16]. During the post-production process, there will be a certain competitive effect among these bacteria to influence each other’s growth. Recently, some microbiological models related to more than one bacterium have been reported, for example, Pseudomonas and Listeria in beef [17], Salmonella and background flora in chicken [8], and Listeria and natural flora in minced tuna [18]. Nevertheless, in pork, there are no reported studies on competitive predictive microbial models of Salmonella with the appearance of dominant background bacteria, and there is no relevant research on using the microbial model to predict product safety.
In this study, the growth kinetic models of Salmonella with the appearance of dominant background bacteria in pork at different temperatures were established, to be used for the prediction of the shelf life of pork, which would provide scientific guidance for pork transportation, storage, and safety monitoring.
2. Results
2.1. Optimal Primary Growth Kinetic Model of Salmonella and Two Background Bacteria in Fresh Pork
According to the experimental observation data of Salmonella Derby (S. Derby), Pseudomonas aeruginosa (P. aeruginosa), and E. coli in fresh pork during storage at 4~37 °C, the primary growth kinetic models of these three bacteria at different temperatures were obtained by data fitting using modified Gompertz, Baranyi, and Logistic equations, respectively, and the coefficient of determination R2 of each fitting growth curve is listed in Table 1. Since the bacterial quantity did not change significantly compared with the initial quantity after storage at 4 °C for 10 d, the curve could not be fitted. The Baranyi model could best fit the growth curves of S. Derby and P. aeruginosa in fresh pork at 37, 30, 22, 16, and 10 °C (Figure 1), and all the values of the coefficient of determination (R2) at different temperatures were higher than the modified Gompertz and Logistic models. The Huang model, with higher R2 values, was better than the other two when fitting the growth curves of E. coli in pork at different temperatures (Figure 1). Therefore, the Baranyi model was chosen as the optimal primary growth model for S. Derby and P. aeruginosa, and the Huang model was chosen as the optimal model for E. coli in this study. The kinetic parameters, Nmax, μmax, and Lag, of each optimal bacterial primary growth model under five temperatures, as well as the observed initial bacterial quantity (N0), are listed in Table 2. With the increase in temperature, the maximum growth rates of the three bacteria in pork all increased, while the lag period decreased.

Table 1.
Decisive coefficient (R2) of fitting curve of each model.
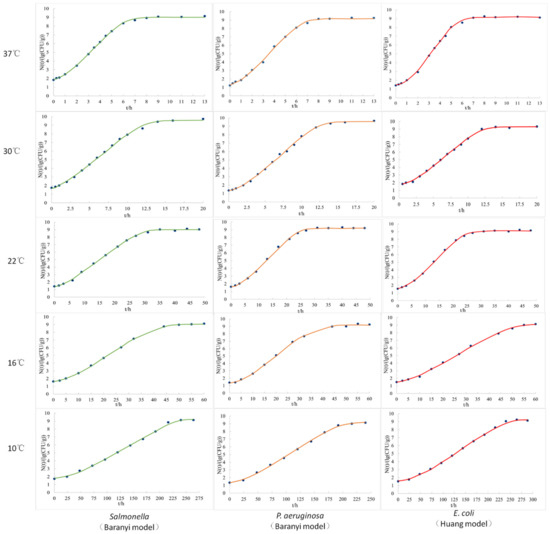
Figure 1.
Growth curves of S. Derby, P. aeruginosa, and E. coli in fresh pork at different temperatures fitted by the corresponding optimal predictive model.

Table 2.
Kinetic parameters of each bacterial optimal primary growth model in pork at different temperatures.
2.2. Secondary Growth Model of S. Derby, P. aeruginosa, and E. coli in Fresh Pork
According to the primary model, the parameters μmax and Lag of S. Derby, P. aeruginosa, and E. coli at different temperatures were separately fitted using the square root model to reflect the relationship between temperature with maximum growth rate and lag phase (Figure 2). The related expression equation of each secondary growth model is also shown in Figure 2. From the results, it was found that the square root of μmax and 1/Lag of each bacterium presented a nice linear relationship with temperature, with most R2 values above 0.98, indicating higher model fitting degrees.
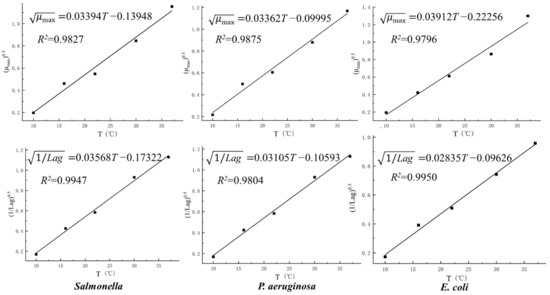
Figure 2.
Secondary square root models of S. Derby, P. aeruginosa, and E. coli in pork.
2.3. Validation of the Bacterial Growth Models
Bias factor Bf and accuracy factor Af are often used as effective tools to evaluate model reliability [19]. A model is considered better and is acceptable when Bf is in the range of 0.90~1.05 and Af is in the range of 1.01~1.15. The closer Af and Bf are to 1, the more reliable the model [20]. The smaller the MSE value, the smaller the variability of the data and the higher the accuracy of the model. Therefore, Bf, Af, and MSE were used to evaluate the constructed secondary model. From the data in Table 3, it was found that the Bf and Af of the secondary model fitted by the temperature with μmax and Lag of the three bacteria are all close to 1, and all the values of MSE are very small, which indicates that the square root model can be used to predict the relationship between temperature and μmax and Lag with high reliability.

Table 3.
Various statistical characteristics of the secondary growth models.
2.4. Pork Safety Prediction by Applying the Constructed Growth Models
The simulated mixed-contamination pork stored at different temperatures was dynamically observed and scored according to the sensory indicators, and the earliest time to reach the corresponding level standard was recorded. It was found that the higher the temperature, the shorter the time for pork from fresh to spoilage (Table 4). At the same time, the time required for S. Derby to reach the unsafe bacterial dose was calculated using the derived shelf life model, and was compared with the sensory change time. The calculated time data at each temperature were found to be relatively close to the elapsed time when the sensory score reached sub-fresh, and earlier than the elapsed time when the sensory score reached nearly fresh. This indicated that the content of pathogenic bacteria in pork might already exceed the safe dose even if the color and smell of the pork are basically normal. Therefore, determining the shelf life of food according to the predicted microbiology could more scientifically and accurately ensure the consumption safety of the product.

Table 4.
Sensory changes in simulated contaminated pork at different temperatures and prediction of shelf life based on S. Derby amount.
3. Discussion
The contamination of pathogenic bacteria and spoilage bacteria affects the quality and safety of pork products [1]. Predictive microbiology provides a more scientific approach to product risk and shelf-life prediction. There are few reports on growth predictive models of multiple bacteria in fresh pork. In this experiment, the growth curves of three bacteria in fresh pork were studied at a common temperature (4~37 °C) during storage and transportation, and the optimal models were selected to simulate their growth, which can dynamically predict the spoilage extent of pork. This study could provide guidance to ensure hygiene throughout the pork industrial chain and to determine pork shelf life more scientifically.
There are many studies and applications of predictive microbiology in food. The research on fresh livestock and poultry products is mostly concentrated on bacteria in poultry or cattle products, and there are also predictive microbiology studies on mixed bacterial contamination [8,17,21,22,23,24]. However, current predictive microbial studies on pork mainly target a single pathogen [25,26]. Given that the microorganisms in swine are exposed during the whole chain from breeding and slaughtering to sale and consumption cannot be single, it is more meaningful to study mixed predictive microbiology in pork. Here, S. Derby, E. coli, and P. aeruginosa were selected to analogously contaminate pork in a laboratory, and the predicted growth kinetics of the three bacteria at 4~37 °C were studied. Although the constructed model was not an integrated competitive growth model, the growth process of the three bacteria in pork was in a competitive mode. Therefore, the model presents a growth curve under a competitive mode.
Although different nonlinear equations, such as modified Gompertz, Baranyi, and Huang models, were used to predict microbial growth, it was not clear which model was inherently better than the other [27]. The determination of the optimal model is inseparable from the culture medium, experimental conditions, bacterial species, and other factors. For a complex meat system, the optimal model, rather than a single model, should be selected to predict the bacterial growth under different temperatures [28]. In this study, Baranyi models could better simulate the growth of both S. Derby and P. aeruginosa at different temperatures, which is consistent with previous research findings, while the optimal model of E. coli is the Huang model. There are recent studies that use the Huang model to fit the growth of E. coli in beef and chicken [29,30], but there is no study using the Huang model to fit the growth of E. coli in fresh pork. Therefore, the results of this study broaden the application of this model in the field of microbiology to a certain extent. Although a mixed inoculation of three bacterial strains were applied in this study, there was no clear competitive inhibition among strains, and all three strains showed an S-shaped growth curve, which may be related to the relatively similar amount of initial inoculum.
According to the growth kinetic parameters obtained by the optimal primary models of the three bacteria in pork, with the increase in temperature, the μmax value increased, while the Lag value decreased, indicating that temperature has a significant effect on bacterial growth in pork. Compared with the predictive growth models of related bacteria in pork in previous studies [31,32], the model obtained through mixed inoculation in this study showed a higher μmax value and lower Lag value, which may be due to the quorum sensing between the inoculated mixed strains. Quorum sensing can cause bacteria to mutually promote growth and shorten the time taken to adapt to external conditions [33,34]. In fact, differences in inoculation method and amount, bacterial characteristics, meat types, etc. during experiments would lead to differences in μmax and Lag.
In addition, it is worth noting that in this study, under the condition of 10 °C, pork could be stored for 192 h and was still nearly fresh. The reason should be that the initial bacterial inoculation amount was quite low (100~300 CFU), and the microbial lag period in pork was longer; therefore, the quorum sensing could not be formed quickly, which meant that the bacteria could not reproduce quickly. In actual production, the total number of bacterial colonies in pork sometimes exceeds 1000 CFU; therefore, the pork may deteriorate within 1 week of storage under refrigeration (4~10 °C).
In summary, the predictive growth kinetic models of Salmonella, Pseudomonas, and E. coli inoculated simultaneously in fresh pork at different temperatures were each constructed and validated, and the shelf life of pork for safe consumption predicted based on the predicted growth model was more secure when compared with the conventional sensory evaluation indicators of pork.
4. Materials and Methods
4.1. Bacterial Cultures and Inoculum Preparation
S. Derby is the most dominant pathogen in pork in Shandong province, P. aeruginosa is one of the dominant spoilage bacteria [35], and E. coli is one of the most common background bacteria. Therefore, we chose these three bacteria to study their predictive growth models in pork at different temperatures. S. Derby, P. aeruginosa, and E. coli were inoculated onto LB agar (Beijing Land Bridge Technology, Beijing, China) and cultured at 37 °C for 16 h. One typical single colony of each strain was selected and transferred into 5 mL of LB liquid medium (Beijing Land Bridge Technology, Beijing, China), and cultured at 37 °C with shaking (200 r/min) for 16 h. Then, three types of bacterial culture were centrifuged at 3000 r/min at 4 °C for 10 min, the supernatant was discarded, sterile saline was added to resuspend the bacteria, and then gradient dilution was carried out to about 102 CFU/mL of final concentration. An equal volume of each bacterial solution was mixed to serve as the bacterial inoculum.
4.2. Preparation of Pork Sample and Inoculation
Fresh pork purchased from a local supermarket was rinsed with sterile water, the surface was wiped with alcohol cotton in sterile biological safety cabinet, and then the whole pork was rinsed with sterile water again. The pork’s surface layer was removed using a sterile scalpel, the pork was divided into small pieces (about 10 g/piece), and then the pork pieces were placed on aseptic trays to be sterilized again with ultraviolet rays for 25–30 min. The sterile pork piece samples were suspended in the bacterial mixed suspension for 15 s for inoculation to yield an initial inoculum of about 10 CFU/g.
4.3. Growth Study and Bacterial Counting
Each piece of inoculated pork sample was placed in one sterile homogenizing bag, and cultured in a constant temperature incubator at 4, 10, 16, 22, 30, and 37 °C, respectively. Before culture, one simulated contaminated pork piece of each temperature was randomly selected to carry out bacterial counting to determine the initial amount of contamination of these three bacteria. At the same time, bacterial counting was conducted for the control pieces without inoculation to ensure no background bacterial contamination. No background bacteria were detected only in the control pieces, and the experiments were continued.
Under each temperature, one sample was taken out in a timely manner and added to 90 mL of sterile normal saline, beaten at a speed of 7 times per second for 2 min. The time points at 37 °C were set as 0, 0.25, 0.5, 1, 1.5, 2, 3, 4, 5, 6, 7, 8, 9, 11, and 13 h; the time points at 30 °C were set as 0, 0.5, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 14, 16, and 20 h; the time points at 22 °C were set as 0, 1.5, 3, 6, 9, 13, 17, 21, 24, 27, 31, 35, 40, 44, and 48 h; the time points at 16 °C were set as 0, 2.5, 5, 10, 15, 20, 27, 32, 44, 50, 55, and 60 h; the time points at 10 °C were set as 0, 24, 48, 72, 96, 120, 14, 168, 192, 216, and 240 h. Then, the liquid portion of each sample was properly diluted, and 100 μL of aliquots of suitable dilution gradients was plated on Salmonella chromogenic medium (CHROMagar™, Paris, France), MacConkey medium (Beijing Land Bridge Technology, Beijing, China), as well as CFC Pseudomonas selective medium (Beijing Land Bridge Technology, Beijing, China) one-by-one. The plates were incubated at 37 °C for 24 h. The bacterial colonies with corresponding colors on each kind of agar plate were counted and converted to log CFU/g of sample. Each experiment was carried out in triplicate.
4.4. Sensory Evaluation of Pork Freshness
The freshness of pork was evaluated based on its color, texture, and smell [36]. The specific evaluation grades and criteria of sensory indicators are shown in Table 5.

Table 5.
Sensory evaluation criteria.
4.5. Establishment of the Primary Bacterial Growth Model
The growth data of the three bacteria obtained in the experiment were fitted by the modified Gompertz model in Origin 2019 software (Equation (1)) [37], by the Huang model in IPMP2013 software (Equations (2) and (3)), and by the Baranyi model in the online Combase database (Equations (4) and (5)) [14,38]. The coefficient of determination (R2) of the three different models was compared, and the best model was selected.
Modified Gompertz model:
where t is the bacterial growth time (h); Nt is the corresponding bacterial concentration at t (log CFU/g); Nmax and N0 are the maximum and initial bacterial concentration (log CFU/g), respectively; μmax is the microbial maximum growth rate (h−1); and Lag is the bacterial growth delay time (h).
Nt = N0 + (Nmax − N0) × exp{−exp [2.718μmax/(Nmax − N0) × (Lag − t)] + 1}
Huang model:
where t is the bacterial growth time (h); Nt is the corresponding bacterial concentration at t (log CFU/g); Nmax and N0 are the maximum and initial bacterial concentration (log CFU/g), respectively; μmax is the microbial maximum growth rate (h−1); Lag is the bacterial growth delay time (h); and α is a constant of 4, which defines the transition from delayed lag phase to logarithmic phase during bacterial growth.
Nt = N0 + Nmax − ln[eN0 + (eNmax − eN0)e−μmax Bt]
Baranyi model:
In the above five equations, t is the bacterial growth time (h); Nt is the corresponding bacterial concentration at t (log CFU/g); Nmax and N0 are the maximum and initial bacterial concentration (log CFU/g), respectively; μmax is the microbial maximum growth rate (h−1); Lag is the bacterial growth delay time (h); and α in Equation (3) is a constant of 4, which defines the transition from delayed lag phase to logarithmic phase during bacterial growth.
4.6. Establishment of the Secondary Bacterial Growth Model
The Ratkowsky square root model can well predict the growth of microorganisms under single-factor condition, which is often used to describe the relationship between temperature, maximum growth rate, and lag period [39,40]. The relationship between temperature and Lag and μmax was fitted using Origin software. The expressions’ square root equation of maximum growth rate and lag period are as in Equations (6) and (7):
where T is the incubation temperature (°C); Tmin is the minimum growth temperature, namely, the temperature at which the microorganism has no metabolic activity, with a maximum growth rate of 0; and b is a constant of the equation.
4.7. Validation of the Bacterial Growth Model
The primary bacterial growth models were verified using decisive coefficient (R2), which was directly obtained from data fitting results by the modified Gompertz model in Origin 2019, the Huang model in IPMP2013, and the Baranyi model in the online Combase database. The secondary bacterial growth models were verified using R2, mean square error (MSE, Equation (8)), bias factor (Bf, Equation (9)), and accuracy factor (Af, Equation (10)) [19,20,41]:
where predicted represents the predicted value of μmax and Lag, which was calculated from the equation of the secondary model; observed represents the observed value of μmax and Lag, which was obtained from the primary model; and n represents the number of observations (here, n = 5).
4.8. Shelf-Life Prediction of Fresh Pork
The safety of fresh pork was mainly considered in terms of shelf-life prediction in this study. Through the constructed bacterial primary and secondary growth models, the shelf life (SL (h)) of fresh pork was predicted based on the S. Derby growth time required from the initial bacterial quantity (N0) to the minimum pathogenic dose (Ns). The shelf-life prediction equation was derived from the simplified Baranyi model [42] (Equation (11)).
Author Contributions
Conceptualization, G.Z.; methodology, G.Z., H.C., L.W., Y.L. and Y.G.; validation, T.Y.; investigation, J.Z., N.L., X.H. and X.Z.; data curation, G.Z. and T.Y.; writing—original draft preparation, G.Z.; writing—review and editing, J.W. (Jun Wang); supervision, J.L., Y.X. and J.W. (Junwei Wang); project administration, J.W. (Junwei Wang); funding acquisition, J.W. (Junwei Wang). All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (2018YFD0500505).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Koutsoumanis, K.; Taoukis, P.S. Meat Safety, Refrigerated Storage and Transport: Modeling and Management. In Improving the Safety of Fresh Meat; Woodhead Publishing: Sawston, UK, 2015; pp. 505–561. [Google Scholar]
- Liu, J.; Bai, L.; Li, W.; Han, H.; Fu, P.; Ma, X.; Bi, Z.; Yang, X.; Zhang, X.; Zhen, S.; et al. Trends of foodborne diseases in China: Lessons from laboratory-based surveillance since 2011. Front. Med. 2018, 12, 48–57. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Burden of Foodborne Diseases is Substantial. 2015. Available online: https://apps.who.int/iris/bitstream/handle/10665/327488/WHO-FOS-FZD-15.3-eng.pdf?sequence=1 (accessed on 30 July 2022).
- Mcmeekin, T.; Bowman, J.; Mcquestin, O.; Mellefont, L.; Ross, T.; Tamplin, M. The future of predictive microbiology: Strategic research, innovative applications and great expectations. Int. J. Food Microbiol. 2008, 128, 2–9. [Google Scholar] [CrossRef]
- McMeekin, T.A.; Ross, T. Predictive microbiology: Providing a knowledge-based framework for change management. Int. J. Food Microbiol. 2002, 78, 133–153. [Google Scholar] [CrossRef]
- Poschet, F.; Vereecken, K.M.; Geeraerd, A.H.; Nicolai, B.M.; Impe, J.F.V. Analysis of a novel class of predictive microbial growth models and application to coculture growth. Int. J. Food Microbiol. 2005, 100, 107–124. [Google Scholar] [CrossRef]
- Verheyen, D.; Baka, M.; Akkermans, S.; Skra, T.; Impe, J. Effect of microstructure and initial cell conditions on thermal inactivation kinetics and sublethal injury of Listeria monocytogenes in fish-based food model systems. Food Microbiol. 2019, 84, 103267. [Google Scholar] [CrossRef] [PubMed]
- Zhen, J.; Peng, Y.; Yan, X.; Zhang, Z.; Fang, T.; Li, C. One-step kinetic analysis of competitive growth of Salmonella spp. and background flora in ground chicken. Food Control 2020, 117, 107103. [Google Scholar]
- Mcmeekin, T.A.; Olley, J.; Ratkowsky, D.A.; Ross, T. Predictive microbiology: Towards the interface and beyond. Int. J. Food Microbiol. 2002, 73, 395–407. [Google Scholar] [CrossRef]
- McDonald, K.; Sun, D.-W. Predictive food microbiology for the meat industry: A review. Int. J. Food Microbiol. 1999, 52, 1–27. [Google Scholar] [CrossRef]
- Juneja, V.K.; Osoria, M.; Purohit, A.S.; Golden, C.E.; Mishra, A.; Taneja, N.K.; Salazar, J.K.; Thippareddi, H.; Kumar, G.D. Predictive model for growth of Clostridium perfringens during cooling of cooked pork supplemented with sodium chloride and sodium pyrophosphate. Meat Sci. 2021, 180, 108557. [Google Scholar] [CrossRef]
- Lee, Y.J.; Jung, B.S.; Yoon, H.J.; Kim, K.-T.; Paik, H.-D.; Lee, J.-Y. Predictive model for the growth kinetics of Listeria monocytogenes in raw pork meat as a function of temperature. Food Control 2014, 44, 16–21. [Google Scholar] [CrossRef]
- Mansur, A.R.; Park, J.H.; Oh, D.H. Predictive Model for Growth of Staphylococcus aureus on Raw Pork, Ham, and Sausage. J. Food Prot. 2016, 79, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Borch, E.; Kantmuermans, M.L.; Blixt, Y. Bacterial spoilage of meat and cured meat products. Int. J. Food Microbiol. 1996, 33, 103–120. [Google Scholar] [CrossRef]
- Dainty, R.H.; Mackey, B.M. The relationship between the phenotypic properties of bacteria from chill-stored meat and spoilage processes. J. Appl. Microbiol. 2010, 73, 103–114. [Google Scholar] [CrossRef]
- Lebert, I.; Robles-Olvera, V.; Lebert, A. Application of polynomial models to predict growth of mixed cultures of Pseudomonas spp. and Listeria in meat. Int. J. Food Microbiol. 2000, 61, 27–39. [Google Scholar] [CrossRef]
- Koseki, S.; Takizawa, Y.; Miya, S.; Takahashi, H.; Kimura, B. Modeling and predicting the simultaneous growth of Listeria monocytogenes and natural flora in minced tuna. J. Food Prot. 2012, 74, 176–187. [Google Scholar] [CrossRef]
- te Giffel, M.C.; Zwietering, M.H. Validation of predictive models describing the growth of Listeria monocytogenes. Int. J. Food Microbiol. 1999, 46, 135–149. [Google Scholar] [CrossRef]
- Ross, T. Indices for performance evaluation of predictive models in food microbiology. J. Bacteriol. 1996, 81, 501–508. [Google Scholar] [CrossRef]
- Huang, L. Thermal resistance of Listeria monocytogenes and background microbiota in unsalted and 10% salted liquid egg yolk. J. Food Saf. 2019, 39, e12665. [Google Scholar] [CrossRef]
- Huang, L. Dynamic analysis of growth of Salmonella spp. in raw ground beef—Estimation of kinetic parameters, sensitivity analysis, and Markov Chain Monte Carlo simulation. Food Control 2020, 108, 106845. [Google Scholar]
- Hwang, C.-A.; Huang, L. Dynamic analysis of competitive growth of Escherichia coli O157:H7 in raw ground beef. Food Control 2018, 93, 251–259. [Google Scholar] [CrossRef]
- Vereecken, K.M.; Dens, E.J.; Impe, J. Predictive Modeling of Mixed Microbial Populations in Food Products: Evaluation of Two-species Models. J. Theor. Biol. 2000, 205, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Huang, L. Dynamic kinetic analysis of growth of Listeria monocytogenes in a simulated comminuted, non-cured cooked pork product. Food Control 2017, 71, 160–167. [Google Scholar] [CrossRef]
- Zhou, B.; Fan, X.; Song, J.; Wu, J.; Pan, L.; Tu, K.; Peng, J.; Dong, Q.; Xu, J.; Wu, J. Growth simulation of Pseudomonas fluorescens in pork using hyperspectral imaging. Meat Sci. 2022, 188, 108767. [Google Scholar] [CrossRef] [PubMed]
- López, S.; Prieto, M.; Dijkstra, J.; Dhanoa, M.S.; France, J. Statistical evaluation of mathematical models for microbial growth. Int. J. Food Microbiol. 2004, 96, 289–300. [Google Scholar] [CrossRef]
- Li, M.; Niu, H.; Zhao, G.; Tian, L.; Huang, X.; Zhang, J.; Wei, T.; Zhang, Q. Analysis of mathematical models of Pseudomonas spp. growth in pallet-package pork stored at different temperatures. Meat Sci. 2013, 93, 855–864. [Google Scholar]
- Huang, L.; Tu, S.I.; Phillips, J.; Fratamico, P. Mathematical Modeling of Growth of Non-O157 Shiga Toxin-Producing Escherichia coli in Raw Ground Beef. J. Food Sci. 2012, 77, 217–225. [Google Scholar] [CrossRef]
- Sommers, C.; Huang, C.-Y.; Sheen, L.-Y.; Sheen, S.; Huang, L. Growth modeling of Uropathogenic Escherichia coli in ground chicken meat. Food Control 2017, 86, 397–402. [Google Scholar] [CrossRef]
- Jiang, Y.; Zou, X.; Peng, Z. Construction of Growth Prediction Model of Escherichia coli in Longissimus Dorsi Muscle. Food Sci. 2008, 29, 115–119. [Google Scholar]
- Zhang, L.; Yin, D.; Zhang, D.; Luo, L. Predictive Models for Salmonella in Fresh Pork at Household Temperatures. Food Ind. 2017, 38, 201–205. [Google Scholar]
- Gram, L.; Christensen, A.B.; Ravn, L.; Molin, S.; Givskov, M. Production of Acylated Homoserine Lactones by Psychrotrophic Members of the Enterobacteriaceae Isolated from Foods. Appl. Environ. Microbiol. 1999, 65, 3458–3463. [Google Scholar] [CrossRef] [PubMed]
- Ravn, L.; Christensen, A.B.; Molin, S.; Givskov, M.; Gram, L. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 2001, 44, 239–251. [Google Scholar] [CrossRef]
- Schirmer, B.C.; Heir, E.; Langsrud, S. Characterization of the bacterial spoilage flora in marinated pork products. J. Appl. Microbiol. 2010, 106, 2106–2116. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Fan, Z.F.; Dong, P.; Di, Y.; Li, J.; Gao, Y.; Song, Z.; Wu, Z. Influence of Microbial Contamination on Quality of Chilled Pork during Storage. China Anim. Health Insp. 2018, 35, 29–32. [Google Scholar]
- Zwietering, M.H. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef]
- Huang, L. Optimization of a new mathematical model for bacterial growth. Food Control 2013, 32, 283–288. [Google Scholar] [CrossRef]
- Davey, K.R. Applicability of the Davey (linear Arrhenius) predictive model to the lag phase of microbial growth. J. Appl. Microbiol. 1991, 70, 253–257. [Google Scholar] [CrossRef]
- Ratkowsky, D.A.; Olley, J.; McMeekin, T.A.; Ball, A. Relationship between temperature and growth rate of bacterial cultures. J. Bacteriol. 1982, 149, 1–5. [Google Scholar] [CrossRef]
- Park, H.S.; Sung, B.Y.; Ryu, K. Predictive Model for Growth of Staphylococcus aureus in Blanched Spinach with Seasoning. J. Korean Soc. Appl. Biol. Chem. 2012, 55, 529–533. [Google Scholar] [CrossRef]
- Gao, L.; Weng, S.; Liu, Q.; Zhang, J.; Chen, E.; Bai, Y.; Han, B. Predictive Models for Salmonella Growth in Fresh Pork. Food Res. Dev. 2011, 32, 143–146. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).