Abstract
Previously reported (S)-5′-C-aminopropyl-2′-arabinofluoro-thymidine (5ara-T) and newly synthesized (S)-5′-C-aminopropyl-2′-arabinofluoro-5-methyl-cytidine (5ara-MeC) analogs were incorporated into a series of antisense gapmers containing multiple phosphorothioate (PS) linkages and locked nucleic acids (LNAs) in their wing regions. The functional properties of the gapmers were further evaluated in vitro. Compared with the positive control, for the LNA-wing full PS gapmer without 5ara modification, it was revealed that each gapmer could have a high affinity and be thermally stable under biological conditions. Although the cleavage pattern was obviously changed; gapmers with 5ara modification could still efficiently activate E. coli RNase H1. In addition, incorporating one 5ara modification into the two phosphodiester linkages could reverse the destabilization in enzymatic hydrolysis caused by fewer PS linkages. In vitro cellular experiments were also performed, and the Lipofectamine® 2000 (LFA)+ group showed relatively higher antisense activity than the LFA-free group. KN5ara-10, which contains fewer PS linkages, showed similar or slightly better antisense activity than the corresponding full PS-modified KN5ara-3. Hence, KN5ara-10 may be the most promising candidate for KNTC2-targeted cancer therapy.
1. Introduction
Antisense oligonucleotides (ASOs), composed of approximately 20-bp DNA-like nucleotides, are classified as a kind of mRNA-targeted oligonucleotide therapeutics [1]. Since the first ASO therapeutic, fomivirsen, was approved by the U.S. Food and Drug Administration (FDA) in 1998, nine ASO therapeutics have been approved [2,3,4,5,6,7,8,9,10]. There is no doubt that further research on ASOs will be developed. ASOs can be divided into the following two types depending on the antisense mechanism: the ribonuclease H (RNase H)-dependent type and the splice-switching type [11,12,13]. Both types of ASOs need to be taken up into cytoplasm or nucleoplasm for binding with the targeted mRNAs or pre-mRNAs, which would raise several challenges for the clinical application of ASOs. For example, nucleotides experience difficulty passing through cellular or nuclear membranes because of the negative charges on their phosphodiester (PO) linkages. Moreover, natural DNA strands are quickly degraded via nuclease-mediated hydrolysis inside the plasm, resulting in low pharmacological effects. To overcome these challenges, chemically modified nucleosides have been developed and utilized in approved ASOs.
Phosphorothioate (PS) linkages, which contain sulphur substitutions of non-bridging oxygens at each PO linkage, are the most popular chemical modification to improve the properties of approved ASOs [3,4,5,6,7,8,9,10]. Due to the high degree of similarity between sulfur and oxygen atoms, PS linkages would not obstruct the recognition of ASO/RNA duplexes by RNase H and might even promote the RNase H-mediated cleavage mechanism [14]. PS linkages also help to stabilize ASOs from nucleolytic degradation. In addition, the anionic sulfur in PS linkages could tightly interact with multiple proteins by electrostatic and hydrophobic interactions [15]. Therefore, the application of PS linkages could enhance the membrane permeability of ASOs via binding to membrane proteins. However, the presence of apoptosis of off-target cells, relating to cytotoxicity, was reported in some cases, possibly due to unfavorable interactions between PS linkages and paraspeckle proteins [15]. As general strategies, coordination with other chemical modifications and reducing the content of PS linkages might be beneficial for developing more effective PS-ASOs with less side effects [16,17,18,19,20].
In previous studies, our group designed and synthesized various chemically modified nucleoside analogs containing an aminoalkyl sidechain, which is expected to strengthen nuclease resistance and improve the intracellular internalization of oligonucleotides [21,22,23,24,25,26,27]. As a result of evaluating (S)-5ʹ-C-aminopropyl- (5deo) and (S)-5ʹ-C-aminopropyl-2ʹ-arabinofluoro- (5ara) modified ASOs, it was revealed that ASOs with several 5ara modifications could significantly enhance the nuclease resistance and show a superior ability to activate the RNase H-dependent antisense mechanism compared with the 5deo-modified ones, although the cleavage of the complementary RNA strands would be obviously impeded compared with the natural DNAs [27].
In this study, we focus on a special formation, named “gapmer”, composed of chemically modified wing regions for high RNA-binding affinity and robust nuclease resistance, and a DNA-based gap region for efficient RNase H-dependent antisense mechanism [3,6,7].
We also focus on locked nucleic acids (LNAs), a class of chemically modified nucleosides, which contain a methylene bridge connecting the 2′ oxygen and 4′ carbon of nucleoside [28,29]. LNA demonstrated high RNA-binding affinity because it locks the ribose conformation into a C3′-endo type, which is beneficial for the A-type formation of ASO/RNA duplex, while the incorporation of LNAs inside oligonucleotides might suppress the recognition of RNase H. Therefore, LNA is often used in the wing regions of gapmers but not in the gap regions. However, even superior properties have been proved in the laboratory level, due to a lack of clinical knowledge, as there are not any cases yet approving LNA-containing ASOs. In this study, an LNA-wing antisense gapmer was selected as an excellent positive control, and the further improvements resulting from the collaboration with 5ara modification were evaluated, as well as the expedition of LNA into clinical usage.
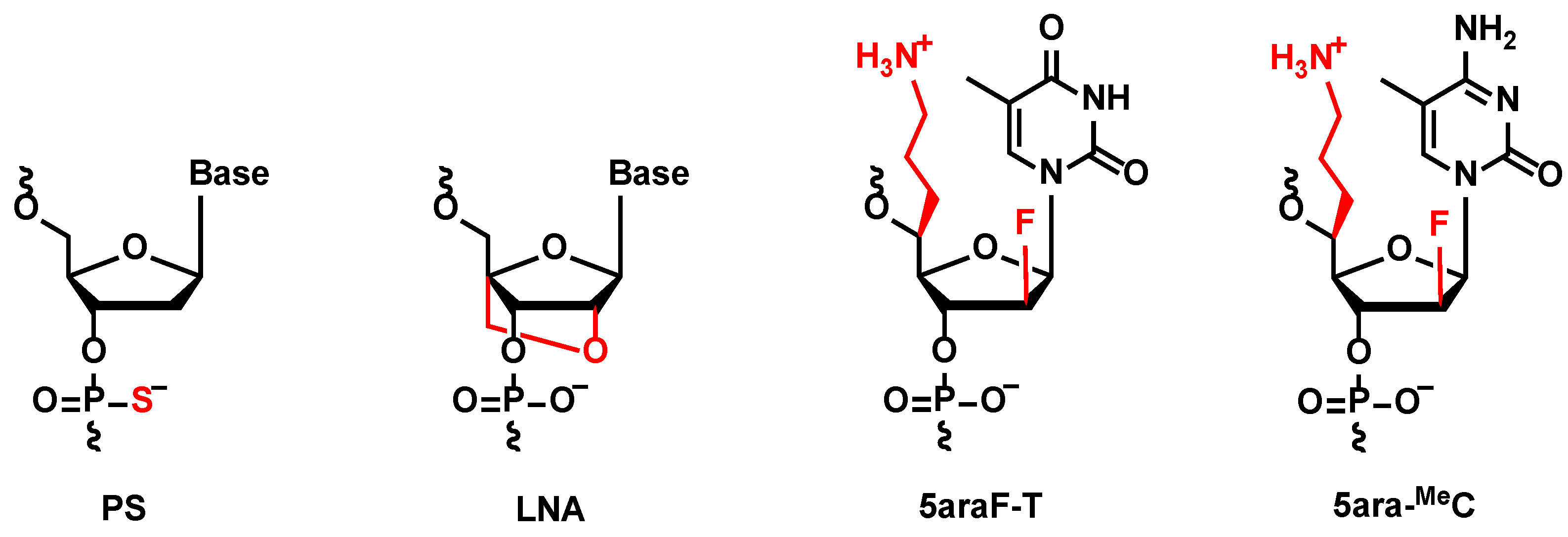
The KNTC2 (Kinetochore associated 2) gene, referred as to KNTC2, was selected to be the target gene in this study, which is known to be highly expressed in various cancer cells. Selective knockdown of KNTC2 was reported as a promising treatment for cancers [30,31,32]. Based on previous findings, we designed a series of KNTC2-targeted antisense gapmers with LNA-wings and 5ara modifications (described as “KN5ara gapmers”) in this study. Each structure of these chemical modifications is shown in Figure 1. The following sections outline the synthesis of KN5ara gapmers by incorporating the reported (S)-5ʹ-C-aminopropyl-2ʹ-arabinofluoro-thymidine (5ara-T) and newly synthesized (S)-5ʹ-C-aminopropyl-2ʹ-arabinofluoro-5-methyl-cytidine (5ara-MeC) analogs, followed by the in vitro evaluations of their functional properties.
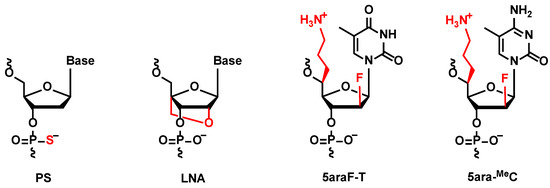
Figure 1.
Chemical structure of phosphorothioate (PS), locked nucleic acid (LNA), (S)-5ʹ-C-aminopropyl-2ʹ-arabinofluoro-thymidine (5ara-T), and (S)-5ʹ-C-aminopropyl-2ʹ-arabinofluoro-5-methyl-cytidine (5ara-MeC).
2. Results and Discussion
2.1. Oligonucleotide Synthesis
The synthesis of phosphoramidite corresponding to 5ara-MeC is shown in Scheme S1 in the Supporting Information. The modified nucleoside analogs 5ara-T and 5ara-MeC were incorporated into a series of LNA-wing antisense gapmers utilizing a DNA/RNA synthesizer via the solid-phase phosphoramidite method. After synthesis, to prevent the additional reaction of acrylonitrile with 5ʹ-C-aminopropyl groups, the controlled-pore glass (CPG) beads were treated with 10% dimethylamine in MeCN at room temperature for 5 min, followed by rinsing with MeCN to selectively remove cyanoethyl groups. The gapmers were then cleaved from CPG beads and deprotected by treatment with a concentrated NH3 solution for 12 h at 55 °C. The corresponding RNA oligomers used in this study were prepared with a DNA/RNA synthesizer. After synthesis, in contrast to antisense gapmers, the RNA oligomers were cleaved from CPG beads and deprotected by treatment with concentrated NH3 solution/40% methylamine (1:1, v/v) for 10 min at 65 °C. Then, 2ʹ-O-TBDMS groups in RNA oligomers were removed using Et3N∙3HF (125 µL) in DMSO (100 µL) for 1.5 h at 65 °C. The reaction was quenched with a 0.1 M TEAA buffer (pH 7.0) and the mixture was desalted using a Sep-Pak C18 cartridge. The modified antisense gapmers and RNA oligomers were finally purified by 20% denaturing polyacrylamide gel electrophoresis (PAGE) containing 7 M urea. The sequences of the oligonucleotides used in this study are shown in Table 1 and Table S1 (Supplementary Material).

Table 1.
Sequence of each gapmer and Tm values of duplexes containing these gapmers.
2.2. RNA-Binding Affinity
The accurate binding of ASOs to target mRNA is necessary for RNase H recognition and the following antisense mechanism. As reported before, although PS linkages negatively affected thermal stability, LNAs could significantly increase RNA binding affinity and a single (S)-5ʹ-C-Aminopropyl-2ʹ-arabinofluoro modification caused few changes to the 50% melting temperature (Tm) values of ASO/RNA duplexes [27,28,29]. Therefore, we hypothesized that the series of KN5ara gapmers would have sufficient RNA binding affinity for therapeutic application. In this study, each KN5ara gapmer was mixed in the same volume of cRNA-1, and then annealed to form ASO/RNA duplexes. Temperature-induced melting was measured by ultraviolet (UV) spectroscopy in a 10 mM sodium phosphate buffer (pH 7.0) containing 100 mM NaCl, and Tm values were obtained from melting curves using the standard method. Each ΔTm was calculated from [Tm (duplex containing each KN5ara gapmers) − Tm (duplex containing KN-pos)].
As shown in Table 1, all KN5ara gapmers maintained a stable duplex structure with complementary RNA strands at 37 °C. The incorporation of 5ara-modified analogs in the gap region showed moderate effects on Tm values, while the replacement of LNAs in the wing region with 5ara-T or 5ara-MeC resulted in thermal destabilization. These results are consistent with previous studies showing that the thermal stability of 5ara modification is comparable to that of natural DNA but lower than that of LNA [27]. Furthermore, the Tm value of KN5ara-9 was similar to the addition of KN5ara-7 and KN5ara-8, indicating that the continuous introduction of modified analogs might not have an additional impact on thermal stability. The gapmers with fewer PS linkages (KN5ara-10 and KN5ara-11) were more stable than the corresponding full PS gapmers (KN5ara-3 and KN5ara-6).
2.3. The Ability for E. coli RNase H1 Activation
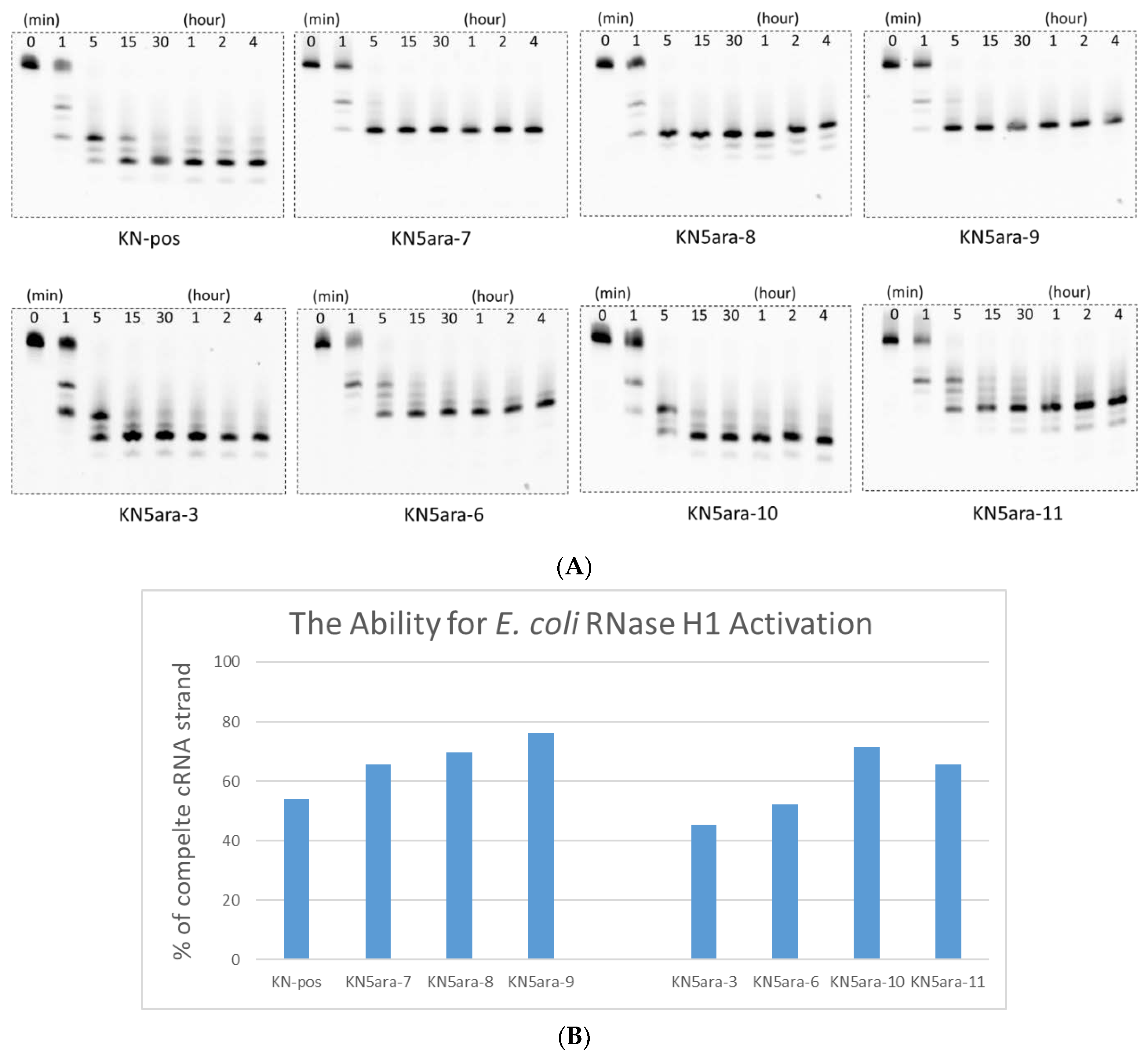
Before the in vitro cell experiments, we established a simple enzymatic reaction system using RNase H1 purified from E. coli to determine whether RNase H-mediated cleavage could be activated by KN5ara gapmers [33,34]. The KN5ara gapmers used in this experiment were mixed with fluorescein-labeled cRNA-2 at 1:5 before annealing. These duplexes were dissolved in a buffer containing 50 mM Tris–HCl (pH 8.0), 75 mM KCl, 3 mM MgCl2, and 10 mM dithiothreitol. Diluted E. coli RNase H1 solution (60 unit/L in H2O) was then added, and the mixture was incubated at 37 °C for the required time (0, 1, 5, 15, and 30 min and 1, 2, and 4 h). The aliquots were analyzed using 20% denaturing PAGE and then quantified using a luminescent image analyzer LAS-4000 (Fujifilm). As shown in Figure 2, even though the initial reaction velocity showed a slight change, it was confirmed that the complete cRNA strand was almost cleaved in KN5ara gapmers as well as in the positive control KN-pos after a 5 min reaction. Despite differences in each cleavage pattern, a single 5ara modification moderately affected the cleavage mechanism of E. coli RNase H1, which was consistent with previous reports, because of the remaining recognition portions [27].
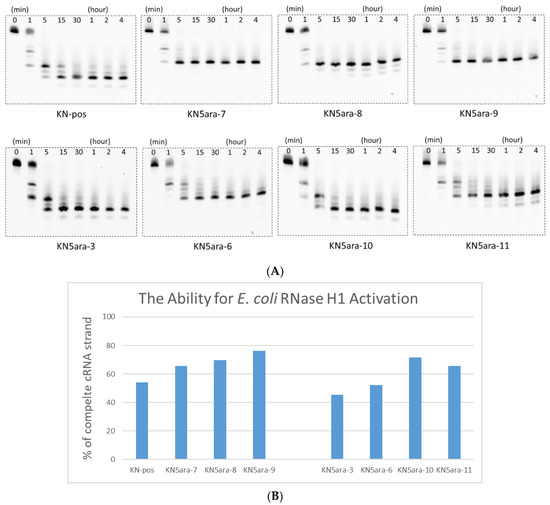
Figure 2.
PAGE analysis of ASO/RNA duplexes treated in a buffer containing 50 mM Tris–HCl (pH 8.0), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, and RNase H1 from E. coli. Sequence of complementary RNA strand (cRNA-2) is shown in Table S1. All duplexes were incubated in a buffer containing 50 mM Tris–HCl (pH 8.0), 75 mM KCl, 3 mM MgCl2, and 10 mM dithiothreitol, and then diluted RNase H1 solution (60 unit/L in H2O) was added; subsequently, the mixture was incubated at 37 °C for the required time. The reaction mixtures at various incubation times (0, 1, 5, 15, and 30 min and 1, 2, and 4 h) were analyzed with 20% PAGE and quantified with a luminescent image analyzer LAS-4000 (Fujifilm). (A) PAGE images obtained from luminescent analysis. (B) Percentage of complete cRNA strands remaining at 1 min, analyzed with ImageJ.2.4. Nuclease Resistance.
Notably, not all KN5ara gapmers were evaluated in this assay. Since the recognition of E. coli RNase H1 would begin with a few nucleotides from the 5ʹ-terminal of ASO, KN5ara-1 and -2, in which the 5ara modification was inserted into 5ʹ-gap region, it might activate E. coli RNase H1 similarly with KN-pos. Meanwhile, as there are no significant differences for KN5ara-6 and -7, KN5ara-4 and -5 were found to be similar to the others. The antisense activity of all KN5ara gapmers is directly evaluated in the following section.
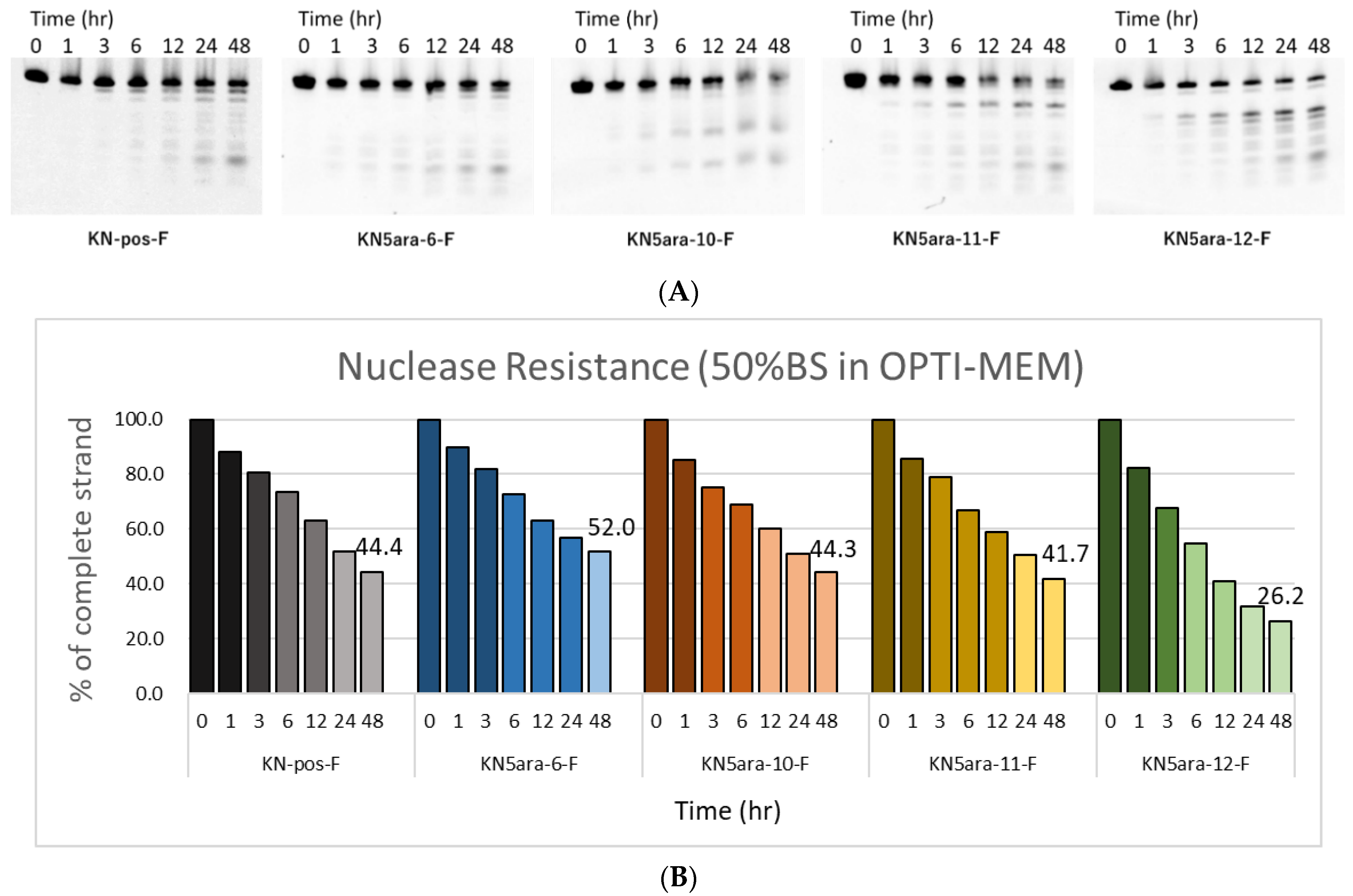
ASOs containing multiple 5ara modifications showed extremely high enzyme tolerance in a 3% bovine serum (BS) experiment system [27]. LNAs and PS linkages are also expected to improve the stability of KN5ara gapmers during enzyme-mediated hydrolysis. However, PS linkages may lead to undesirable apoptosis because of their high affinity with plasma proteins, such as paraspeckle proteins [15]. We hypothesized that the application of the 5ara modification could reduce the number of PS linkages while maintaining sufficient nuclease resistance. In this research, a series of fluorescein-labeled KN5ara gapmers were synthesized (Table S1), including positive control (KN-pos-F), and gapmers with full-PS (KN5ara-6-F) or with less PS linkages (KN5ara-10/11-F). For comparison, the gapmer without any 5ara modification corresponding to KN5ara-11-F was prepared as well. The gapmers described above were dissolved in OPTI-MEM and incubated with 50% BS at 37 °C. During incubation, aliquots from the reactions were taken for the required time (0, 1, 3, 6, 12, 24 and 48 h), then analyzed with 20% PAGE containing 7 M urea and quantified using the Luminescent Image analyzer LAS-4000 (Fujifilm). The results are shown in Figure 3.
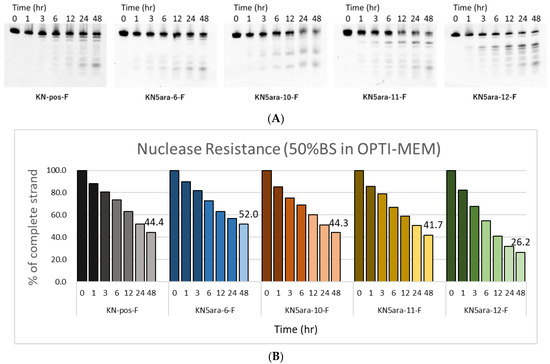
Figure 3.
PAGE analysis of single-stranded KN5ara gapmers treated with OPTI-MEM containing 50% bovine serum (BS). Fluorescein-labeled KN5ara gapmers (300 pmol) were incubated in OPTI-MEM containing 50% BS. The reaction mixtures at various incubation time (0, 1, 3, 6, 12, 24, and 48 h) were analyzed by 20% PAGE containing 7 M urea and quantified by luminescent image analyzer LAS-4000 (Fujifilm). (A) PAGE images obtained from luminescent analysis. (B) Percentage of complete gapmer strands remaining at each time point, analyzed via ImageJ.
After 48 h incubation, approximately 44% of the KN-pos-F strands remained, while only 26% of KN5ara-12-F strands persisted, apparently owing to the decrease in the two PS linkages. A comparison of KN-pos-F and KN5ara-6-F showed that 5ara modification could further increase the nuclease resistance of gapmers, resulting in a half-life of >48 h with 50% BS treatment. Meanwhile, 44% and 42% of the complete strands of KN5ara-10-F and KN5ara-11-F remained, respectively, indicating the same nuclease resistance as KN-pos-F. It is presumed that incorporating one 5ara modification into the two PO linkages could reverse destabilization in enzymatic hydrolysis, which is caused by fewer PS linkages.
2.4. Antisense Activity
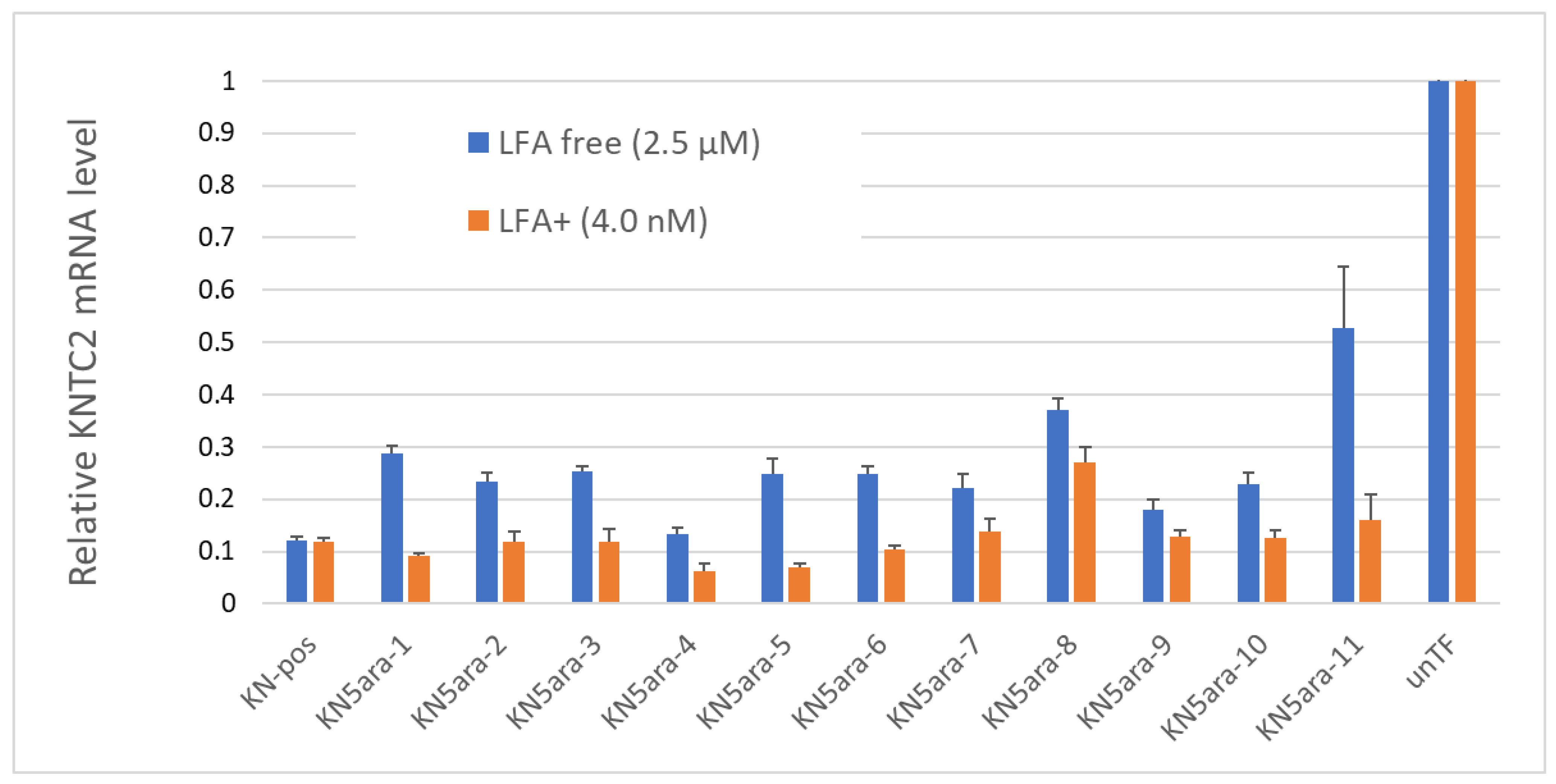
In addition to the physical experiments and simple in vitro enzymatic assays, in vitro cellular experiments were performed to knock down KNTC2 in A549tGFP cells to evaluate the antisense activity of each KN5ara gapmer. The pre-cultured A549tGFP cells were treated with KN5ara gapmers at a final concentration of 4.0 nM or 2.5 µM, and were transfected with 0.3% Lipofectamine® 2000 (LFA) or not, respectively. After incubation, total mRNA inside the cells was extracted, followed by reverse transcription of the targeted KNTC2 mRNA. A quantitative real-time polymerase chain reaction (qRT-PCR) was performed in duplicate, and relative KNTC2 mRNA levels were calculated, as shown in Figure 4.
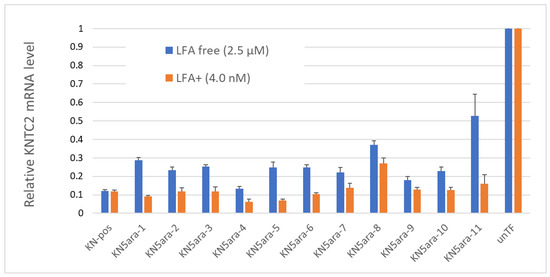
Figure 4.
Relative KNTC2 mRNA levels of each KN5ara gapmer. A549tGFP cells were treated with KN5ara gapmers at different final concentrations: 2.5 µM for lipofection-free group and 4.0 nM for 0.3% Lipofectamine® 2000 (LFA) treated group. LFA-free groups were incubated with the gapmers for 48 h, and further incubated for 24 h after exchanging medium. LFA+ groups were incubated with the gapmers for 24 h, and further incubated for 24 h after exchanging medium. Treatment without gapmers was used as a control (unTF). The total mRNA inside cells were extracted, followed by the reverse transcription of the targeted KNTC2 mRNA. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed in duplicate, and the relative KNTC2 mRNA levels were calculated via the ddCT method.
The results suggested that there was no obvious change in the antisense activity of KN5ara gapmers with a single 5ara modification when LFA was used. KN5ara-4/5 showed much higher KNTC2 knockdown efficiency than the positive control KN-pos, although KN5ara-8 showed a 2-fold decrease. Interestingly, compared to KN5ara-8, KN5ara-9, which has another 5ara modification at the same site as KN5ara-7, maintained antisense activity similar to that of KN5ara-7, suggesting that continuous 5ara modifications may induce further improvement. Contrastingly, KN5ara-3 and KN5ara-10 exhibited similar antisense activity, whereas KN5ara-11 had slightly lower activity than KN5ara-6.
In the LFA-free group, KN-pos showed antisense activity comparable to that in the LFA+ condition. All KN5ara gapmers were clearly less active, indicating that cellular uptake might be reduced, although the extent of the reduction varied greatly depending on the site of the 5ara modification. Meanwhile, KN5ara-4 efficiently knocked down KNTC2 mRNA, and the decrease in antisense activity of KN5ara-8 was also reversed by continuous 5ara modifications (KN5ara-9). Notably, significant property degradation was observed in KN5ara-11, as compared to KN5ara-6, whereas KN5ara-10 showed similar or slightly better antisense activity than KN5ara-3.
In more detail, it is obvious that KN5ara-4 showed the most effective antisense activity in both methods, when used with lipofection or not. The relative KNTC2 mRNA level tended to increase according to the shift of 5ara modification to the 3ʹ-terminal, which suggests 5ara modification might be accepted well in the center of the gap region. Further investigation is need, however, of the synthesis of 5ara modified adenosine and guanosine analogs. Moreover, according to previous studies, the incorporation of a single 2ʹ-OMe modification at gap position 2 could reduce the PS-derived toxicity while maintaining enough antisense activity [16]. In this study, KN5ara-3 and -10 obtained a 5ara modification at gap position 2; additionally, KN5ara-10 retained even less antisense activity for PS linkages. Therefore, KN5ara-10 might be the most promising candidate for reducing PS-derived cytotoxicity. Further experiments studying the cytotoxicity are planned.
3. Conclusions
In summary, the novel synthesis of (S)-5ʹ-C-Aminopropyl-2ʹ-arabinofluoro-5-methyl-cytidine (5ara-MeC) was accomplished in this study. The properties of a series of LNA-wing antisense gapmers containing (S)-5ʹ-C-Aminopropyl-2ʹ-arabinofluoro (5ara) modification, named KN5ara gapmers, were evaluated. It was revealed that each KN5ara gapmer could bind to the complementary RNA strand with high affinity and could be thermally stable under biological condition. The ability for E. coli RNase H1 activation was moderately affected by the incorporation of a single 5ara modification, although the cleavage pattern was obviously changed, which is consistent with previous reports. To determine whether the 5ara modification could be an alternative to PS linkage, the nuclease resistance of several KN5ara gapmers was evaluated using a simple enzymatic reaction system. As a result, incorporating one 5ara modification inside the two PO linkages could reverse the destabilization in enzymatic hydrolysis caused by fewer PS linkages. In addition, in vitro cellular experiments were performed to knockdown KNTC2 in A549tGFP cells. The LFA+ group showed a relatively higher antisense activity than the LFA-free group, while KN5ara-4 showed superior antisense activity in both methods. Compared to KN5ara-7, KN5ara-8, and KN5ara-9, continuous 5ara modifications at certain sites might induce further improvement. KN5ara-10, which contained fewer PS linkages, showed similar or slightly better antisense activity than the corresponding KN5ara-3. Hence, given the possibility of lower PS-derived cytotoxicity, KN5ara-10 might be the best candidate for KNTC2-targeted ASO for cancer therapy in this study. However, the further cytotoxicity assay of these KN5ara gapmers should be performed in future studies, and gapmers with fewer PS linkages than KN5ara-10 should also be synthesized and evaluated as well.
4. Experimental Sections
4.1. Solid-Phase Oligonucleotide Synthesis
The synthesis was carried out with a DNA/RNA synthesizer by the phosphoramidite method. After the synthesis, the RNA oligomers were cleaved from CPG beads and deprotected by treatment with concentrated NH3 solution/40% methylamine (1:1, v/v) for 10 min at 65 °C, while the DNA-based oligomers were treated with concentrated NH3 solution for 12 h at 55 °C. Notably, before the treatment of the NH3 solution, the CPG beads were treated with 10% dimethylamine in MeCN for 5 min followed by rinsing with MeCN to selectively remove cyanoethyl groups, if there are analogs introduced in oligomers. Then, 2ʹ-O-TBDMS groups in RNA oligomers were removed by Et3N∙3HF (125 µL) in DMSO (100 µL) for 1.5 h at 65 °C. The reaction was quenched with 0.1 M TEAA buffer (pH 7.0), and the mixture was desalted using a Sep-Pak C18 cartridge. The oligomers were purified by 20% PAGE containing 7 M urea to give highly purified oligonucleotides.
4.2. MALDI-TOF/MS Analysis of ONs
The spectra were obtained with a time-of-flight mass spectrometer equipped with a nitrogen laser (337 nm, 3 ns pulse). A solution of 3-hydroxypicolinic acid (3-HPA) and diammonium hydrogen citrate in H2O was used as a matrix. Data of synthetic ONs: KN-pos: m/z = 5346.49 (calcd for C165H202N58O88P15S15 [M-H]−, 5348.32); KNpos-F: m/z = 5884.62 (calcd for C192H226N59O87P16S15 [M-H]−, 5882.91); KN5ara-1: m/z = 5395.54 (calcd for C167H208N59O87FP15S15 [M-H]−, 5393.54); KN5ara-2: m/z = 5396.54 (calcd for C167H207N59O87FP15S15 [M-H]−, 5397.39); KN5ara-3: m/z = 5428.03 (calcd for C168H208N59O88FP15S15 [M-H]−, 5425.40); KN5ara-4: m/z = 5438.57 (calcd for C169H210N59O88FP15S15 [M-H]−, 5439.43); KN5ara-5: m/z = 5424.63 (calcd for C168H208N59O88FP15S15 [M-H]−, 5425.40); KN5ara-6: m/z = 5425.59 (calcd for C168H208N59O88FP15S15 [M-H]−, 5425.40); KN5ara-6-F: m/z = 5959.67 (calcd for C195H232N60O97FP16S15 [M-H]−, 5960.54); KN5ara-7: m/z = 5427.77 (calcd for C168H208N59O88FP15S15 [M-H]−, 5425.40); KN5ara-8: m/z = 5393.92 (calcd for C167H208N59O87FP15S15 [M-H]−, 5393.54); KN5ara-9: m/z = 5472.64 (calcd for C170H214N60O87F2P15S15 [M-H]−, 5468.59); KN5ara-10: m/z = 5392.62 (calcd for C168H208N59O90FP15S13 [M-H]−, 5389.59); KN5ara-10-F: m/z = 5927.72 (calcd for C195H232N60O99FP16S13 [M-H]−, 5928.59); KN5ara-11: m/z = 5391.73 (calcd for C168H208N59O90FP15S13 [M-H]−, 5389.59); KN5ara-11-F: m/z = 5927.72 (calcd for C195H232N60O99FP16S13 [M-H]−, 5928.94); KN5ara-12-F: m/z = 5852.67 (calcd for C192H226N59O99P16S13 [M-H]−, 5853.65); cRNA-1: m/z = 5046.38 (calcd for C152H190N61O107P15 [M-H]−, 5045.74); cRNA-2: m/z = 5585.96 (calcd for C179H215N62O116P16 [M-H]−, 5583.87).
4.3. Thermal Denaturation Study
The solution containing 3.0 µM ASO/RNA duplexes were prepared by mixing the ASOs (600 pmol) with complementary target cRNA-1 (600 pmol) in a buffer of 10 mM sodium phosphate (pH 7.0) containing 100 mM NaCl, and then heated at 90–100 °C, followed by being cooled gradually to room temperature. Thermally induced transitions were monitored at 260 nm with a UV/vis spectrometer fitted with a temperature controller in quartz cuvettes with a path length of 1.0 cm. The sample temperature was increased by 0.5 °C/min.
4.4. RNase H Assay
The ASO/RNA duplexes used for RNase H assay were prepared by mixing the ASOs (600 pmol) with fluorescein labeled complementary target cRNA-2 (3000 pmol) in 75 µL of 50 mM Tris–HCl (pH 8.0) containing 75 mM KCl, 3 mM MgCl2 and 10 mM dithiothreitol, followed by heating at 90–100 °C for 5 min and cooling gradually to room temperature. Then, 70 µL diluted RNase H solution (60 unit/L in H2O) was added, and subsequently the mixture was incubated at 37 °C for the required time. Aliquots of 5 µL were diluted with 100% formamide (10 µL). Samples were subjected to electrophoresis in 20% PAGE containing 7M urea and quantified by Luminescent Image analyzer LAS-4000 (Fujifilm).
4.5. Nuclease Resistance of Single-Stranded ASO
Fluorescein labeled ASOs (300 pmol) were dissolved in OPTI-MEM (37 µL) and used for the serum stability test. 1.0 µL of the oligomer solution was diluted in 10 µL stop solution (10% formamide in 10 mM EDTA) as the control sample (0 min). Then, 36 µL bovine serum was added to achieve a final concentration of 50% (v/v), and subsequently the mixture was incubated at 37 °C for the required time. Aliquots of 2.0 µL were diluted with 10 µL stop solution. Samples were subjected to electrophoresis in 20% PAGE containing 7M urea and quantified by Luminescent Image analyzer LAS-4000 (Fujifilm).
4.6. ASOs In Vitro Activity Assay
A549tGFP cells were established by transfection with pGIPZ (Horizon Discovery) into a human lung cancer cell line A549 (ATCC). For LFA-free (LFA-) group, A549tGFP cells were plated into 96-well plates at 2 × 103 cells/well, followed by the cultivation in D-MEM (Nissui) containing 10% fetal bovine serum (FBS), streptomycin (100 µg/mL) and penicillin (100 U/mL). Cells were incubated at 37 °C with 5% carbon dioxide for 24 h prior to the ASO administration. Then, the medium was exchanged into OPTI-MEM containing 2% FBS, and the KNTC2-targeting antisense gapmers were added into specified wells at the final concentration of 2.5 µM. After a 48-h incubation in 2% FBS_OPTI-MEM and a further 24-h incubation in 10% FBS_D-MEM, cells were retrieved, and the mRNAs were extracted using CellAmp Direct RNA Prep Kit for RT-PCR (Takara Bio). For LFA+ group, A549tGFP cells (ATCC) were plated into 96-well plates at 4 × 103 cells/well and cultured in 10% FBS_D-MEM for 24 h prior to the ASO administration. After exchanging the medium into 10% FBS_OPTI-MEM, the KNTC2-targeting antisense gapmers, mixed with 0.3% Lipofectamine® 2000 (Invitrogen) in advance, were added into specified wells at the final concentration of 4.0 nM and incubated for 24 h. Then, as the same as LFA- group, the medium was replaced by 10% FBS_D-MEM and cells were incubated for further 24 h, followed by the extraction of mRNAs. Treatment without ASOs was used as a control, named “unTF”.
Quantitative real-time polymerase chain reaction (qRT-PCR) of the targeted KNTC2 mRNA was performed using One Step TB Green PrimeScript RT-PCR kit II (Takara), and Thermal Cycler Dice Real Time System (Takara). Each PCR reaction was performed in duplicate, and the relative KNTC2 mRNA levels were calculated with the ddCT method, using beta-actin (ACTB) as a reference. The sequences of primers used in the qRT-PCR for Human KNTC2 are 5ʹ-CCTCTCCATGCAGGAGTTAAGA-3ʹ for the forward primer, 5ʹ-GGTCTCGGGTCCTTGATTTTCT-3ʹ for the reverse primer. For Human ACTB, the sequences of forward primer and reverse primer are 5ʹ-GGAGCAATGATCTTGATCTT-3ʹ and 5ʹ-CCTTCCTGGGCATGGAGTCCT-3ʹ, respectively.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27217384/s1, Scheme S1: Novel synthesis of (S)-5ʹ-C-aminopropyl-2ʹ-arabinofluoro-5-methyl-cytidine phosphoramidites, and General remark and details of the synthesis of compounds 2-4; Figure S1: Thermal Stability of Duplexes: the UV melting profiles of duplex composed of each KN5ara gapmer and cRNA-1; Table S1. The sequences of all oligonucleotides used in this research; 1H-, 13C- and 19F-NMR spectra of compounds 2-4 and 31P-NMR spectra of compound 4.
Author Contributions
Conceptualization, Y.U.; synthesis of nucleosides and nucleotides, Y.Z.; evaluation and data analysis, Y.Z. and S.S.; writing—original draft preparation, Y.Z.; writing—review and editing, Y.Z., S.S. and Y.U.; supervision, Y.U.; project administration, Y.Z. and Y.U.; funding acquisition, Y.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All data involved in this paper was shown in the text or “Supplementary Materials”.
Acknowledgments
This work was supported by the Japan Agency for Medical Research and Development (AMED) through its funding program for the research project number: 21ae0121029h0001, and the Convolution of Informatics and Biomedical Sciences on Glocal Alliances (CIBoG) in Doctoral Programfor World-leading Innovative & Smart Education (WISE Program). The authors would like to thank Yasuko Kohda (BIKAKEN) for her technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds 1-4, KN-pos, KN5ara 1-11 (including the fluorescein labeled ones) and cRNA 1-2 are available from the authors.
References
- Anais, M.Q.; Maria, L.C.; Scott, L.S.; Riccardo, P. Small Drugs, Huge Impact: The Extraordinary Impact of Antisense Oligonucleotides in Research and Drug Development. Molecules 2022, 27, 536. [Google Scholar]
- Roehr, B. Fomivirsen approved for CMV retinitis. J. Int. Assoc. Physicians AIDS Care 1998, 4, 14–16. [Google Scholar]
- Bell, D.A.; Hooper, A.J.; Burnett, J.R. Mipomersen, an antisense apolipoprotein B synthesis inhibitor. Expert Opin. Investig. Drugs 2011, 20, 265–272. [Google Scholar] [CrossRef]
- Yahiya, Y.S. Eteplirsen: First Global Approval. Drugs 2016, 76, 1699–1704. [Google Scholar]
- Gidaro, T.; Servais, L. Nusinersen treatment of spinal muscular atrophy: Current knowledge and existing gaps. Dev. Med. Child Neurol. 2019, 61, 19–24. [Google Scholar] [CrossRef]
- Susan, J.K. Inotersen: First Global Approval. Drugs 2018, 78, 1371–1376. [Google Scholar]
- Witztum, J.L.; Gaudet, D.; Freedman, S.D.; Alexander, V.J.; Digenio, A.; Williams, K.R.; Yang, Q.; Hughes, S.G.; Geary, R.S.; Arca, M.; et al. Volanesorsen and Triglyceride Levels in Familial Chylomicronemia Syndrome. N. Engl. J. Med. 2019, 381, 531–542. [Google Scholar] [CrossRef]
- Heo, Y.A. Golodirsen: First Approval. Drugs 2020, 80, 329–333. [Google Scholar] [CrossRef]
- Sohita, D. Viltolarsen: First Approval. Drugs 2020, 80, 1027–1031. [Google Scholar]
- Matt, S. Casimersen: First Approval. Drugs 2021, 81, 857–879. [Google Scholar]
- Manoharan, M. Oligonucleotide Conjugates as Potential Antisense Drugs with Improved Uptake, Biodistribution, Targeted Delivery, and Mechanism of Action. Antisense Nucl. Acid Drug Dev. 2002, 12, 103–128. [Google Scholar] [CrossRef]
- Aboul-Fadl, T. Antisense Oligonucleotides: The State of the Art. Curr. Med. Chem. 2005, 12, 2193–2214. [Google Scholar] [CrossRef]
- Crooke, S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucl. Acid Ther. 2017, 27, 70–77. [Google Scholar] [CrossRef]
- Seth, P.P.; Swayze, E.E. The Medicinal Chemistry of RNase H-activating Antisense Oligonucleotides. In Advances in Nucleic Acid Therapeutics; Royal Society of Chemistry: Cambridge, UK, 2019; pp. 32–61. [Google Scholar]
- Crooke, S.T.; Vickers, T.A.; Liang, X.-H. Phosphorothioate modified oligonucleotide—Protein interactions. Nucl. Acids Res. 2020, 48, 5235–5253. [Google Scholar] [CrossRef]
- Shen, W.; De Hoyos, C.L.; Migawa, M.T.; Vickers, T.A.; Sun, H.; Low, A.; Bel, T.A., 3rd; Rahdar, M.; Mukhopadhyay, S.; Hart, C.E.; et al. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 2019, 37, 640–650. [Google Scholar] [CrossRef]
- Vasquez, G.; Freestone, G.C.; Wan, W.B.; Low, A.; De Hoyos, C.L.; Yu, J.; Prakash, T.P.; Ostergaard, M.E.; Liang, X.H.; Crooke, S.T.; et al. Site-specific incorporation of 5’-methyl DNA enhances the therapeutic profile of gapmer ASOs. Nucl. Acids Res. 2021, 49, 1828–1839. [Google Scholar] [CrossRef]
- Prakash, T.P.; Yu, J.; Shen, W.; De Hoyos, C.L.; Berdeja, A.; Gaus, H.; Liang, X.-H.; Crooke, S.T.; Seth, P.P. Site-specific incorporation of 2ʹ,5ʹ-linked nucleic acids enhances therapeutic profile of antisense oligonucleotides. ACS Med. Chem. Lett. 2021, 12, 922–927. [Google Scholar] [CrossRef]
- Migawa, M.T.; Shen, W.; Wan, W.B.; Vasquez, G.; Oestergaard, M.E.; Low, A.; De Hoyos, C.L.; Gupta, R.; Murray, S.; Tanowitz, M.; et al. Site-specific replacement of phosphorothioate with alkyl phosphonate linkages enhances the therapeutic profile of gapmer ASOs by modulating interactions with cellular proteins. Nucl. Acids Res. 2019, 47, 5465–5479. [Google Scholar] [CrossRef]
- Anderson, B.A.; Freestone, G.C.; Low, A.; De-Hoyos, C.L.; Iii, W.J.D., 3rd; Østergaard, M.E.; Crooke, S.T.; Seth, P.P.; Berdeja, A.; Scandalis, E.; et al. Towards next generation antisense oligonucleotides: Mesylphosphoramidate modification improves therapeutic index and duration of effect of gapmer antisense oligonucleotides. Nucl. Acids Res. 2021, 49, 9026–9041. [Google Scholar] [CrossRef]
- Kanazaki, M.; Ueno, Y.; Shuto, S.; Matsuda, A. Highly Nuclease-Resistant Phosphodiester-Type Oligodeoxynucleotides Containing 4’-α-C-Aminoalkylthymidines Form Thermally Stable Duplexes with DNA and RNA. A Candidate for Potent Antisense Molecules. J. Am. Chem. Soc. 2000, 122, 2422–2432. [Google Scholar] [CrossRef]
- Koizumi, K.; Maeda, Y.; Kano, T. Synthesis of 4’-C-aminoalkyl-2’-O-methyl modified RNA and their biological properties. Bioorg. Med. Chem. 2018, 26, 3521–3534. [Google Scholar] [CrossRef] [PubMed]
- Kano, T.; Katsuragi, Y.; Maeda, Y.; Ueno, Y. Synthesis of 4′-C-aminoalkyl-2′-O-methyl modified RNA and their biological properties. Bioorg. Med. Chem. 2018, 26, 4574–4582. [Google Scholar] [CrossRef] [PubMed]
- Tsuchihira, T.; Kajino, R.; Maeda, Y.; Ueno, Y. 4’-C-Aminomethyl-2’-deoxy-2’-fluoroarabinonucleoside increases the nuclease resistance of DNA without inhibiting the ability of a DNA/RNA duplex to activate RNase, H. Bioorg. Med. Chem. 2020, 28, 115611. [Google Scholar] [CrossRef]
- Kajino, R.; Maeda, Y.; Yoshida, H.; Yamagishi, K.; Ueno, Y. Synthesis and biophysical characterization of RNAs containing ®- and (S)-5’-C-aminopropyl-2’-O-methyluridines. J. Org. Chem. 2019, 84, 3388–3404. [Google Scholar] [CrossRef] [PubMed]
- Kajino, R.; Ueno, Y. (S)-5’-C-Aminopropyl-2’-O-methyl nucleosides enhance antisense activity in cultured cells and binding affinity to complementary single-stranded RNA. Bioorg. Med. Chem. 2021, 30, 115925. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Kajino, R.; Ishii, S.; Yamagishi, K.; Ueno, Y. Synthesis and evaluation of (S)-5′-C-aminopropyl and (S)-5′-C-aminopropyl-2′-arabinofluoro modified DNA oligomers for novel RNase H-dependent antisense oligonucleotides. RSC Adv. 2020, 10, 41901. [Google Scholar] [CrossRef] [PubMed]
- Wahlestedt, C.; Salmi, P.; Good, L.; Kela, J.; Johnsson, T.; Wengel, J.; Hökfelt, T.; Broberger, C.; Porreca, F.; Lai, J.; et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. USA 2000, 97, 5633–5638. [Google Scholar] [CrossRef]
- Hagedorn, P.H.; Persson, R.; Funder, E.D.; Albæk, N.; Diemer, S.L.; Hansen, D.J.; Møller, M.R.; Papargyri, N.; Christiansen, H.; Hansen, B.R.; et al. Locked nucleic acid: Modality, diversity, and drug discovery. Drug Discov. Today 2018, 23, 101–114. [Google Scholar] [CrossRef]
- Hayama, S.; Daigo, Y.; Kato, T.; Ishikawa, N.; Yamabuki, T.; Miyamoto, M.; Ito, T.; Tsuchiya, E.; Kondo, S.; Nakamura, Y. Activation of CDCA1-KNTC2, Members of Centromere Protein Complex, Involved in Pulmonary Carcinogenesis. Cancer Res. 2006, 66, 10339–10348. [Google Scholar] [CrossRef]
- Kaneko, N.; Miura, K.; Gu, Z.; Karasawa, H.; Ohmuma, S.; Sasaki, H.; Horii, A.; Tsukamoto, N.; Yokoyama, S.; Yamamura, A.; et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem. Biophys. Res. Commun. 2009, 390, 1235–1240. [Google Scholar] [CrossRef]
- Makita, Y.; Teratani, M.; Murata, S.; Hoashi, Y.; Matsumoto, S.; Kawamata, Y. Antitumor activity of kinetochore-associated protein 2 siRNA against lung cancer patient-derived tumor xenografts. Oncol. Lett. 2018, 15, 4676–4682. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Ghirlando, R.; Cerritelli, S.M.; Crouch, R.J.; Yang, W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: Insight into HIV reverse transcription. Mol. Cell 2007, 28, 264–276. [Google Scholar] [CrossRef]
- Nakamura, H.; Oda, Y.; Iwai, S.; Inoue, H.; Ohtsuka, E.; Kanaya, S.; Kimura, S.; Katsuda, C.; Katayanagi, K.; Morikawa, K. How does RNase H recognize a DNA. RNA hybrid? Proc. Natl. Acad. Sci. USA 1991, 88, 11535–11539. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).