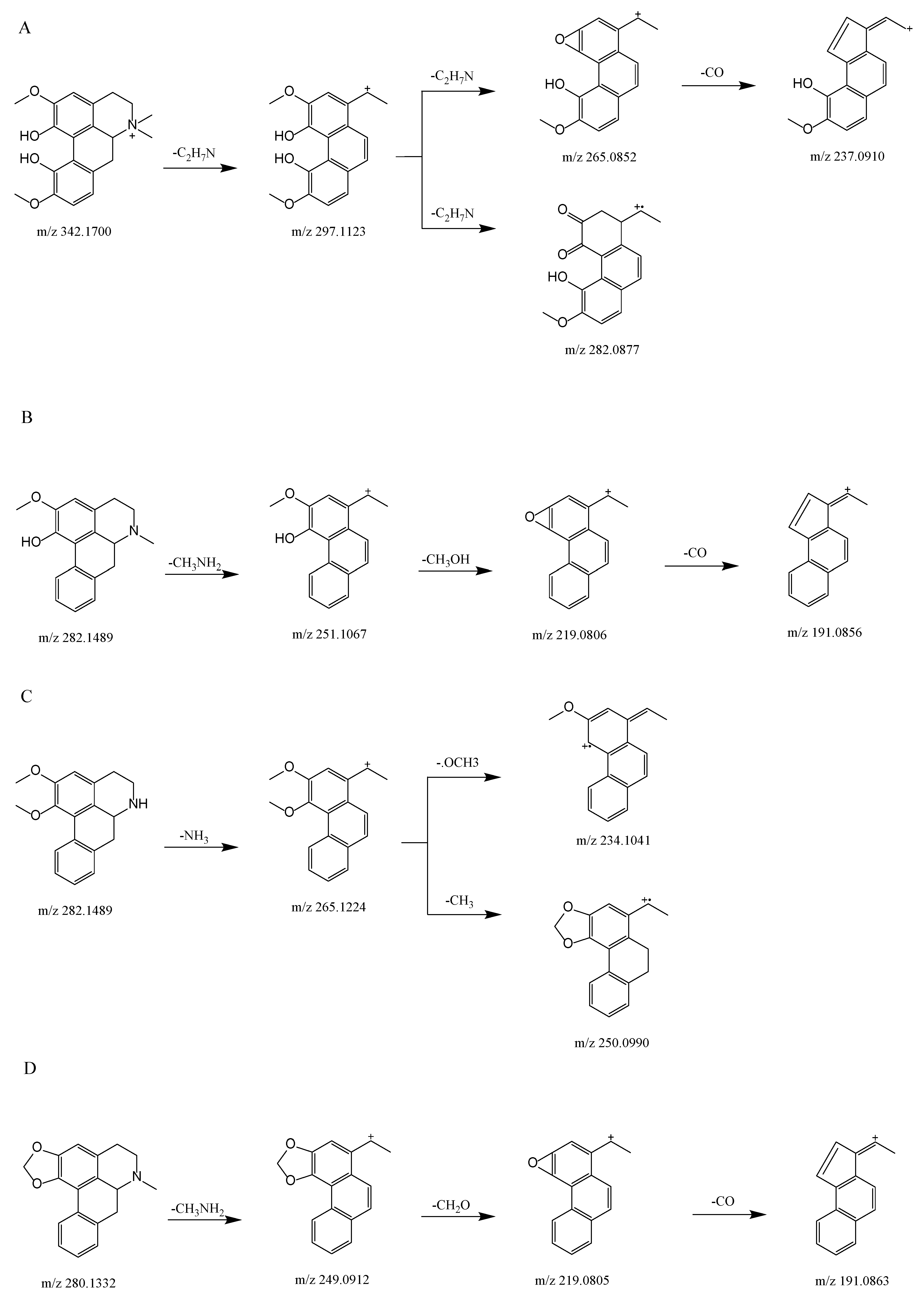Abstract
Sabia schumanniana Diels (SSD) is a plant whose stems are used in traditional folk medicine for the treatment of lumbago and arthralgia. Previous studies have revealed chemical constituents of SSD, including triterpenoids and aporphine alkaloids. Aporphine alkaloids contain a variety of active components, which might facilitate the effective treatment of lumbago and arthralgia. However, only 5-oxoaporphine (fuseine) has been discovered in SSD to date. In this study, we sought to systematically identify the aporphine alkaloids in SSD. We established a fast and reliable method for the detection and identification of these aporphine alkaloids based on ultra-high-performance liquid chromatography (UHPLC)-Q-Exactive-Orbitrap/mass spectrometry combined with parallel reaction monitoring (PRM). We separated all of the analyzed samples using a Thermo Scientific Hypersil GOLD™ aQ C18 column (100 mm × 2.1 mm, 1.9 μm). Finally, we identified a total of 70 compounds by using data such as retention times and diagnostic ions. No fewer than 69 of these SSD aporphine alkaloids have been reported here for the first time. These findings may assist in future studies concerning this plant and will ultimately contribute to the research and development of new drugs.
1. Introduction
Sabia schumanniana Diels (SSD) is a deciduous climbing woody vine of the genus Sabia in the family Sabiaceae and is widely distributed in the Sichuan and Guizhou provinces of China. The stems of SSD are used in traditional folk medicine for the treatment of lumbago and arthralgia [1,2]. The main active constituents in the genus Sabia are alkaloids [3]; however, only triterpenoids and 5- oxoaporphine (fuseine) have been identified in Sabia schumanniana Diels before now [4]. Aporphine alkaloids are natural chemical compounds that are highly biologically active and play an important role in plants. In recent studies, aporphine alkaloids have been shown to exhibit potent anti-diabetic, anti-cancer [5], anti-inflammatory [6], and antivirus properties [7]. Further studies on the aporphine alkaloid components of SSD are therefore warranted.
Ultra-high-performance liquid chromatography-Q-Exactive Orbitrap/mass spectrometry is a process which can be used for chemical constitution identification and offers high selectivity, high sensitivity, and high efficiency [8,9,10]. The fragment information obtained through MS combined with advanced post-processing technology data enables the determination of the diagnostic fragment ions and neutral losses. Typically, sample data acquisition involves a full scan with data-dependent MS2 (full MS/dd-MS2). However, MS2 data cannot be detected in this mode if the relative abundance of MS1 ions does not reach a required level. As a result, any desired compounds that are present only in trace amounts are disregarded because of the limitations of the analytical method. Recently, this tool has been used to conveniently acquire MS2 data using the parallel reaction monitoring (PRM) detection mode, which allows for the isolation of the targeted precursor ions and product fragment ions from the precursor and enables the detection of the resulting product ions based on the preset isolation window width and collision energy, eliminating most interference. By such means, researchers have achieved the accurate detection and quantification of confirmed and targeted fragments [11,12].
In this study, we systematically characterized SSD constituents using UHPLC-Q-Exactive Orbitrap MS combined with PRM. We putatively identified 70 aporphine alkaloids based on their precise mass measurement, chromatographic retention, MSn spectra analysis, and bibliographical data. No fewer than 69 of these aporphine alkaloids were identified in SSD for the first time in this study. These results may contribute to a better understanding of the medicinal effects of SSD and help to lay the groundwork for the future quality control of SSD-derived medicines in a clinical setting.
2. Results and Discussion
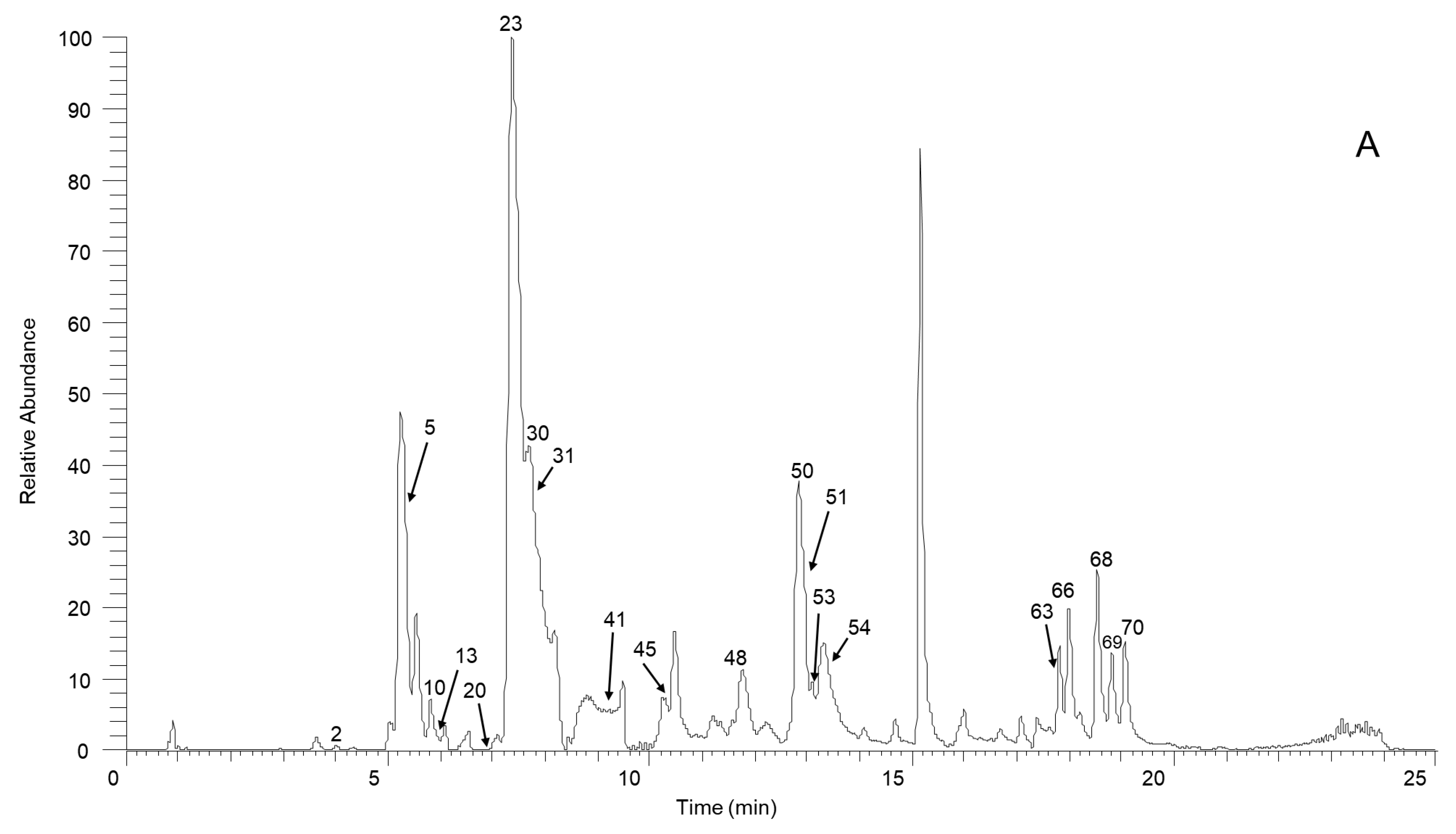
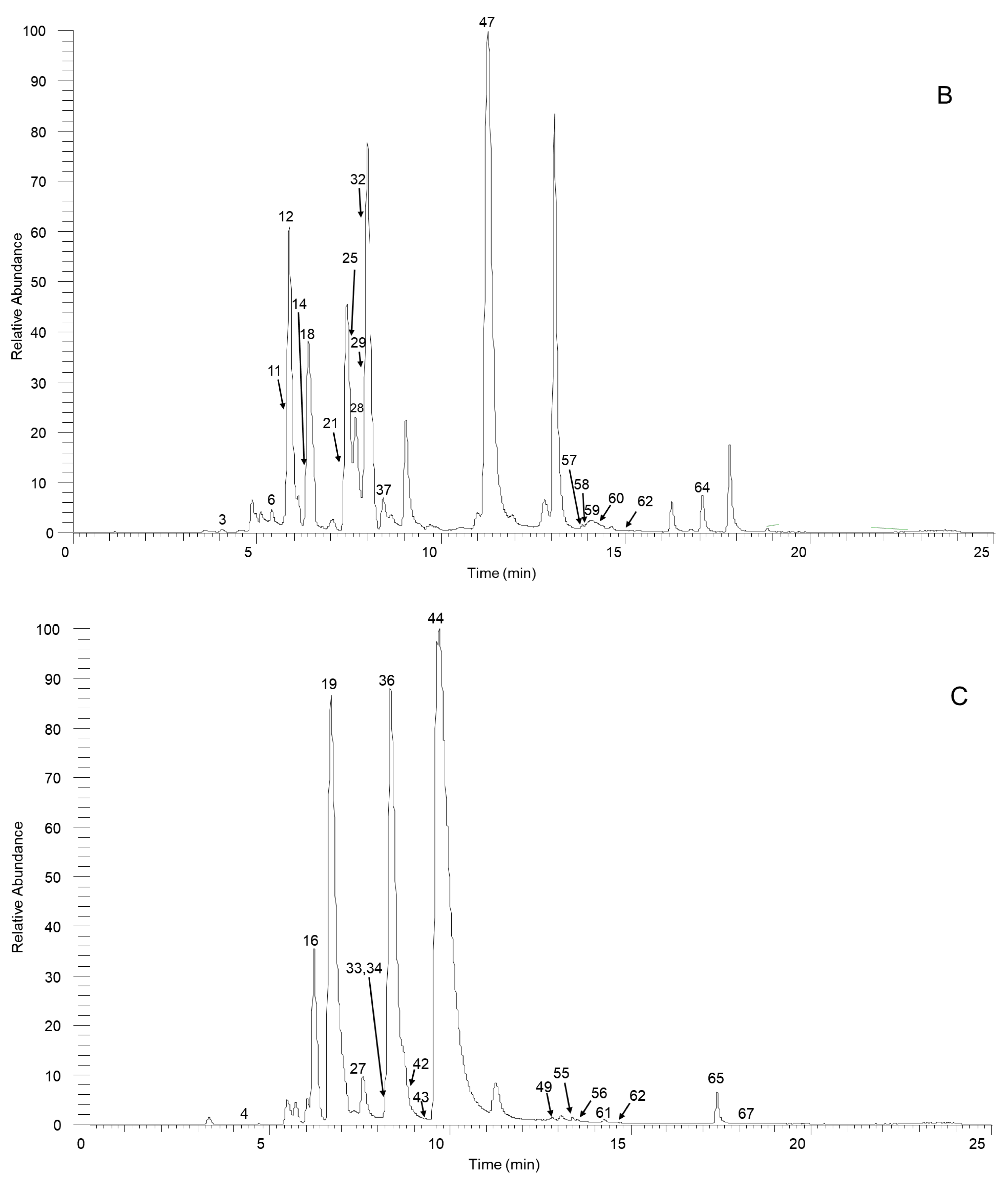
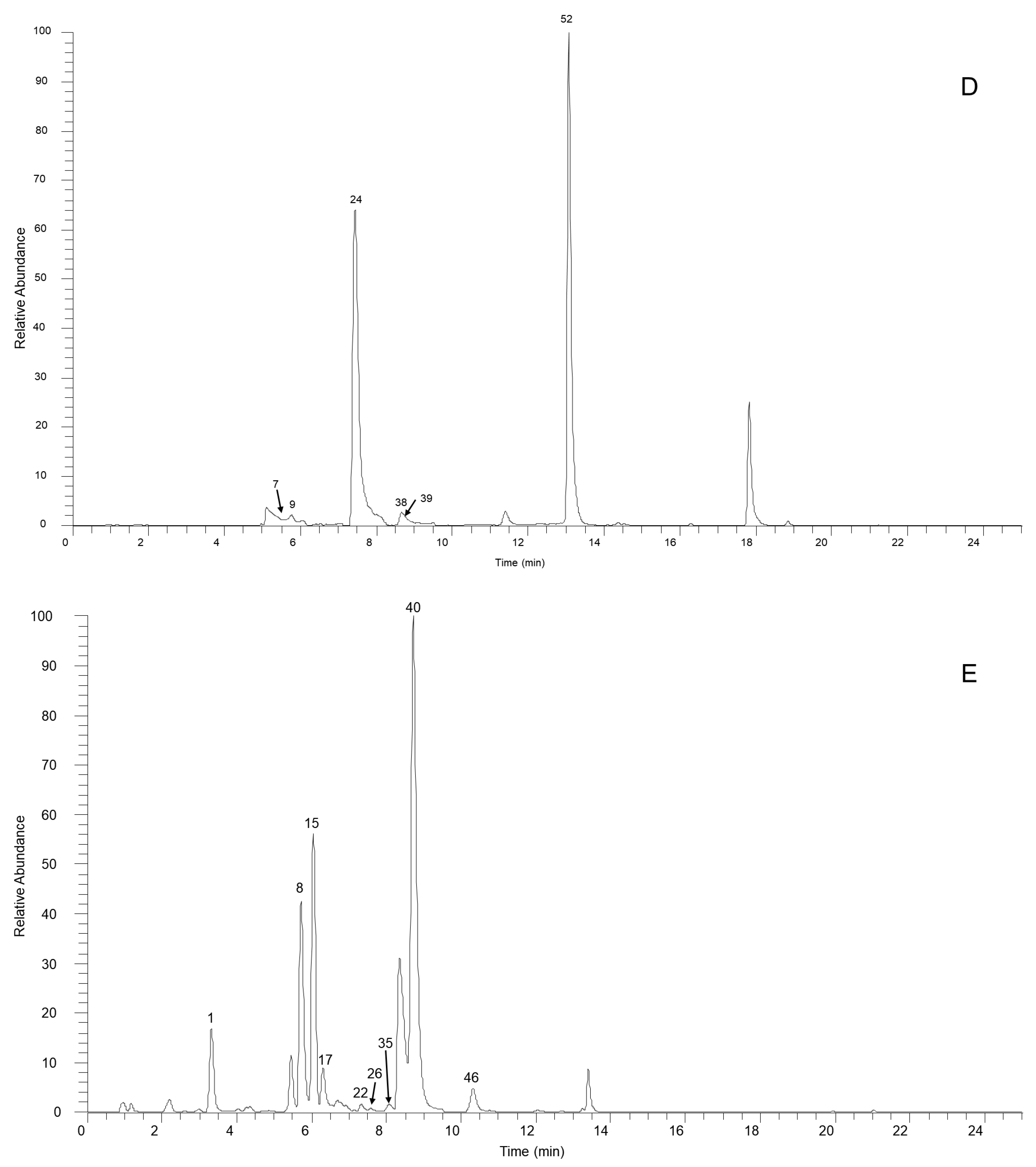
From the experimental data for our SSD sample and a summarized fragmentation pattern, we identified a total of 70 aporphine alkaloids. Table 1 and Table S1 give the chromatographic and mass data for these detected constituents and includes retention times (tR), experimental masses, and the discrepancies between the theoretical and experimental masses (in ppm), in addition to molecular formulas for all the aporphine alkaloids as well as MS/MS fragment ions. Figure 1 illustrates the high-resolution extracted ion chromatogram from the SSD extract in the positive ion mode. All compounds are numbered according to their order of elution.

Table 1.
The chromatographic and mass data for the components detected from Sabia schumanniana Diels though UHPLC-Q-Exactive Orbitrap MS.
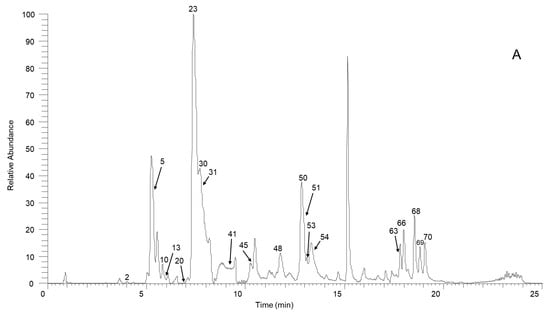
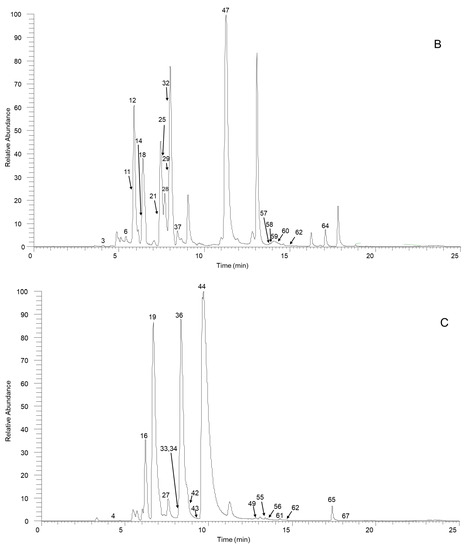
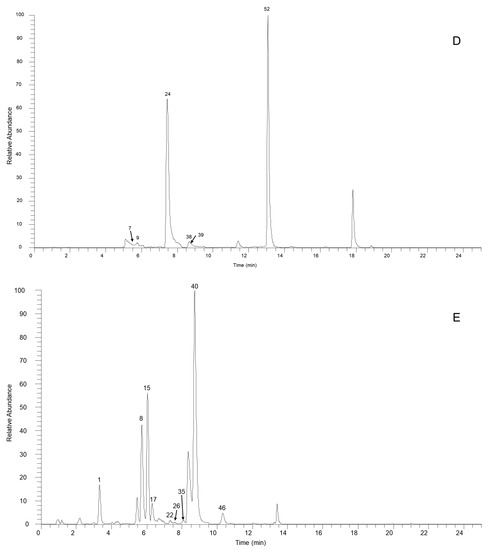
Figure 1.
The high-resolution extracted ion chromatogram (HREIC) in 5 ppm for the multiple compounds in Sabia schumanniana Diels. (A) Peak 2, 13: m/z 490.2072; 5: m/z 278.1175; 10: m/z 298.1437; 20: m/z 374.1598; 23, 31, 45, 50: m/z 282.1489; 30: m/z 354.17; 41: m/z 400.1755; 48: m/z 384.1805; 51: m/z 266.1176; 53: m/z 280.1332; 54, 68, 69: m/z 292.0968; 63: m/z 294.1124; 66: m/z 308.1281; 70: m/z 338.1386; (B) Peak 3, 11, 21, 29, 32, 37, 58: m/z 312.1594; 6, 12, 18, 25: m/z 328.1543; 14: m/z 340.1543; 47, 62: m/z 294.1488; 57: m/z 312.1230; 59, 64: m/z 310.1438; 60: m/z 310.1437; (C) Peak 4: m/z 358.1649; 16: m/z 344.1856; 19, 27, 42: m/z 342.1700; 33: m/z 344.1492; 34: m/z 374.1598; 36, 43, 49: m/z 356.1856; 44, 61: m/z 296.1645; 55, 65, 67: m/z 324.1230; 56: m/z 356.1492; (D) Peak 7, 14, 24, 38, 39: m/z 340.1543; 9: m/z 314.1386; 52: m/z 294.1489; (E) Peak 1: m/z 358.1649; 8: m/z 358.1649; 15: m/z 358.1649; 17: m/z 328.1908; 22, 26: m/z 358.2013; 36: m/z 356.1856; 40: m/z 328.1907; 46: m/z 374.1598.
2.1. Establishment of the Analytical Method
For this study, we established an analytical strategy based on utilizing UHPLC-Q-Exactive Orbitrap MS combined with parallel reaction monitoring (PRM) to identify diagnostic fragment ions (DFIs) and neutral losses (NLs) in order to comprehensively screen for and detect the aporphine alkaloids present in SSD. First, we injected SSD samples into a UHPLC-Q-Exactive Orbitrap MS to obtain full mass raw data via use of the full-mass scanning mode. Second, we predicted the potential chemical compounds using Compound Discoverer 3.0 and Metabolite Workflow. We determined parameters in line with [13]. The drug was set to magnoflorine, while roemerine and the added group were assigned to a list of substituents including -CH3, -OH, -OCH3, C=O, and -OCH2O-. Third, we collected fragmentation ions using UHPLC-Q-Exactive Orbitrap MS based on the parallel reaction monitoring mode activated by inclusion ions from the list described above. Finally, we performed an accurate full-scan mass spectrometry and MS2. We also extracted the retention time information and incorporated relevant database and literature data. By such means, we obtained our SSD identification results.
2.2. Identifification and Analysis of Aporphine Alkaloids in SSD
We used UHPLC-Q-Exactive Orbitrap MS to examine the fragmentation patterns of four reference standards in positive mode in order to establish the neutral loss and the diagnoses for fragmentation ions.
Figure 2A shows the proposed fragmentation pathway for magnoflorine. This generated a fragment ion at m/z 297.1123 (C18H17O4+) via the neutral loss at m/z 45 [C2H7N] when the isoquinoline ring was opened and the amino group along with two methyl groups were removed, this being an essential characteristic of aporphine alkaloids [14,15]. We then obtained a product ion at m/z 265.0852 (C17H13O3+) by the precursor-ion neutral loss of CH3OH. The presence of a fragment ion at m/z 282.0877 (C17H14O4+) manifested the parallel loss of CH3. The base peak for the fragment ions was obtained at m/z 265.0852, and the loss of CO was obtained at m/z 237.0910 (C16H13O2+).
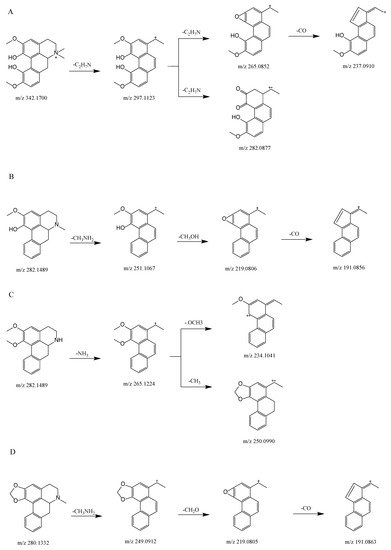
Figure 2.
Proposed selected fragmentation pattern for components identified from SSD: magnoflorine (A); lirindine (B); N-nornuciferine (C); roemerine (D).
Figure 2B shows the proposed fragmentation pathway for lirinidine. The neutral loss of CH3NH2 and the production of the ion at m/z 251.1067 (C17H14O2+), in addition to the consequent neutral loss of CH3OH and CO, yielded fragment ions at m/z 219.0806 (C16H10O+) and 191.0856 (C15H10+).
Figure 2D shows the proposed fragmentation pathway for roemerine. This yielded a fragment ion at m/z 249.0912 (C17H13O2+) because of the characteristic elimination of CH3NH2, and also involved the expulsion of CH3O, which produced a fragment ion at m/z 219.0805 (C16H10O+). The consequent neutral loss of CO generated a fragment ion at m/z 191.0856 (C15H10+).
Figure 2C shows the proposed fragmentation pathway for N-nornuciferine. The fragment ion at m/z 265.1224 (C18H17O2+) resulted from the characteristic elimination of NH3. Subsequently, the product ion at m/z 234.1041 (C17H14O+) was produced by the precursor-ion loss of OCH3. The fragment ion at m/z 250.0990 (C17H14O2+) was observed because of the parallel loss of CH3.
At m/z 45 [C2H7N], m/z 31 [CH3NH2], and 17 [NH3], the types of nitrogen substituted in aporphine alkaloids could be distinguished, representing quaternary, tertiary and secondary aporphine alkaloids, respectively; m/z 32 [CH3OH], 31 [CH3O], 28 [CO], and 18 [H2O] Da were all neutral loss fragments of aporphine alkaloids. At m/z 58 [C3H8N], four standards exhibited this characteristic peak in diagnosis fragmentation ions. As a result, the diagnostic product ion and the neutral loss are important in determining the fracture process for each chemical.
2.2.1. Fragmentation Pattern for Quaternary Aporphine Alkaloids
We accurately identified Compound 27 as magnoflorine by comparing the retention time and the MS and MS2 spectra with the reference-standard data. We also found that Compounds 19 and 42 were eluted at 6.70 and 8.97 min, respectively, and they possessed the same MS1 at 342.1670 [M]+. They also exhibited five distinct fragment ion peaks at m/z 58.0658, 237.0905, 265.0852, 282.0877, and 297.1123. We identified these as magnoflorine isomers.
Compound 1 had a molecular formula of C20H24NO5 and a retention time of 3.32 min. This compound produced the precursor ion at 358.1649 [M]+ and four fragment ion peaks at m/z 58.0660, 227.0703, 255.0667, and 287.0917 in the positive ion mode. Based on secondary fragmentation data, we identified Compound 1 as C6a-hydroxylation of magnoflorine [16].
For Compounds 3, 11, 21, and 29, we determined a molecular design based on the structure of magnoflorine with one methoxy group removed, giving a molecular formula of C19H22NO3. We obtained the precursor ion at m/z 312.1594 [M]+ and observed four characteristic fragment ion peaks at m/z 58.0660, 217.0650, 207.0808, and 267.1018 in the positive ion mode. We tentatively identified these four compounds as C2-O-demethylation of magnoflorine isomers [16].
The isomeric Compounds 4, 8, 15, and 35 exhibited identical fragment ions and molecular ions. The precursor ion at m/z 358.1649 [M]+ was formed using the chemical formula C20H24NO5. We observed five characteristic fragment ion peaks at m/z 58.0659, 253.0863, 281.0809, 285.0740, and 313.1071 in the positive ion mode. We identified these compounds as isomers of trilobinine [17].
Compounds 17 and 40 had a molecular formula of C20H26NO3 and produced a precursor ion at m/z 328.1907 [M]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 58.0659, 251.1067, 253.1226, and 283.1328. On the basis of the above information, we tentatively identified these compounds as isomers of N-ring-opening C1-dehydroxylation of magnoflorine [16].
Compound 16 had a chemical formula of C20H26NO4, was eluted at 6.22 min, and produced a precursor ion at m/z 344.1856 [M]+ in the positive ion mode. We observed five characteristic fragment ion peaks at m/z 58.0659, 137.0598, 143.0493, 175.0754, and 299.1278. On the basis of the above MS and previous findings in the literature, we identified this compound as zizyphusine+ 2H [18].
For Compounds 20, 34, and 46, we designed a molecular structure from dihydroxylation of magnoflorine, giving it the molecular formula of C20H24NO6, and produced a precursor ion at m/z 374.1599 [M]+ in the positive ion mode. We observed three characteristic fragment ion peaks at m/z 58.0659, 297.0758, and 329.1022. We identified these compounds as isomers of di-hydroxylation of magnoflorine [19].
Compounds 22 and 26 had the chemical formula of C21H28NO4 and generated a precursor ion at m/z 358.20128 [M]+ in the positive ion mode. We observed three characteristic fragment ion peaks at m/z 58.0660, 281.0813, and 313.1446. We compared these data with previous findings in the literature and identified these compounds as isomers of pareirarinea [20].
Compounds 7, 14, 24, and 39 had the formula C20H22NO4, with the same quasi- molecular ions [M]+ at m/z 340.1543 in the positive ion mode. We observed five characteristic fragment ion peaks at m/z 189.0692, 217.0644, 235.0754, 263.0703, and 295.0966. Drawing on the findings from previous research, we identified these compounds as isomers of N-methylbulbocapnine [21].
Compound 33 had a molecular formula of C19H22NO5 and a retention time of 8.04 min; it produced the precursor ion at m/z 344.1492 [M]+ in the positive ion mode. We observed four characteristic fragment ions at m/z 58.0660, 237.0907, 265.0860, and 283.0926. We tentatively identified Compound 33 as N-CH3-hydroxylation and C2-O-demethylation of magnoflorine [16].
Compounds 36, 43, and 49 were obtained form a molecular design in which one of the hydroxyl groups that is in magnoflorine becomes methoxy. These compounds had a molecular formula of C21H26NO4 and produced a precursor ion at m/z 356.1856 [M]+ in the positive ion mode. We observed characteristic fragment ion peaks at m/z 58.0660, 236.0833, 264.0785, 251.1066, 279.1018, 280.1082, 296.1038, and 311.1280. On the basis of the above molecular design and fragmentation information, we identified Compounds 36, 43, and 49 as menisperine isomers [22].
Compound 41 had a molecular formula of C22H26NO6 and a retention time of 8.83 min; it produced a precursor ion at m/z 400.1755 [M]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 58.0660, 295.0961, 323.0918, and 355.1180. We identified this compound as C10-OCH3-hydroxylation and C11-O-acetylation of magnoflorine [16].
Compounds 44 and 61 had a molecular design based on the structure of magnoflorine with one hydroxyl group and one methoxy group removed, giving a molecular formula of C19H22NO2, and it produced a precursor ion at m/z 296.1645 [M]+. We observed six characteristic fragment ion peaks at m/z 58.0660, 219.0807, 220.0842, 221.0957, 236.0826, and 251.1068 in the positive ion mode. On the basis of the above information, we identified Compounds 44 and 61 as isomers of C1-demethoxy-C2-dehydrox of magnoflorine [16].
Compound 48 had a molecular formula of C22H26NO5 and produced the precursor ion at m/z 384.18054 [M]+ in the positive ion mode. In addition, we observed seven characteristic fragment ion peaks at m/z 58.0659, 251.1067, 279.1019, 292.0738, 307.0953, 325.1070, and 339.1230. We therefore identified Compound 48 as C1- O-acetylation of magnoflorine [16].
The molecular design of Compound 52 was based on the structure of magnoflorine, from which one adjacent hydroxyl group and one methoxy group were removed, and one adjacent hydroxyl group and one methoxy group were changed to dioxolane, giving a molecular formula of C19H20NO2. This compound had a retention time of 13.07 min and produced a precursor ion at m/z 294.1489 [M]+. We observed four characteristic fragment ion peaks at m/z 58.0659, 191.0862, 219.0805, and 249.0911. On the basis of the above molecular design and fragmentation information, we identified Compound 52 as roemrefidine [23].
Compound 56 had a molecular formula of C20H22NO5 and a retention time of 13.53 min, and it produced a precursor ion at m/z 356.1492 [M]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 58.0659, 251.0703, 279.1028, and 311.0918. On the basis of the above fragmentation patterns, we tentatively identified Compound 56 as C5-methylene to ketone of magnoflorine [16].
For Compounds 59 and 64, we obtained a molecular design based on the structure of magnoflorine, with an adjacent hydroxyl group in which one methoxy group was changed to 1,3-dioxolane and from which one methoxy group was removed, giving a molecular formula of C19H20NO3. These compounds had retention times of 14.11 and 17.08 min, respectively, and produced a precursor ion at m/z 310.1438 [M]+. We observed five characteristic fragment ions at m/z 58.0659, 177.0555, 205.0648, 233.0598, and 265.0859. On the basis of the above information, we identified Compounds 59 and 64 as isomers of C1-demethoxy -C2-dehydrox- C10, C11- ethyl epoxide of magnoflorine [16].
2.2.2. Fragmentation Pattern of Tertiary Aporphine Alkaloid
We definitively identified Compound 45 as lirinidine by comparing its retention time and MS and MS2 spectra with reference standard data. Furthermore, Compounds 23 and 31 were eluted at 7.38 and 7.72 min, respectively, and exhibited the same MS1 at m/z 282.1489 [M+H]+. We observed five distinct fragment ion peaks at m/z 58.0660(90), 191.0855(5), 219.0806(23), 237.0911(100), and 251.1063, as with lirinidine. We therefore identified Compounds 23 and 31 as lirinidine isomers.
We precisely identified Compound 53 as roemerine by comparing its retention time and its MS and MS2 spectra with those in the reference standard data.
Compound 5 had a molecular formula of C18H15NO2, was eluted at 5.33 min, and produced a precursor ion at m/z 278.1175 [M+H]+ in the positive ion mode. We observed three characteristic fragment ion peaks at m/z 107.0497, 246.0928, and 262.0858. We therefore identified this compound as dehydroroemerine [24].
For Compounds 6, 12, 18, and 25, the molecular design was based on the structure of lirinidine with an additional set of adjacent hydroxyls and methoxy groups, giving a molecular formula of C19H22NO4. These were eluted at 5.40, 5.88, 6.40, and 7.53 min, respectively, and they produced a precursor ion at m/z 328.1543 [M+H]+ in the positive ion mode. In addition, we observed five characteristic fragment ion peaks at m/z 58.0660, 177.0551, 222.1118, 265.0862, and 283.0967. On the basis of the above fragment characteristics, we identified these compounds as isomers of bolidine [25].
Compound 10 had a molecular formula of C18H19NO4, was eluted at 5.81 min, and produced a precursor ion at m/z 298.1437 [M+H]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 58.0659, 192.1022, 254.0953, and 283.1197. We identified Compound 10 as apoglaziovine [26].
Compounds 28, 47, and 62 had a molecular formula of C19H19NO2 and produced a precursor ion at m/z 294.1488 [M+H]+ in the positive ion mode. We observed six characteristic fragment ion peaks at m/z 58.0658, 217.0650, 236.0831, 250.0946, 263.1286, and 279.1256. On the basis of the above fragmentation patterns, we tentatively identified these compounds as dehydronuciferine isomers [27].
We produced Compound 30 by adding two adjacent methoxy groups and N-methyl to the structure of roemerine, giving a molecular formula of C21H24NO4. This compound exhibited a retention time of 7.69 min and produced a precursor ion at m/z 354.1670 [M+H]+. We observed three fragment ion peaks at m/z 58.0660, 251.1074, and 309.1119 in the positive ion mode. On the basis of these results, and previously reported findings in the literature [28], we identified Compound 30 as N-methylnantenine.
Compounds 32, 37, and 58 had a chemical formula of C19H21NO3 and produced a precursor ion at m/z 358.20128 [M+H]+ in the positive ion mode. We observed five fragment ion peaks at m/z 58.0659, 217.0650, 267.1016, 280.1064, and 294.1487. On the basis of this information, we identified these compounds as isomers of isothebaine [29].
Compound 38 had a molecular formula of C20H21NO4 and exhibited a retention time of 8.66 min. This compound produced a precursor ion at m/z 340.1543 [M+H]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 58.0660, 220.0526, 264.0755, and 309.1354. Hence, Compound 38 was tentatively identified as crebanine [26].
Compounds 55, 65 and 67 had a molecular formula of C19H17NO4 and produced a precursor ion at m/z 324.1230 [M+H]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 58.0659, 177.0554, 263.0940, and 293.1054. We identified these compounds as isomers of neolitsine [14].
Compounds 54, 68, and 69 had a molecular formula of C18H13NO3 and produced a precursor ion at m/z 292.0968 [M+H]+ in the positive ion mode. We observed three characteristic fragment ion peaks at m/z 248.0712, 264.1024, and 277.1039. The product ion at m/z 277.1039 [M+H−NH]+ was obtained via the neutral loss of NH. We ascribed the loss of this fragment to NH serving as a different substituent for nitrogen. On the basis of the above fragmentation patterns, we tentatively identified these compounds as lysicamine isomers [30].
Compound 60 had a molecular formula of C19H19NO3, exhibited a retention time of 14.22 min, and produced a precursor ion at m/z 310.1437 [M+H]+ in the positive ion mode. We observed three characteristic fragment ion peaks at m/z 58.0660, 279.1008, and 264.0792. On the basis of the above fragmentation patterns, we tentatively identified Compound 60 as stephanine [31].
Compound 63 had a molecular formula of C18H15NO3, exhibited a retention time of 15.99 min, and produced a precursor ion at m/z 294.1124 [M+H]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 58.0658, 239.0951, 257.1901, and 262.0863. On the basis of the above fragmentation patterns, we tentatively identified Compound 63 as N-formyl-annonain [32].
Compound 66 had a molecular formula of C19H17NO3, exhibited a retention time of 18.00 min, and produced a precursor ion at m/z 308.1281 [M+H]+ in the positive ion mode. We observed three characteristic fragment ion peaks at m/z 191.0859, 219.0806, and 249.0914. The product ion at m/z 219.0806 [M+ H−C2H5NO]+ was obtained via the neutral loss of C2H5NO. We ascribed the loss of this fragment to NHCOCH3 serving as a different substituent for nitrogen. Based on the secondary fragmentation data and mass spectral fragmentation behavior, we identified Compound 66 as N-acetylanonaine [33].
2.2.3. Fragmentation Pattern of Secondary Aporphine Substituted
We unambiguously identified Compound 50 as N-nornuciferine by comparing its retention time and its MS and MS2 spectra with the reference standard data.
Compounds 2 and 13 had a molecular formula of C25H31NO9, and they produced a precursor ion at m/z 490.2072 [M+H]+ in the positive ion mode. We observed five characteristic fragment ion peaks at m/z 192.1019, 237.0901, 265.0861, 297.1122, and 328.1544. On the basis of the above fragments, we identified Compounds 2 and 13 as isomers of 11-glc- norisocorydine [18].
Compound 9 had a molecular formula of C18H19NO4, was eluted at 5.77 min, and produced a precursor ion at m/z 314.1386 [M+H]+ in the positive ion mode. We observed six characteristic fragment ions at m/z 58.0660, 165.0913, 205.0658, 237.0910, 265.0861, and 297.1124. We identified this compound as laurolitsine [34].
Compound 51 had a molecular formula of C17H15NO2, exhibited a retention time of 12.80 min, and produced a precursor ion at m/z 266.1176 [M+H]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 131.0494, 191.0855, 219.0804, and 249.0912 in the positive ion mode. On the basis ofn the above information, we identified Compound 51 as anonaine [33].
Compound 57 had a molecular formula of C18H17NO4, was eluted at 13.83 min, and produced a precursor ion at m/z 312.1230 [M+H]+ in the positive ion mode. We observed five characteristic fragment ion peaks at m/z 58.0659, 264.1164, 265.0865, 280.1095, and 295.1328. We identified Compound 57 as nandigerine [21].
Compound 70 had a molecular formula of C18H17NO4, was eluted at 19.08 min, and produced a precursor ion at m/z 338.13860 [M+H]+ in the positive ion mode. We observed four characteristic fragment ion peaks at m/z 279.1258, 307.1201 308.1265, and 323.1153. We identified Compound 70 as sinomendine [35].
2.3. Pharmacological Activity of Aporphine Alkaloids in SSD
Natural aporphine alkaloids exhibit a wide range of biological properties, including antioxidant, antiplatelet-aggregation, anticonvulsant, antispasmodic, anti-cancer, antimalarial, antiprotozoal, anti-poliovirus, anticytotoxicity, and anti-Parkinson effects. Natural products and their synthetic derivatives from the mainstay of research can be made into new medications for a wide range of disorders [36].
Aporphine alkaloids are widely distributed in various medicinal plants and are the active ingredients in many traditional Chinese medicines. Magnoflorine is one of the most important pharmacologically active compounds in the quaternary aporphine alkaloid, with reported anti-diabetic and anti-inflammatory effects [37]. Lirinidine is a tertiary aporphine alkaloid which greatly inhibits the production of collagen and arachidonic acid and reduces the aggregation of platelet-activating factor-induced platelets [38]. Among the secondary aporphine alkaloids, norisocorydine can help regulate transporters in the small intestine [39] and N-nornuciferine exhibits anti-inflammatory effects [40].
3. Material and Methods
3.1. Chemicals and Materials
We obtained acetonitrile and LC grade methanol from MACKIN Company. We acquired MS grade formic acid from Thermo Fisher Scientific Co., Ltd. (New Jersey, NJ, USA). We obtained purified water from Guangzhou Watsons Food & Beverage Co., Ltd. (China). We acquired SSD samples from Liuzhi Special Zone, Liupanshui City, Guizhou Province, with an altitude of 1367M and a latitude and longitude 105°28′ E, 26°13′ N by Kunming Plant Biotechnology Co., Ltd. We obtained roemerine (purity ≥98%) and lirinidine (purity ≥98%) from Wuhan ChemFaces Biochemical Co., Ltd (Wuhan, China). We acquired N-nornuciferine from Chengdu HerbSubstance Co.,Ltd. Magnoflorine (purity ≥98%) from Sichuan Weiqi Biotechnology Co., Ltd. (Sichuan, China).
3.2. Standard and Solution Preparation
We pulverized an SSD stem and accurately weighed 1 g of sample powder. We transferred this to a flask containing 10 mL of 70% aqueous methanol (v/v) and performed ultrasonic extraction for 60 min at room temperature. We obtained supernatant after filtrating (nylon needle filter, 0.45 μm) and centrifuging at 13,523 g for 20 min at 10 °C.
We prepared reference-standard stock solutions of magnoflorine, lirinidine, N-nornuciferine, and roemerine at concentrations of 0.1 mg/mL with methanol. These were stored at 4 °C.
3.3. Instruments and UHPLC-MS Conditions
We achieved a full characterization of aporphine alkaloid in SSD using a Dionex Ultimate 3000 RS UHPLC equipped with a quaternary pump and LPG-3400SD vacuum degasser unit (Thermo Fisher Scientific, California, CA, USA). We also used a Q-Exactive Orbitrap MS mass spectrometer with an electrospray ionization (ESI) source. We separated all the analyzed samples using a Thermo Scientific Hypersil GOLD™ aQ C18 column (100 mm × 2.1 mm, 1.9 μm) at 40 °C with a flow rate of 0.3 mL/min. The mobile phases consisted of 0.1% formic acid aqueous solution (solvent A) and acetonitrile (solvent B) with a flow rate of 0.30 mL/min. The gradient program was as follows: 0–2 min, 95–90% A; 2–5 min, 90–85% A; 5–10 min, 85–80% A; 10–12 min, 80–65% A; 12–20 min, 65–30% A; 20–22 min, 30–5% A; 22–22.1 min, 5–95%A; 22.1–25 min, 95%A, The sample injection volume was 2 μL.
All samples were examined in the positive mode using the following tune approach. We used full-scan mode to produce high-resolution mass spectra with a resolution of 70 000 and a mass range of m/z 120–1000. PRM parameters were set as follows: the resolution was 35,000; the isolation window was 3.0 m/z; and the NEC (normalized collision energy) was set to 35, with 5.0× e4 of automatic gain control (AGC) target. We processed data using Xcalibur™ version 4.1 (Thermo Fisher Scientific, California, CA, USA) and Compound Discovery version 3.0 (Thermo Fisher Scientific, California, CA, USA). ESI source parameters were set as follows: the spray voltage was 3.5 kV; flow rates of 30 and 10 (arbitrary units) were used for the sheath gas and auxiliary gas, respectively; nitrogen was ≥99.99%; capillary temperature and the heater temperature were set to 320 °C and 350 °C, respectively; the S-lens RF level was 50.
3.4. Data Processing
We used the Thermo Xcalibur software version 4.1 and Compound Discover software version 3.0 (Thermo Fisher Scientific, California, CA, USA) to process all the raw data, including full-scan MS and MS2 data. We set the minimum peak intensity to 10,000 and calculated detailed chemical formula parameters from accurate masses for all the parent and fragment ions of selected peaks using a formula predictor, as follows: the maximum element counts were C30, H60, O20, and N10; the MS and MS2 mass tolerances were set to 5 and 10 ppm, respectively.
4. Conclusions
Using UHPLC Q-Exactive MS, we established an effective method to fully identify the aporphine alkaloids in SSD. We identified a total of 70 aporphine alkaloid constituents in SSD based on their chromatographic retention, MS and MS2, and bibliographic data. Sixty-nine of these are here reported as constituents of SSD for the first time. Some of these compounds have previously been shown to exhibit good pharmacological properties, including anti-cancer and anti-diabetic effects. Our findings lay the groundwork for more in-depth investigations of the pharmacodynamic substance basis for SSD.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules27217643/s1, Table S1: The chromatographic and mass data of detected components from Sabia schumanniana Diels though UHPLC-Q-Exactive Orbitrap MS.
Author Contributions
S.E., writing—original draft, investigation, and data curation; Z.-C.S., conceptualization and formal analysis, S.-h.Q., validation; K.-l.L., data curation; Y.-n.L., investigation; J.-L.W., formal analysis; W.C. and F.Y., writing—review and editing, supervision, and conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the science and technology innovation Program of Hunan Province (no. 2022RC1228) and the Scientific Research of Hunan Provincial Education Department (no. 19A353) and the Hunan University of Medicine High-Level Talent Introduction Startup Funds (no. 15001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be provided upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Chen, Q.Y.; Sun, Q.W.; Zhang, Y.P.; Xu, W.F.; Huang, Y.; Chen, C.L. The Research Progress on Genus Sabia. J. Guizhou Univ. Tradit. Chin. Med. 2022, 44, 71–80. [Google Scholar] [CrossRef]
- Wuhan Institute of Botany; Chinese Academy of Sciences. Flora of Hubei 2; Hubei People’ Press: Wuhan, China, 1979; Volume 480. [Google Scholar]
- Liu, X.; Sun, Y.; Dou, L.M.; Wu, Y.N.; Tan, Y.F.; Dong, L.; Zhang, X.B. Research Progress in Anti-diabetic Activities of Apophine Alkaloids. Pharm. Sci. 2017, 41, 704–709. [Google Scholar] [CrossRef]
- Liang, G.Y.; Zhou, Y.; Cao, P.X.; Xu, B.X. Studies on chemical constituents of Sabia schumanniana. Chin. Pharm. J. 2005, 40, 900. [Google Scholar] [CrossRef]
- Qing, Z.X.; Huang, J.L.; Yang, X.Y.; Liu, J.H.; Cao, H.L.; Xiang, F.; Cheng, P.; Zeng, J.G. Anticancer and Reversing Multidrug Resistance Activities of Natural Isoquinoline Alkaloids and their Structure-activity Relationship. Curr. Med. Chem. 2018, 25, 5088–5114. [Google Scholar] [CrossRef]
- Wei, C.Y.; Wang, S.W.; Ye, J.W.; Hwang, T.L.; Cheng, M.J.; Sung, P.J.; Chang, T.H.; Chen, J.J. New Anti-Inflammatory Aporphine and Lignan Derivatives from the Root Wood of Hernandia nymphaeifolia. Molecules 2018, 23, 2286. [Google Scholar] [CrossRef]
- Zhao, Q.Z.; Zhao, Y.M. Progress in Biologlcal Activities of Aporphinoid Alkaloids. Nat. Prod. Res. Dev. 2006, 18, 316–324. [Google Scholar] [CrossRef]
- Chandradevan, M.; Simoh, S.; Mediani, A.; Ismail, N.H.; Ismail, I.S.; Abas, F. UHPLC-ESI-Orbitrap-MS Analysis of Biologically Active Extracts from Gynura procumbens (Lour.) Merr. and Cleome gynandra L. Leaves. Evid. Based Complement. Altern. Med. 2020, 2020, 3238561. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Zheng, B.; Guan, Y.; Wang, L.; Chen, L.; Cai, W. Rapid characterization of chlorogenic acids in Duhaldea nervosa based on ultra-high-performance liquid chromatography-linear trap quadropole-Orbitrap-mass spectrometry and mass spectral trees similarity filter technique. J. Sep. Sci. 2018, 41, 1764–1774. [Google Scholar] [CrossRef]
- Ye, X.; Wang, Y.; Zhao, J.; Wang, M.; Avula, B.; Peng, Q.; Ouyang, H.; Lingyun, Z.; Zhang, J.; Khan, I.A. Identification and Characterization of Key Chemical Constituents in Processed Gastrodia elata Using UHPLC-MS/MS and Chemometric Methods. J. Anal. Methods Chem. 2019, 2019, 4396201. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, H.; Liu, Y.; Liu, J.; Zhao, X.; Yin, Y. Development and Evaluation of a Parallel Reaction Monitoring Strategy for Large-Scale Targeted Metabolomics Quantification. Anal. Chem. 2016, 88, 4478–4486. [Google Scholar] [CrossRef]
- Xiang, L.; Wei, J.; Tian, X.Y.; Wang, B.; Chan, W.; Li, S.; Tang, Z.; Zhang, H.; Cheang, W.S.; Zhao, Q.; et al. Comprehensive Analysis of Acylcarnitine Species in db/db Mouse Using a Novel Method of High-Resolution Parallel Reaction Monitoring Reveals Widespread Metabolic Dysfunction Induced by Diabetes. Anal. Chem. 2017, 89, 10368–10375. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Li, K.L.; Xiong, P.; Gong, K.Y.; Zhu, L.; Yang, J.B.; Wu, W.H. A systematic strategy for rapid identification of chlorogenic acids derivatives in Duhaldea nervosa using UHPLC-Q-Exactive Orbitrap mass spectrometry. Arab. J. Chem. 2020, 13, 3751–3761. [Google Scholar] [CrossRef]
- Stévigny, C.; Jiwan, J.L.; Rozenberg, R.; de Hoffmann, E.; Quetin-Leclercq, J. Key fragmentation patterns of aporphine alkaloids by electrospray ionization with multistage mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Staack, R.F.; Fritschi, G.; Maurer, H.H. Studies on the metabolism and toxicological detection of the new designer drug N-benzylpiperazine in urine using gas chromatography-mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 773, 35–46. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, Y.; Li, Z.; Hu, P.; Chen, M.; Sun, Z.; Lin, Y.; Pan, G.; Huang, C. Systematic and comprehensive strategy for metabolite profiling in bioanalysis using software-assisted HPLC-Q-TOF: Magnoflorine as an example. Anal. Bioanal. Chem. 2016, 408, 2239–2254. [Google Scholar] [CrossRef]
- Cornélio, M.L.; Barbosa-Filho, J.M.; Côrtes, S.F.; Thomas, G. Tracheal relaxant activity of cissaglaberrimine and trilobinine, two aporphinic alkaloids from Cissampelos glaberrima. Planta Med. 1999, 65, 462–464. [Google Scholar] [CrossRef]
- Li, M.; Zhang, F.X.; Wei, Z.C.; Li, Z.T.; Zhang, G.X.; Li, H.J. Systematically characterization of in vivo substances of Ziziphi Spinosae Semen in rats by ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry analysis. J. Pharm. Biomed. Anal. 2021, 193, 113756. [Google Scholar] [CrossRef]
- Xue, B.; Zhao, Y.; Miao, Q.; Miao, P.; Yang, X.; Sun, G.; Su, J.; Ye, J.; Wei, B.; Zhang, Y.; et al. In vitro and in vivo identification of metabolites of magnoflorine by LC LTQ-Orbitrap MS and its potential pharmacokinetic interaction in Coptidis Rhizoma decoction in rat. Biomed. Chromatogr. 2015, 29, 1235–1248. [Google Scholar] [CrossRef]
- Bhatt, V.; Kumari, S.; Upadhyay, P.; Agrawal, P.; Anmol; Sahal, D.; Sharma, U. Chemical profiling and quantification of potential active constituents responsible for the antiplasmodial activity of Cissampelos pareira. J. Ethnopharmacol. 2020, 262, 113185. [Google Scholar] [CrossRef]
- Yan, R.; Wang, W.; Guo, J.; Liu, H.; Zhang, J.; Yang, B. Studies on the alkaloids of the bark of Magnolia officinalis: Isolation and on-line analysis by HPLC-ESI-MS(n). Molecules 2013, 18, 7739–7750. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, T.; Zhang, X.; Xu, H.; Liu, C. Chemical fingerprint analysis of Phellodendri Amurensis Cortex by ultra performance LC/Q-TOF-MS methods combined with chemometrics. J. Sep. Sci. 2010, 33, 3347–3353. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, Y.; He, J.; Kang, Y.; Wang, Y.; Yang, P.; Guo, J.; Huang, J. Structural Characterisation of Alkaloids in Leaves and Roots of Stephania kwangsiensis by LC-QTOF-MS. Phytochem. Anal. 2018, 29, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Thuy, T.; Van Sung, T.; Franke, K.; Wessjohann, L. Aporphine and proaporphine alkaloids from Stephania rotunda. TAP CHI HOA HOC 2005, 43, 0378–2336. [Google Scholar]
- Zhang, S.; Zhang, Q.; Guo, Q.; Zhao, Y.; Gao, X.; Chai, X.; Tu, P. Characterization and simultaneous quantification of biological aporphine alkaloids in Litsea cubeba by HPLC with hybrid ion trap time-of-flight mass spectrometry and HPLC with diode array detection. J. Sep. Sci. 2015, 38, 2614–2624. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y.; Jiang, L.; Li, C.; Sun, Z.; Zhang, Y.; Lin, T.; Jiang, Y.; Liu, B. A triple combination strategy of UHPLC-MS(n), hypolipidemic activity and transcriptome sequencing to unveil the hypolipidemic mechanism of Nelumbo nucifera alkaloids. J. Ethnopharmacol. 2022, 282, 114608. [Google Scholar] [CrossRef]
- Wu, X.L.; Wu, M.J.; Chen, X.Z.; Zhang, H.M.; Ding, L.Q.; Tian, F.Y.; Fu, X.M.; Qiu, F.; Zhang, D.Q. Rapid characterization of the absorbed chemical constituents of Tangzhiqing formula following oral administration using UHPLC-Q-TOF-MS. J. Sep. Sci. 2018, 41, 1025–1038. [Google Scholar] [CrossRef]
- Conceição, R.S.; Reis, I.M.A.; Cerqueira, A.P.M.; Perez, C.J.; Junior, M.; Branco, A.; Ifa, D.R.; Botura, M.B. Rapid structural characterisation of benzylisoquinoline and aporphine alkaloids from Ocotea spixiana acaricide extract by HPTLC-DESI-MS(n). Phytochem. Anal. 2020, 31, 711–721. [Google Scholar] [CrossRef]
- Lockwood, G.B. Orientalidine and isothebaine from cell cultures of Papaver bracteatum. Phytochemistry 1981, 20, 1463–1464. [Google Scholar] [CrossRef]
- Lima, B.R.D.; Silva, F.; Soares, E.R.; Almeida, R.A.D.; Silva-Filho, F.A.D.; Barison, A.; Costa, E.V.; Koolen, H.H.; de Souza, A.D.; Pinheiro, M.L.B. Integrative approach based on leaf spray mass spectrometry, HPLC-DAD-MS/MS, and NMR for comprehensive characterization of isoquinoline-derived alkaloids in leaves of Onychopetalum amazonicum RE Fr. J. Braz. Chem. Soc. 2020, 31, 79–89. [Google Scholar] [CrossRef]
- Le, P.M.; Srivastava, V.; Nguyen, T.T.; Pradines, B.; Madamet, M.; Mosnier, J.; Trinh, T.T.; Lee, H. Stephanine from Stephania venosa (Blume) Spreng Showed Effective Antiplasmodial and Anticancer Activities, the Latter by Inducing Apoptosis through the Reverse of Mitotic Exit. Phytother. Res. 2017, 31, 1357–1368. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, T.; Yuan, C.M.; Gu, Y.; Hao, X.J.; Huang, L.J.; Mu, S.Z.; Zhang, J.X. Chemical constituents of Sabia parviflora. Chin. Tradit. Herb. Drugs 2015, 46, 0253–2670. [Google Scholar]
- Guo, K.; Tong, C.; Fu, Q.; Xu, J.; Shi, S.; Xiao, Y. Identification of minor lignans, alkaloids, and phenylpropanoid glycosides in Magnolia officinalis by HPLCDADQTOF-MS/MS. J. Pharm. Biomed. Anal. 2019, 170, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Castro-Saavedra, S.; Fuentes-Barros, G.; Tirapegui, C.; Acevedo-Fuentes, W.; Cassels, B.K.; Barriga, A.; Vilches-Herrera, M. Phytochemical analysis of alkaloids from the chilean endemic tree Cryptocarya alba. J. Chil. Chem. Soc. 2016, 61, 3076–3080. [Google Scholar] [CrossRef]
- Ng, K.M.; Liang, Z.; Lu, W.; Tang, H.W.; Zhao, Z.; Che, C.M.; Cheng, Y.C. In vivo analysis and spatial profiling of phytochemicals in herbal tissue by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 2007, 79, 2745–2755. [Google Scholar] [CrossRef]
- Zhang, A.; Zhang, Y.; Branfman, A.R.; Baldessarini, R.J.; Neumeyer, J.L. Advances in development of dopaminergic aporphinoids. J. Med. Chem. 2007, 50, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Kuang, T.; Du, H.; Li, Q.; Feng, T.; Zhang, Y.; Fan, G. Magnoflorine: A review of its pharmacology, pharmacokinetics and toxicity. Pharmacol. Res. 2020, 152, 104632. [Google Scholar] [CrossRef]
- Chang, F.R.; Wei, J.L.; Teng, C.M.; Wu, Y.C. Two new 7-dehydroaporphine alkaloids and antiplatelet action aporphines from the leaves of Annona purpurea. Phytochemistry 1998, 49, 2015–2018. [Google Scholar] [CrossRef]
- Lin, C.J.; Chen, C.H.; Liu, F.W.; Kang, J.J.; Chen, C.K.; Lee, S.L.; Lee, S.S. Inhibition of intestinal glucose uptake by aporphines and secoaporphines. Life Sci. 2006, 79, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, G.; Zhang, Y.; Yang, M.; Chen, J.; Guo, M. Potential hypoglycemic, hypolipidemic, and anti-inflammatory bioactive components in Nelumbo nucifera leaves explored by bioaffinity ultrafiltration with multiple targets. Food Chem. 2022, 375, 131856. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).