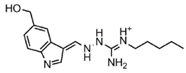
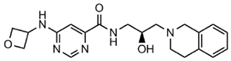
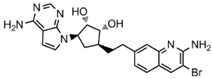
Abstract
Protein arginine methyltransferase 5 (PRMT5) is a popular anticancer target that regulates histone or nonhistone methylation and is linked to the development and poor prognosis of non-small cell lung cancer. PRMT5 inhibitors have shown great promise in clinical trials as a cancer therapy. However, most inhibitors reported recently act in a SAM-competitive mode and lack structural diversity. In this paper, a novel non-SAM inhibitor, 3039-0164, was discovered by the structure-based virtual screening method. The binding mechanism of 3039-0164 to PRMT5 was revealed via molecular docking and molecular dynamics simulations. 3039-0164 inhibited PRMT5 enzymatic activity, downregulated the expression of PRMT5 downstream target genes (FGFR3 and eIF4E), and blocked the activation of the PI3K/AKT/mTOR and ERK signaling pathways. The discovery of 3039-0164 provides precise and creative hit compounds for the design optimization of PRMT5 lead compounds in non-small cell lung cancer.
1. Introduction
Non-small cell lung cancer (NSCLC) is the most common and lethal primary lung cancer, which is difficult to detect and requires systemic treatment [1]. Despite recent advances in surgery, radiotherapy, and targeted molecular therapies, the prognosis of NSCLC remains very poor. There is an urgent need to identify new therapeutic targets to alleviate the disease symptoms.
Epigenetics is closely linked to cancer development, affecting gene expression and cellular function by regulating DNA or histone methylation, acetylation, phosphorylation, ubiquitination modifications, and chromatin remodeling [2]. It is significant to note that epigenetic dysregulation is a characteristic of various human malignancies [3]. Recently, researchers have paid a lot of attention to the novel idea of using epigenetic treatments to generate focused anticancer medications. In eukaryotes, protein post-translational modification (PTM) is a sort of chemical modification that enhances protein function by adding covalent chemical groups and is engaged in a range of epigenetic regulatory mechanisms [4]. One of the most common PTMs is arginine methylation, which occurs mainly on nuclear and cytoplasmic proteins, and this process requires the regulation of protein arginine methyltransferases (PRMTs) that target arginine residues [5,6]. Therefore, PRMTs have recently attracted a lot of attention. PRMTs are involved in several cellular life processes in cancers, including transcriptional activation, RNA splicing, DNA damage, and other responses [7]. Human-derived PRMTs have been classified into three types based on their catalytic activity: type I, II, and III. PRMT5 is a type II PRMT that catalyzes monomethylation or symmetric dimethylation of protein substrates on arginine residues [8] and participates in tumor cell proliferation, growth, development, and Golgi assembly [9,10,11]. PRMT5 is highly expressed in human lung cancer cells and tissues [11]. Sheng and Wang et al. [12] concluded that PRMT5 could promote lung cancer cell proliferation by regulating multiple signaling pathways. A growing body of evidence suggests that targeting PRMT5 has therapeutic value in the treatment of human lung cancer [11,13]. Even though various PRMT5 inhibitors have been reported, most of them are S-adenosyl methionine (SAM) competitive, which means that they are located at the SAM site [14,15,16]. The typical PRMT5 inhibitors representing different scaffolds and their pharmacological mechanisms are shown in Table 1. LLY-283 and AMI-1 are both SAM-competitive inhibitors. The former has the inhibitory effect by blocking MDM4 splicing regulation, which interacts with the PRMT5 residue Phe327 by its phenyl group, making it highly selective for PRMT5; the latter possesses a double anion structure that binds to the SAM site, whose interaction poly activity is established by its sulfonic acid group [14,17,18]. T1551 is a new non-SAM inhibitor. The benzene ring in its indole scaffold forms a cation–π interaction with SAM, which along with hydrogen bonding and π–π stacking forces explains the inhibitory activity of T1551 against PRMT5 [19]. JNJ64619178 as a dual SAM/substrate competition inhibitor binds to the PRMT5/MEP50 complex’s SAM and substrate pockets [20]. One of the proteolysis targeting chimera (PROTAC) degraders, MS4322, competes with the PRMT5 substrate and breaks down PRMT5 through PROTAC protein hydrolysis, inhibiting PRMT5 production [10]. EPZ015666 has an IC50 value of 22 nM, and its THIQ moiety forms the cation–π interaction with the positively charged methyl group in SAM, which makes a fundamental linkage, reduces SmD3 methylation and cell death, and shows a dose-dependent anticancer effect [21,22]. Additionally, a growing number of clinical investigations have proven that EPZ015666 can exhibit antitumor properties by inactivating PRMT5 in acute myeloid leukemia [23], lung cancer [19,22], and retinoblastoma [24,25], and is a novel and potential therapeutic target independent of the SAM binding site. These findings have greatly stimulated our interest in exploring novel non-SAM PRMT5 inhibitors.

Table 1.
Typical PRMT5 inhibitors representing different scaffolds and their pharmacological mechanisms.
In this paper, we used the virtual screening method based on the receptor structure to choose 158 candidates acting at the PRMT5 substrate binding site (at the location of EPZ015666). Through molecular docking and molecular dynamics simulations, it could be found that the identified 3039-0164 bound to PRMT5 in a stable manner mainly by hydrogen bonding and π–π interactions. The MTT experiment showed that 3039-0164 could inhibit cell viability of the A549 non-small cell lung cancer cell line. Furthermore, protein function assays showed that 3039-0164 downregulated the expression of two PRMT5 target genes, FGFR3 and eIF4E, and impeded the activation of the PI3K/AKT/mTOR and ERK signaling pathways. The discovery of 3039-0164 opens up new avenues for research and design optimization of PRMT5 inhibitors, which can help improve the prognosis of non-small cell lung cancer and other cancers in the clinic.
2. Materials and Methods
2.1. Virtual Screening Based on Protein Structure
The Schrödinger Maestro package (Schrödinger, LLC, New York, NY, USA; Schrödinger, 2015) [26] was used for the virtual screening. The protein receptor PRMT5 was derived from the Protein Data Bank (PDB ID: 4X61, https://www.rcsb.org/, accessed: 25 December 2021). The complex was first processed in the Protein Preparation Wizard module. All water molecules were removed, and the protonation state of all charged residues was adjusted to neutral pH. Then the docking grid was generated with EPZ015666 as the center. Since SAM has a cation–π interaction with EPZ015666 and affects PRMT5 inhibitor binding [21], the substrates are divided into two groups with/without SAM for enrichment factors’ (EFs) control assessment for subsequent screening. The sixteen EPZ015666 active derivatives selected in the previous study [27] were generated by DUD-E [28] in a 1:50 ratio of 800 decoys so that the relevant active factor and decoy molecules were docked near the EPZ015666 binding site. The electric field intensity was then calculated for the two groups of models 1 and 10% with/without SAM, respectively.
To eliminate the compounds with false-positive chemical and non-drug-like functional groups, before the virtual screening, the 1,671,908 compounds from the ChemDiv, Specs, and TargetMol databases were preliminarily screened based on the three pan-assay interference structures (PAINS) and one rapid elimination of swill (REOS) rules in Schrödinger Suite’s Canvas software. Meanwhile, the compounds meeting Lipinski’s rule of five were retained. The obtained compounds were then used for the virtual screening based on molecular docking. The top 10% (1706) of small molecules were clustered based on the Glide docking score, and the group size was set to 200. Finally, 158 compounds were selected for subsequent experimental examination by observing the binding pose of each ligand. All compounds were purchased from TopScience (Shanghai, China).
2.2. Molecular Docking and MD Simulations
Molecular docking was performed using the Schrödinger software package (Schrödinger, LLC, New York, NY, USA; Schrödinger, 2015). PRMT5 was selected from the Protein Data Bank (PDB ID: 4X61, https://www.rcsb.org/, accessed: 25 December 2021) and prepared using the Protein Preparation Wizard module, for example, hydrogenation and refinement of the structure, with the OPLS3 force field. Then, with EPZ015666 as the docking center, a docking grid box was generated. Next, the 3039-0164 small molecule structure file was downloaded from pubchem.ncbi.nlm.nih.gov/ and prepared in the Schrödinger software. Finally, molecular docking analysis was performed by the Glide module.
The Amber20 [29] software was used for MD simulations. For the PRMT5-SAM-3039-0164 complex, the FF19SB [30] and GAFF [31] force fields were employed, respectively. Steepest-descent/conjugate-gradient minimizations were initially applied to the systems for every 2500 steps, and then the systems were gradually heated in NVT ensemble from 0 to 310 K in 100 ps with a 10 kcal/(mol Å2) restrain on protein backbone atoms. The restrain was gradually decreased within 0.9 ns from 10 to 0.01 kcal/(mol Å2). Finally, without applying any constraints, three 200 ns MD simulations were run at 310 K and 1 atm. The Cpptraj [32] module of Amber20 was used to calculate the root mean square deviations (RMSD) and the contact analysis.
2.3. Cell Culture and Cytotoxicity Test
The non-small cell lung cancer cell line A549 (purchased from ATCC) was selected and cultured in RPMI 1640 medium containing 10% FBS (Gibco Products, Big Cabin, OK, United States) and 1% penicillin–streptomycin solution in a CO2 incubator at 37 °C containing 5% CO2. A total of 158 compounds obtained from the virtual screening were dissolved in DMSO and placed in a −40 °C environment. To rapidly identify compounds with strong inhibitory activity, the A549 cell line was first treated with 20 μM of each compound for 72 h. For the MTT assay, cells were first inoculated in 96-well 3000 cells/well microplates and incubated overnight to allow cell adhesion, and DMSO (10.0 µM) and different concentrations (2.5, 5.0, and 10.0 µM) were compared with compounds, which were studied at 24, 48, and 72 h. Then, 10 µL of MTT (5 mg/mL) was added to each for 4 h incubation at 3 °C, followed by 100 µL of acid isopropanol (10% SDS and 0.01 mol/L hydrochloric acid). Finally, the absorbance of the samples at 570 nm was measured with a microplate reader (Tecan US, Inc., Morrisville, NC, USA). Cell viability was calculated relative to untreated controls, and data conclusions were based on at least three independent experiments.
2.4. In Vitro Enzymatic Assays
PRMT5 enzymatic assay was performed as previously done by Ji et al. [33] at ChemPartner Shanghai (998 Harley Road, Pudong New Area, Shanghai, 201203, China). 3039-0164 was diluted to 10 concentrations to obtain IC50 values. PRMT5 and SAM/SAH were derived from BPS Bioscience (Cat. No. 51045) and Sigma (Cat. No. a7007-100 mg, and No. A9384-25MG). 3039-0164 was prepared as 10 mM of backup material in DMSO and diluted in DMSO to the final concentration. PRMT5 and substrate were incubated at the indicated concentrations of 3039-0164 in 384-well plates for 60 min at room temperature. Then, the remaining substrate of PRMT5 was labeled by adding the receptor and ligand solutions. The labeling procedure was reacted at room temperature for 60 min, and then the endpoint values were read using EnSpire Alpha mode. In the in vitro enzymatic assay, 1% DMSO was used as the control.
2.5. Western Blotting
Proteins were extracted from A549 cells using RIPA lysis buffer containing protease and phosphatase inhibitors. After two washes with cold PBS, the cell lysate (12,000× g, 4 °C) was centrifuged for 5 min, and the supernatant fraction was collected. The protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad, Philadelphia, PA, USA). The extracted proteins were separated in 10% SDS-PAGE gels (50 μg) and transferred to nitrocellulose (NC) membranes at 300 mA for 1 h at 4 °C. The proteins were then incubated with a primary antibody (1:1000) and a fluorescence-coupled secondary antibody (1:10,000). Additionally, eIF4E antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX, USA); antibodies against total/phospho-AKT, total/phospho-ERK, and total/phospho-mTOR were obtained from Cell Signaling Technology (Danvers, MA, USA).
2.6. Statistics
Analyzed data were expressed through mean ± standard deviation. All data were statistically analyzed using GraphPad Prism 5.0. One-way analysis of variance (ANOVA) was used for multiple comparisons. p < 0.05 was considered statistically significant.
3. Results
3.1. Screening of Candidate Compounds by Structure-Based Virtual Screening
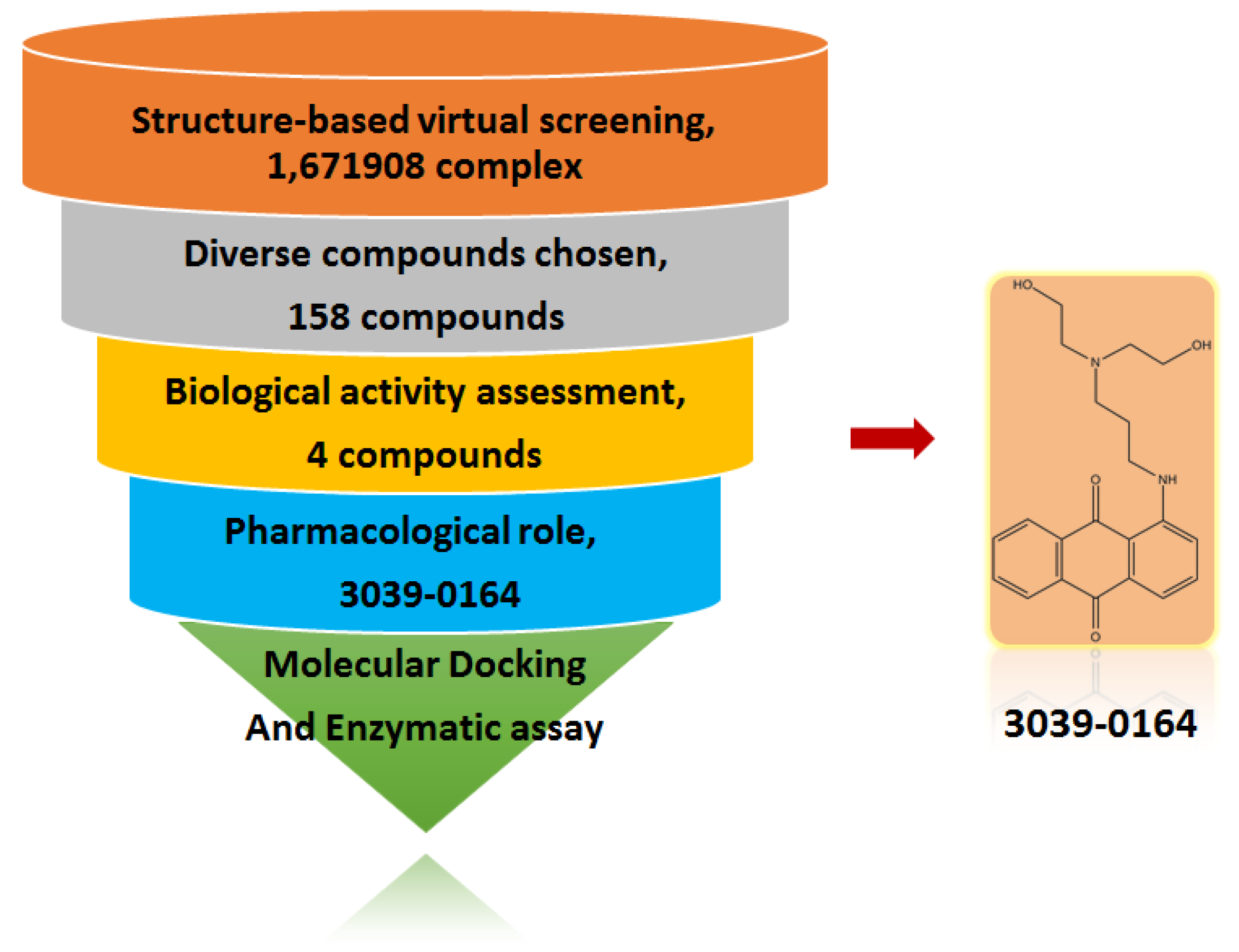
Structure-based virtual screening (SBVS) plays a key role in drug discovery research [34,35]. The method eliminates low-affinity candidate compounds by protein-ligand binding patterns and selects features of interest to further reduce the number of tested ligands for screening [36]. The SBVS workflow was indicated in Figure 1 in this paper. To identify the novel non-SAM PRMT5 inhibitors, the EPZ015666 pocket (Figure 2a) was chosen as the binding site. Since SAM has a partial interaction with EPZ015666 [16,37] and its effect is unknown, the enrichment factor was calculated after the docking preparation by setting the two complex modes with/without SAM. Additionally, the results showed that the 1% and 10% enrichment factors of the PRMT5-EPZ015666-SAM group were 44.6 and 8.7, respectively, which were higher than that of the PRMT5-EPZ015666 group (38.3 and 6.8). This shows that the presence of SAM increases the PRMT5’s affinity for the active compounds; hence, the PRMT5-EPZ015666-SAM complex was chosen for the screening.
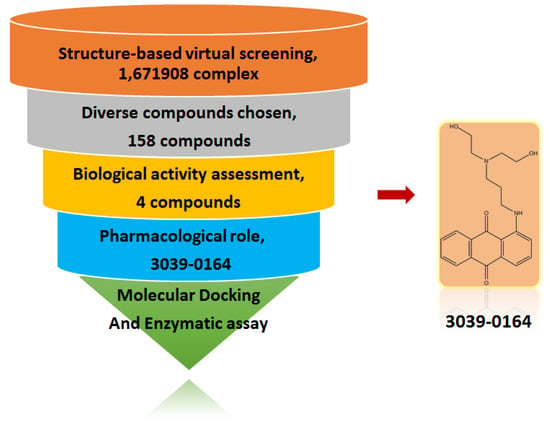
Figure 1.
Schematic diagram of identifying 3039-064 by a structure-based virtual screening method.
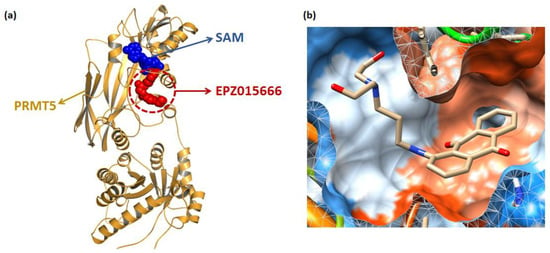
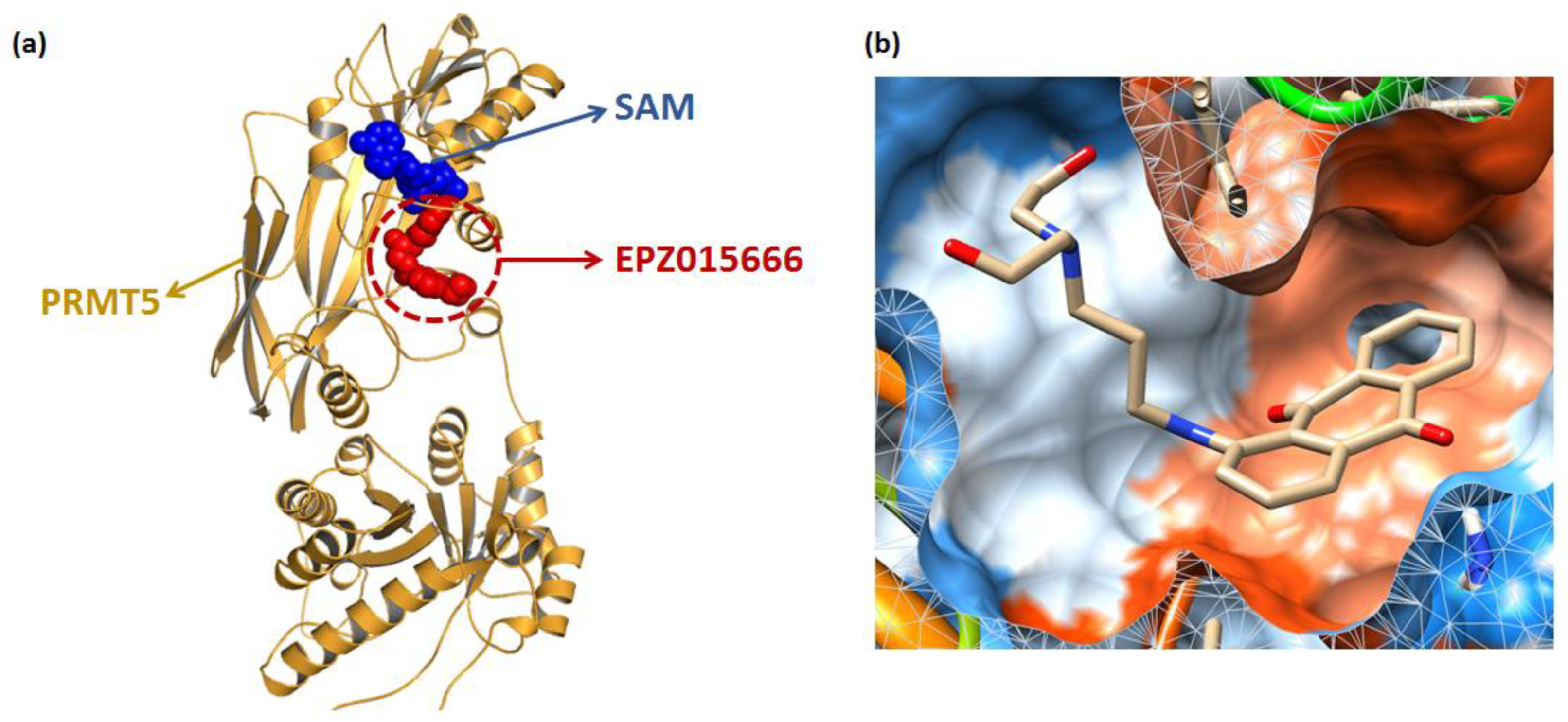
Figure 2.
3039-0164 molecular docking. (a) Distribution of two co-crystalline compounds, SAM and EPZ015666, in PRMT5. The blue spherical part is SAM, and the red spherical model indicates EPZ015666. (b) The state of 3039-0164 is bound in the PRMT5 pocket. The blue region of the pocket is the hydrophilic part, and the orange region indicates hydrophobicity.
Virtual screening was carried out for 1,671,908 chemical ligands with the previously established grid boxes. Following that, 1706 candidate compounds in the top 10% of the Glide score were submitted to 200-group cluster analysis. Compounds with more diversity, more interactions, and plausible binding patterns were chosen. Candidates with smaller molecular weights or lower ∆GMMGBSA values were preferred, when structurally similar candidates were discovered. Based on these conditions, 158 small-molecule candidates were reserved for the further biological test. We performed MTT assays on these 158 candidate compounds. Results showed that in the A549 non-small cell lung cancer cell line, 3039-0164 showed a strong inhibitory effect. Meanwhile, its ∆GMMGBSA value of 3039-0164 with PRMT5 was lower than the others (−76.2 kcal/mol). Therefore, 3039-0164 was selected for the following studies.
3.2. Molecular Mechanism of 3039-0164 Binding to PRMT5
Molecular docking [38] is a mature analytical method focusing on the analysis of protein ligands, which predicts the binding mode and affinity based on the 3D protein structure, and has recently been used to study new inhibitors [39]. The binding mode of 3039-0164 with PRMT5-SAM is shown in Figure 2b, with a docking score of −8.51 kcal/mol. In Figure 2b, it is shown that 3039-0164 is a U-shape embedded in the protein pocket in a complementary shape, with its 9, 10-anthraquinone moiety occupying the hydrophobic end of the pocket and the two hydroxyl ends near the hydrophilic region.
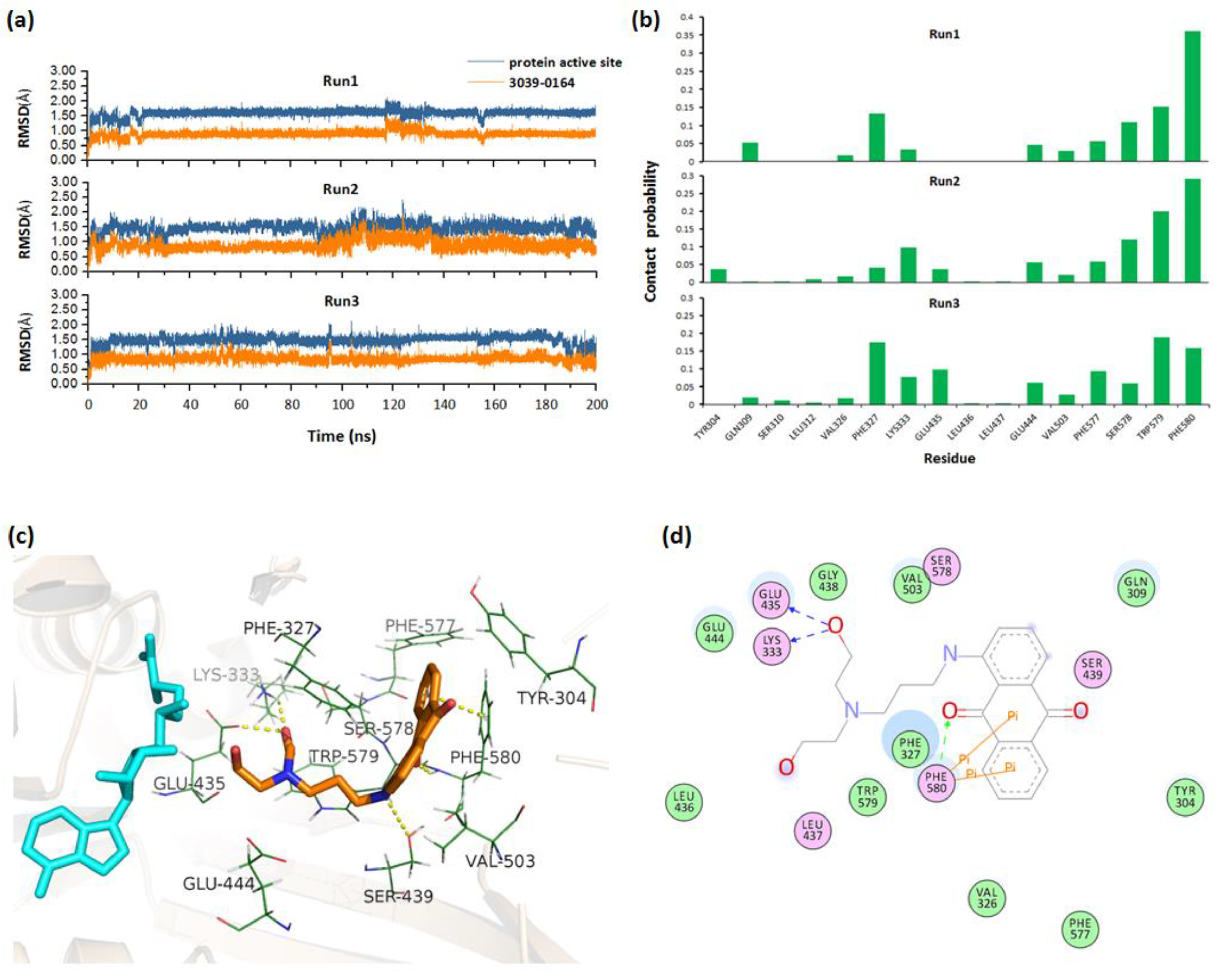
To obtain the stable complex, three parallel 200 ns molecular dynamics simulations were performed for the docked PRMT5-SAM-3039-0164 complex. By monitoring the RMSD values of 3039-0164 and PRMT5 active site along the simulation, we could see that the system fluctuated stably after 140 ns (Figure 3a). Based on the stable trajectory, the atomic contact numbers of PRMT5 active site residues with the inhibitors were calculated. Figure 3b shows that the residues Phe580/327, Trp579/304, Ser578, Glu444, and Leu333 had relatively more contact with 3039-0164, indicating their importance for protein-inhibitor binding. Then, the stable PRMT5-SAM-3039-0164 structure was extracted to analyze the detailed binding mode. Figure 3c,d shows that 3039-0164 formed strong hydrogen bonds with the residues Lys333, Glu435, and Ser439. Additionally, the π–π stacking interactions were formed between the residue Phe580 and 3039-0164. These interacting factors work together to make the inhibitor bind to PRMT5 stably.
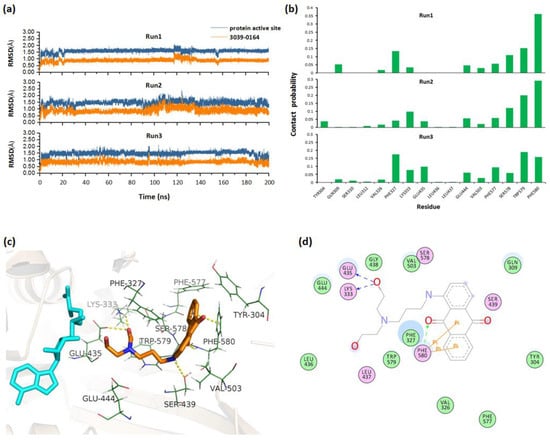
Figure 3.
Molecular mechanism of 3039-0164 binding to PRMT5 based on MD simulations. (a) RMSDs for 3039-0164 and protein active residues along three parallel 200 ns MD simulations. (b) Contact probability of residues with 3039-0164 for three trajectories. (c) The 3D binding mode of 3039-0164 and PRMT5. Cyan and orange indicate the SAM substrate and 3039-0164 inhibitor, respectively. (d) The 2D binding mode of 3039-0164 and PRMT5.
3.3. 3039-0164 Possesses Strong Cytotoxicity for A549 Cells
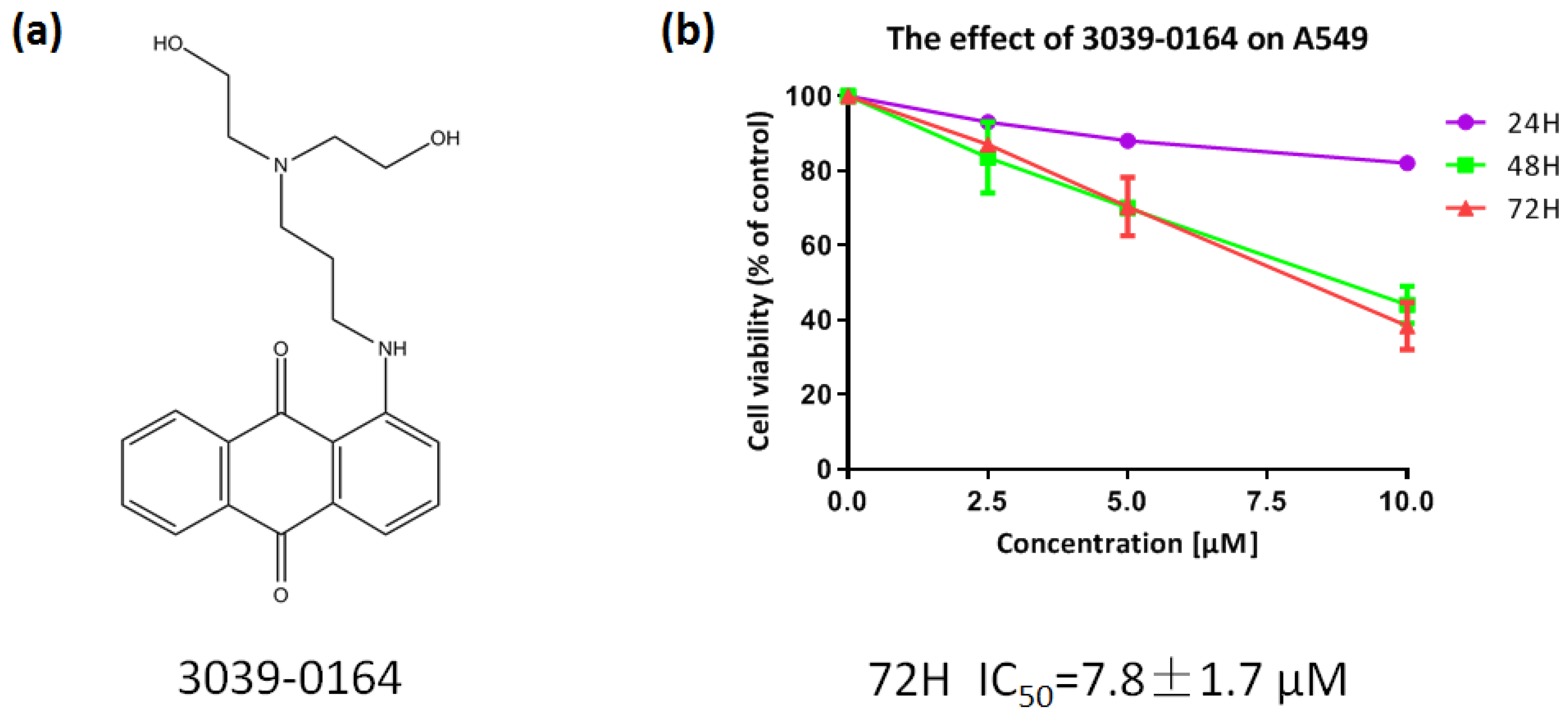
PRMT5 was shown to be highly elevated in human lung cancer cells and tissues, and inhibiting its expression decreased the proliferation of tissue-cultured lung adenocarcinoma A549 cells significantly [40,41]. As a result, we chose the A549 non-small cell lung cancer cell line to investigate the inhibitory effect of 3039-0164 (Figure 4a). After preparing the compound into a range of concentrations (2.5, 5.0, 7.5, 10.0 µM), the survival rates of A549 cells were calculated after being treated for 24, 48, and 72 h (Figure 4b). With an IC50 value of 7.8 ± 1.7 μM and concentration dependence, 3039-0164 demonstrated the most significant antiproliferative effect on A549 cells in the 72 h continuous effect group. In summary, A549 was discovered to be extremely hazardous to 3039-0164.
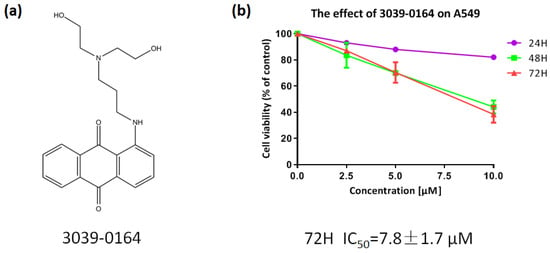
Figure 4.
(a) Molecular structure of 3039-0164. (b) The cytotoxic effect of 3039-0164 on A549 cells and its IC50 value were detected by MTT assay.
3.4. 3039-0164 Inhibits PRMT5 Methyltransferase Activity and the Expression of Its Downstream Target Genes
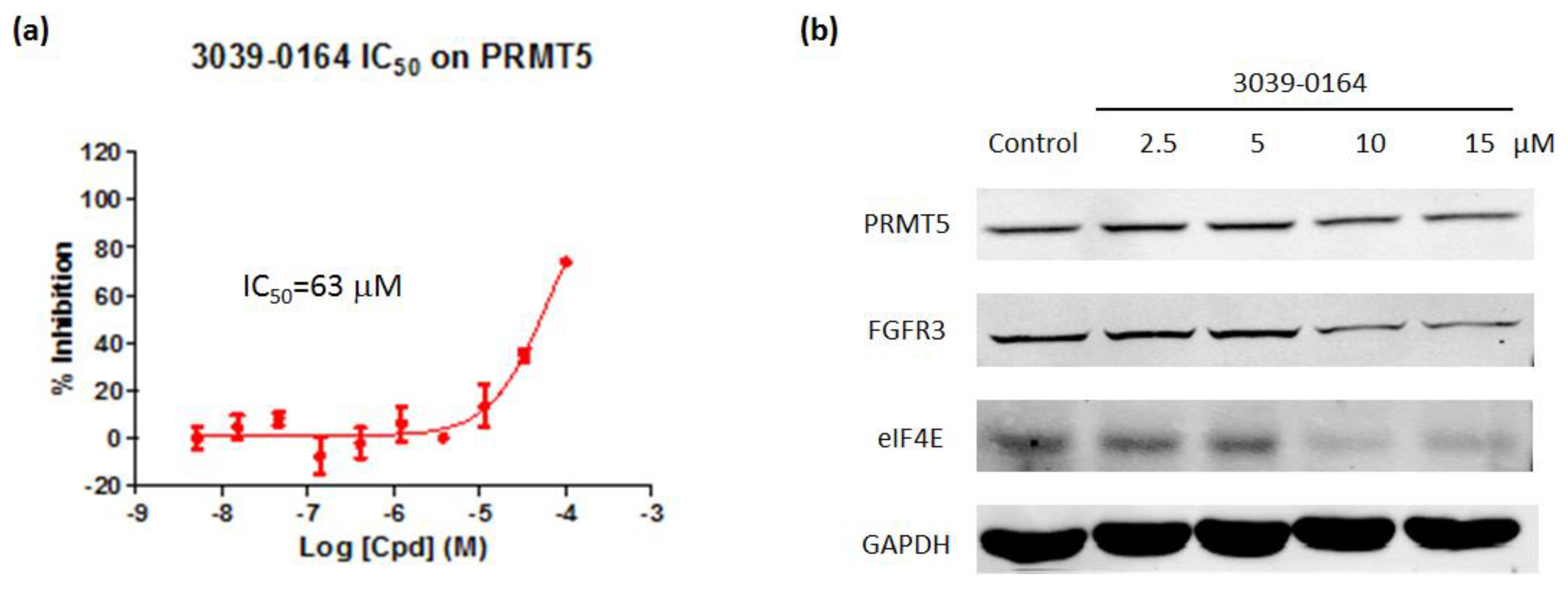
AlphaLISA assays were then used to investigate the inhibitory effect of 3039-0164 on PRMT5 activity. The results revealed that 3039-0164 inhibited this protein with an IC50 of 63 μM and a concentration-dependent impact (Figure 5a). This means that the PRMT5 methyltransferase activity was inhibited by this compound.
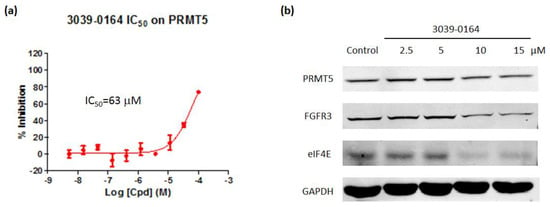
Figure 5.
(a) 3039-0164 inhibition of PRMT5 methyltransferase activity and its IC50 value. (b) Protein expression levels of the oncogenes FGFR3 and eIF4E after treatment of A549 cells with different concentrations of 3039-0164 (2.5, 5.0, 10.0, and 15 µM). Western blot analysis was performed in 24 h with at least three independent experiments. Data are expressed as mean ± SEM (n ≥ 3), with * p < 0.05 for the control (DMSO-treated group) compared with the 3039-0164-treated group.
Many target genes are involved in PRMT5-induced lung small cell carcinoma. For example, knocking down the PRMT5 gene reduces the expression of FGFR3 and eIF4E [42]. Furthermore, FGFR3 and eIF4E have been found to be actively expressed in a variety of malignancies, including lung, prostate, and colorectal tumors, which cause disease [42,43,44]. The Western blot was performed to determine whether 3039-0164 inhibited the expression of FGFR3 and eIF4E target genes. The results are shown in Figure 5. The expression levels of FGFR3 and eIF4E proteins in A549 cells were significantly reduced by 3039-0164 at 10 and 15 μM doses. This indicates that 3039-0164 downregulates the expression of two downstream target genes, FGFR3 and eIF4E, by inhibiting PRMT5.
3.5. 3039-0164 Blocks the Activation of the FGFR3 Downstream Signaling Pathway
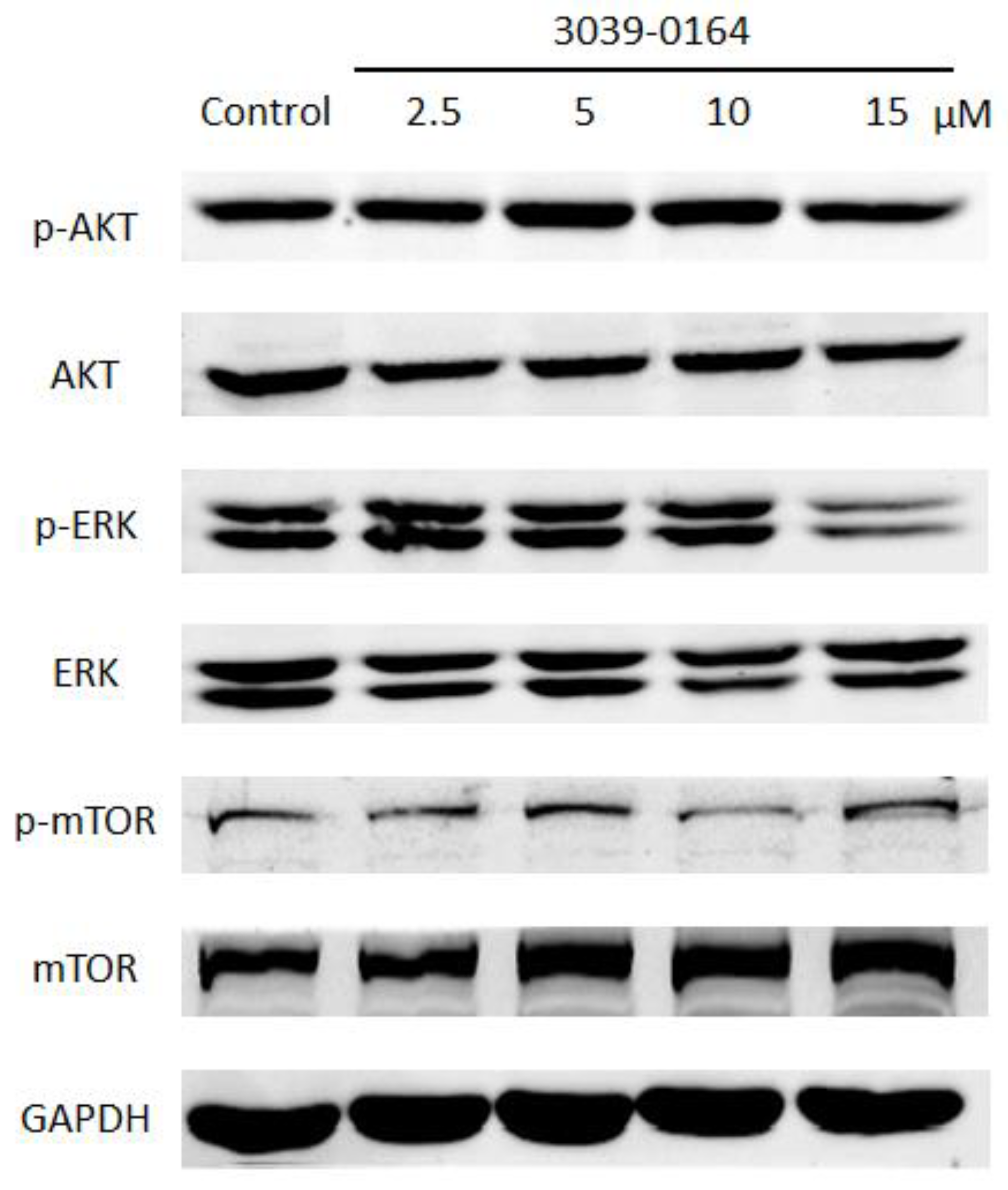
PRMT5 exhibits FGFR3-dependent regulation through the activation of AKT, ERK, and mTOR, and the activated FGFR signaling pathway plays an important role in promoting lung cancer cell proliferation [13,41,42]. We investigated whether the inhibitory effect of 3039-0164 on PRMT5 affected the activation of the AKT, ERK, and mTOR signaling pathways to learn more about the mechanism of cancer inhibition and the inhibitory effect of 3039-0164. When the concentration of 3039-0164 was 15 μM, phosphorylated AKT and ERK proteins were significantly suppressed, as shown in Figure 6. The expression of phosphorylated mTOR protein was considerably reduced at 10 μM. These findings suggest that 3039-0164 suppresses PRMT5 activity while suppressing the activation of the AKT, ERK, and mTOR signaling pathways downstream of PRMT5’s target gene FGFR3.
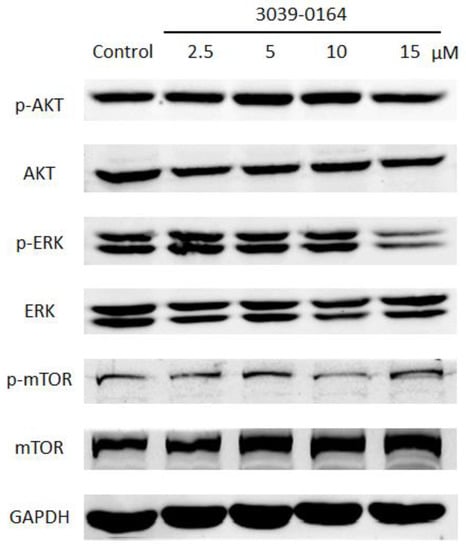
Figure 6.
Protein expression levels of AKT, ERK, and mTOR among oncogene FGFR3 downstream signaling pathway factors after treatment of A549 cells with different concentrations of 3039-0164 (2.5, 5.0, 10.0, and 15 µM).
4. Discussion
Most advanced non-small cell lung cancer patients are prone to metastasis and have a poor prognosis, resulting in a high fatality rate [45]. Epigenetics regulates gene expression, leading to individual phenotypic variation, and its reversible regulatory nature makes it a popular candidate target when it comes to epigenetic therapy for various cancers [46,47]. In a previous study, our group identified the first BPTF bromodomain inhibitor, C620-0696, through protein expression abnormalities in the epigenetic dimension of lung cancer, and demonstrated its ability to inhibit the proliferation and migration of lung cancer cells [48]. PRMT5 is a promising predictive biomarker for lung cancer patients with high expression in lung tissues, according to numerous pieces of research published in recent years [49,50,51]. In this work, a novel non-SAM PRMT5 inhibitor, 3039-0164, was discovered, and it was shown that 3039-0164 inhibited PRMT5 methyltransferase activity and the activation of PRMT5 downstream target genes and pathways.
The PRMT5 has two binding pockets, one for the SAM methyl donor-binding site and the other for the EPZ015666 substrate-binding site. The majority of PRMT5 inhibitors discovered so far compete with SAM [16,37], and their inhibitory activity is not as potent as the original SAM molecule. Here, we concentrated our efforts on identifying novel PRMT5 inhibitors capable of binding to the EPZ015666 binding pocket. The potential inhibitor 3039-0164 was identified by a structure-based virtual screening approach. Following that, hydrogen bonds and π–π stacking interactions play a key role in the PRMT5-30390164 binding. Among them, PHE580, GLU444, SER578, and PHE327 are also the key active residues around EPZ015666 when binding to PRMT5 [19]. The binding conformation similarities between 3039-0164 and EPZ015666, as well as the mechanism of PRMT5 inhibition, were demonstrated to some extent by comparing the binding modes.
In in vitro studies, three features of 3039-0164’s anticancer effect against NSCLC were investigated: PRMT5 enzyme activity; expression of two target genes, FGFR3 and elF4E; and FGFR3-mediated downstream signaling pathways. The kinase experiment revealed that 3039-0164 inhibited PRMT5, but its IC50 value was high at 63 μM, which might be due to the fact that 3039-0164 was not directly targeting to inhibit this protein, and this process could entail the role of other components, which has yet to be investigated. Additionally, we found that the identified 3039-0164 could reduce the protein expression of the oncogenes FGFR3 and eIF4E by inhibiting PRMT5. Furthermore, when the inhibitor concentration was 10 μM, Western blotting results revealed a decrease in phosphorylated mTOR protein expression, whereas phosphorylated AKT and ERKs were simultaneously lowered at a 15 μM concentration of 3039-0164. This could be due to the PI3K signaling pathway’s signaling cascade amplification effect, which causes mTOR expression instability, but the combined phosphorylated AKT and ERK inhibition results could already indicate that 3039-0164 inhibited the activation of the PI3K/AKT/mTOR and ERK pathways in NSCLC cells. For the studied 3039-0164, it contains the anthraquinone substructure, which has been considered as a privileged scaffold in the drug discovery of various diseases, especially cancers, as reviewed in recent studies [52,53]. Some compounds with anthraquinone nucleus are also on the market, such as mitoxantrone, diacerein, and amrubicin [53]. Recently, with a series of biological evaluations, Volodina et al. [54] reported a thiophene-2-carboxamide derivative of anthraquinone (compound 8) as a new potent antitumor chemotype, showing the promising anticancer effects of anthraquinone compounds. Despite this, the toxic assessment of this compound or the identification of toxic moieties, such as anthraquinone and the bis (2-hydroxyethyl) amino group, will be meaningful for drug safety. Simultaneously, in the next plan, further structural optimization is considered to improve its efficiency and selectivity for PRMT5. In the meanwhile, X-ray crystallography and biophysical techniques, such as microscale thermophoresis, isothermal titration calorimetry, or surface plasmon resonance, are expected to be used for verifying the direct targeted effects of promising inhibitors against PRMT5.
In conclusion, based on the virtual screening of a PRMT5-EPZ015666 substrate-binding pocket structure, this work finds a novel PRMT5 inhibitor, 3039-0164, with an anthraquinone moiety and demonstrates the influence of this inhibitor on cellular function through a series of biological tests. We hope that our research will spark some new ideas for improving the prognosis regimen of non-small cell lung cancer patients by optimizing the PRMT5 inhibitor design and developing drug-lead compounds.
Author Contributions
X.Y. and Q.W. designed the study. Q.W. and M.Z. carried out the experiments and data analysis. Y.C. and M.Z. wrote the manuscript. Q.W. and A.W. revised the paper. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the National Natural Science Foundation of China, (grant No. 82000074), Scientific Research Funding Project of the Education Department of Liaoning Province (jyt-dldxjc202005, LJKQZ2021167), Doctoral Start-Up Foundation of Liaoning Province (2021-BS-282), Science and Technology Innovation Fund Project of Dalian (2020JJ27SN071), and Dalian Youth Science and Technology Star Research Project (2020RQ080).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, T.; Hassanabad, M.F.; Hassanabad, A.F. Non-small cell lung cancer: Emerging molecular targeted and immunotherapeutic agents. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188636. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, H.; Barbash, O.; Creasy, C. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Pfister, S.; Ashworth, A. Marked for death: Targeting epigenetic changes in cancer. Nat. Rev. Drug Discov. 2017, 16, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wen, C.; Jiang, H.; Ma, S.; Liu, X. Protein Arginine Methyltransferase 5 Functions via Interacting Proteins. Front. Cell Dev. Biol. 2021, 9, 725301. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1786. [Google Scholar] [CrossRef]
- Wu, Q.; Schapira, M.; Arrowsmith, C.; Barsyte-Lovejoy, D. Protein arginine methylation: From enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 2021, 20, 509–530. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef]
- Blanc, R.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef]
- Gao, J.; Liu, R.; Feng, D.; Huang, W.; Huo, M.; Zhang, J.; Leng, S.; Yang, Y.; Yang, T.; Yin, X.; et al. Snail/PRMT5/NuRD complex contributes to DNA hypermethylation in cervical cancer by TET1 inhibition. Cell. Death Differ. 2021, 28, 2818–2836. [Google Scholar] [CrossRef]
- Shen, Y.; Gao, G.; Yu, X.; Kim, H.; Wang, L.; Xie, L.; Schwarz, M.; Chen, X.; Guccione, E.; Liu, J.; et al. Discovery of First-in-Class Protein Arginine Methyltransferase 5 (PRMT5) Degraders. J. Med. Chem. 2020, 63, 9977–9989. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, Y.; Hu, X.; Zheng, Y.; Chen, X. Targeting PRMT5/Akt signalling axis prevents human lung cancer cell growth. J. Cell. Mol. Med. 2019, 23, 1333–1342. [Google Scholar] [CrossRef]
- Sheng, X.; Wang, Z. Protein arginine methyltransferase 5 regulates multiple signaling pathways to promote lung cancer cell proliferation. BMC Cancer 2016, 16, 567. [Google Scholar] [CrossRef]
- Yin, S.; Liu, L.; Brobbey, C.; Palanisamy, V.; Ball, L.; Olsen, S.; Ostrowski, M.; Gan, W. PRMT5-mediated arginine methylation activates AKT kinase to govern tumorigenesis. Nat. Commun. 2021, 12, 3444. [Google Scholar] [CrossRef]
- Bonday, Z.Q.; Cortez, G.S.; Grogan, M.J.; Antonysamy, S.; Weichert, K.; Bocchinfuso, W.P.; Li, F.; Kennedy, S.; Li, B.; Mader, M.M.; et al. LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity. ACS Med. Chem. Lett. 2018, 9, 612–617. [Google Scholar] [CrossRef]
- Janisiak, J.; Kopytko, P.; Tkacz, M.; Rogińska, D.; Perużyńska, M.; Machaliński, B.; Pawlik, A.; Tarnowski, M. Protein Arginine Methyltransferase (PRMT) Inhibitors-AMI-1 and SAH Are Effective in Attenuating Rhabdomyosarcoma Growth and Proliferation in Cell Cultures. Int. J. Mol. Sci. 2021, 22, 8023. [Google Scholar] [CrossRef]
- Mao, R.; Shao, J.; Zhu, K.; Zhang, Y.; Ding, H.; Zhang, C.; Shi, Z.; Jiang, H.; Sun, D.; Duan, W.; et al. Potent, Selective, and Cell Active Protein Arginine Methyltransferase 5 (PRMT5) Inhibitor Developed by Structure-Based Virtual Screening and Hit Optimization. J. Med. Chem. 2017, 60, 6289–6304. [Google Scholar] [CrossRef]
- Bajbouj, K.; Ramakrishnan, R.K.; Saber-Ayad, M.; Omar, H.A.; Sharif-Askari, N.S.; Shafarin, J.; Elmoselhi, A.B.; Ihmaid, A.; Ali, S.A.; Alalool, A.; et al. PRMT5 Selective Inhibitor Enhances Therapeutic Efficacy of Cisplatin in Lung Cancer Cells. Int. J. Mol. Sci. 2021, 22, 6131. [Google Scholar] [CrossRef]
- Sachamitr, P.; Ho, J.; Ciamponi, F.; Ba-Alawi, W.; Coutinho, F.; Guilhamon, P.; Kushida, M.; Cavalli, F.; Lee, L.; Rastegar, N.; et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat. Commun. 2021, 12, 979. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, J.; Li, Y.; Huang, J.; Jiang, Z.; Wang, Y.; Liu, L.; Leung, E.L.H.; Yao, X. Identification of a Novel Protein Arginine Methyltransferase 5 Inhibitor in Non-small Cell Lung Cancer by Structure-Based Virtual Screening. Front. Pharmacol. 2018, 9, 173. [Google Scholar] [CrossRef]
- Brehmer, D.; Beke, L.; Wu, T.; Millar, H.; Moy, C.; Sun, W.; Mannens, G.; Pande, V.; Boeckx, A.; van Heerde, E.; et al. Discovery and Pharmacological Characterization of JNJ-64619178, a Novel Small-Molecule Inhibitor of PRMT5 with Potent Antitumor Activity. Mol. Cancer Ther. 2021, 20, 2317–2328. [Google Scholar] [CrossRef]
- Chan-Penebre, E.; Kuplast, K.; Majer, C.; Boriack-Sjodin, P.; Wigle, T.; Johnston, L.; Rioux, N.; Munchhof, M.; Jin, L.; Jacques, S.; et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015, 11, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shao, X.; Zhao, X.; Ji, Y.; Liu, X.; Li, P.; Zhang, M.; Wang, Q. Targeting protein arginine methyltransferase 5 in cancers: Roles, inhibitors and mechanisms. Biomed. Pharmacother. 2021, 144, 112252. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Liu, F.; Veazey, K.; Gao, G.; Das, P.; Neves, L.; Lin, K.; Zhong, Y.; Lu, Y.; Giuliani, V.; et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia 2018, 32, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Song, C.; Wang, X.; Zhang, G.; Wang, Y.; Jiang, X.; Sun, Q.; Huang, L.; Xiang, R.; Hu, Y.; et al. Discovery of New SIRT2 Inhibitors by Utilizing a Consensus Docking/Scoring Strategy and Structure-Activity Relationship Analysis. J. Chem. Inf. Model 2017, 57, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; He, J.; Mao, L.; Zhang, Y.; Cui, W.; Duan, S.; Jiang, A.; Gao, Y.; Sang, Y.; Huang, G. EPZ015666, a selective protein arginine methyltransferase 5 (PRMT5) inhibitor with an antitumour effect in retinoblastoma. Exp. Eye Res. 2021, 202, 108286. [Google Scholar] [CrossRef]
- Schrödinger. Maestro Version 10.2., New York. 2015. Available online: https://www.schrodinger.com/products/maestro (accessed on 20 January 2022).
- Duncan, K.W.; Rioux, N.; Boriack-Sjodin, P.A.; Munchhof, M.J.; Reiter, L.A.; Majer, C.R.; Jin, L.; Johnston, L.D.; Chan-Penebre, E.; Kuplast, K.G.; et al. Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666. ACS Med. Chem. Lett. 2016, 7, 162–166. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, I.T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. Amber2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Tian, C.; Kasavajhala, K.; Belfon, K.; Raguette, L.; Huang, H.; Migues, A.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.; Caldwell, J.; Kollman, P.; Case, D. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Roe, D.; Cheatham, T. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Ji, S.; Ma, S.; Wang, W.; Huang, S.; Wang, T.; Xiang, R.; Hu, Y.; Chen, Q.; Li, L.; Yang, S. Discovery of selective protein arginine methyltransferase 5 inhibitors and biological evaluations. Chem. Biol. Drug Des. 2017, 89, 585–598. [Google Scholar] [CrossRef]
- Sahihi, M.; Gaci, F.; Navizet, I. Identification of new alpha-synuclein fibrillogenesis inhibitor using in silico structure-based virtual screening. J. Mol. Graph. Model 2021, 108, 108010. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Y.; Zhong, L.; Yu, N.; Ouyang, L.; Fang, R.; Wang, Y.; He, Q. Structure-based discovery of neoandrographolide as a novel inhibitor of Rab5 to suppress cancer growth. Comput. Struct. Biotechnol. J. 2020, 18, 3936–3946. [Google Scholar] [CrossRef]
- Crampon, K.; Giorkallos, A.; Deldossi, M.; Baud, S.; Steffenel, L. Machine-learning methods for ligand-protein molecular docking. Drug Discov. Today 2022, 27, 151–164. [Google Scholar] [CrossRef]
- Smil, D.; Eram, M.; Li, F.; Kennedy, S.; Szewczyk, M.; Brown, P.; Barsyte-Lovejoy, D.; Arrowsmith, C.; Vedadi, M.; Schapira, M. Discovery of a Dual PRMT5-PRMT7 Inhibitor. ACS Med. Chem. Lett. 2015, 6, 408–412. [Google Scholar] [CrossRef]
- Sulimov, V.; Kutov, D.; Sulimov, A. Advances in Docking. Curr. Med. Chem. 2019, 26, 7555–7580. [Google Scholar] [CrossRef]
- He, Z.; Jiao, H.; An, Q.; Zhang, X.; Zengyangzong, D.; Xu, J.; Liu, H.; Ma, L.; Zhao, W. Discovery of novel 4-phenylquinazoline-based BRD4 inhibitors for cardiac fibrosis. Acta Pharm. Sin. B 2022, 12, 291–307. [Google Scholar] [CrossRef]
- Cai, C.; Gu, S.; Yu, Y.; Zhu, Y.; Zhang, H.; Yuan, B.; Shen, L.; Yang, B.; Feng, X. PRMT5 Enables Robust STAT3 Activation via Arginine Symmetric Dimethylation of SMAD7. Adv. Sci. 2021, 8, 2003047. [Google Scholar] [CrossRef]
- Gu, Z.; Gao, S.; Zhang, F.; Wang, Z.; Ma, W.; Davis, R.; Wang, Z. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem. J. 2012, 446, 235–241. [Google Scholar] [CrossRef]
- Zhang, B.; Dong, S.; Zhu, R.; Hu, C.; Hou, J.; Li, Y.; Zhao, Q.; Shao, X.; Bu, Q.; Li, H.; et al. Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3. Oncotarget 2015, 6, 22799–22811. [Google Scholar] [CrossRef]
- D’Abronzo, L.; Ghosh, P. eIF4E Phosphorylation in Prostate Cancer. Neoplasia 2018, 20, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Qi, F.; Lu, S.; Li, Y.; Han, W. Nicotine upregulates FGFR3 and RB1 expression and promotes non-small cell lung cancer cell proliferation and epithelial-to-mesenchymal transition via downregulation of miR-99b and miR-192. Biomed. Pharmacother. 2018, 101, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; An, X.; Xu, J.; Wang, Y.; Liu, L.; Leung, E.L.; Yao, X. Classical molecular dynamics and metadynamics simulations decipher the mechanism of CBP30 selectively inhibiting CBP/p300 bromodomains. Org. Biomol. Chem. 2018, 16, 6521–6530. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Shao, X.; Leung, E.L.H.; Chen, Y.; Yao, X. Selectively targeting individual bromodomain: Drug discovery and molecular mechanisms. Pharmacol. Res. 2021, 172, 105804. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Q.; Leung, E.L.H.; Li, Y.; Fan, X.; Wu, Q.; Yao, X.; Liu, L. Compound C620-0696, a new potent inhibitor targeting BPTF, the chromatin-remodeling factor in non-small-cell lung cancer. Front. Med. 2020, 14, 60–67. [Google Scholar] [CrossRef]
- Ibrahim, R.; Matsubara, D.; Osman, W.; Morikawa, T.; Goto, A.; Morita, S.; Ishikawa, S.; Aburatani, H.; Takai, D.; Nakajima, J.; et al. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum. Pathol. 2014, 45, 1397–1405. [Google Scholar] [CrossRef]
- Jing, P.; Zhao, N.; Ye, M.; Zhang, Y.; Zhang, Z.; Sun, J.; Wang, Z.; Zhang, J.; Gu, Z. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett. 2018, 427, 38–48. [Google Scholar] [CrossRef]
- Xiao, W.; Chen, X.; Liu, L.; Shu, Y.; Zhang, M.; Zhong, Y. Role of protein arginine methyltransferase 5 in human cancers. Biomed. Pharmacother. 2019, 114, 108790. [Google Scholar] [CrossRef]
- Baqi, Y. Anthraquinones as a privileged scaffold in drug discovery targeting nucleotide-binding proteins. Drug Discov. Today 2016, 21, 1571–1577. [Google Scholar] [CrossRef]
- Siddamurthi, S.; Gutti, G.; Jana, S.; Kumar, A.; Singh, S. Anthraquinone: A promising scaffold for the discovery and development of therapeutic agents in cancer therapy. Future Med. Chem. 2020, 12, 1037–1069. [Google Scholar] [CrossRef]
- Volodina, Y.; Tikhomirov, A.; Dezhenkova, L.; Ramonova, A.; Kononova, A.; Andreeva, D.; Kaluzhny, D.; Schols, D.; Moisenovich, M.; Shchekotikhin, A.; et al. Thiophene-2-carboxamide derivatives of anthraquinone: A new potent antitumor chemotype. Eur. J. Med. Chem. 2021, 221, 113521. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).