Abstract
Chemical investigation of the total extract of the Egyptian soft coral Heteroxenia fuscescens, led to the isolation of eight compounds, including two new metabolites, sesquiterpene fusceterpene A (1) and a sterol fuscesterol A (4), along with six known compounds. The structures of 1–8 were elucidated via intensive studies of their 1D, 2D-NMR, and HR-MS analyses, as well as a comparison of their spectral data with those mentioned in the literature. Subsequent comprehensive in-silico-based investigations against almost all viral proteins, including those of the new variants, e.g., Omicron, revealed the most probable target for these isolated compounds, which was found to be Mpro. Additionally, the dynamic modes of interaction of the putatively active compounds were highlighted, depending on 50-ns-long MDS. In conclusion, the structural information provided in the current investigation highlights the antiviral potential of H. fuscescens metabolites with 3β,5α,6β-trihydroxy steroids with different nuclei against SARS-CoV-2, including newly widespread variants.
Keywords:
sterol; virus; sesquiterpene; soft coral; in silico; molecular dynamic simulation; docking 1. Introduction
The global pandemic caused by coronavirus 2 (SARS-CoV-2) has taken many lives [1,2] from 2019 until the present. Several variants and outbreaks are still present in countries such as China and Korea [3].
Coronaviruses are a family of viruses that can cause a wide array of enteric, hepatic, neurological, and respiratory diseases [1]. The four subfamilies of coronavirus are α, β, γ, and δ, and are sorted based on genotype and serotype data [4]. Six species of coronaviruses cause human diseases. The most pathogenic and fatal species are Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV). SARS-CoV-2 is the seventh type, which infects humans and is related to the β lineage of the beta-coronaviruses, which are known to cause severe disease and fatalities [4]. Based on genomic analysis, SARS-CoV-2 belongs to two bat-derived SARS-like coronaviruses with 96% similarity, whereas its similarity to SARS-CoV is 79% [5]. The lung is the primary site and the most affected organ of SARS-CoV-2 infection. The clinical manifestations range from asymptomatic to severe respiratory disease. The most common symptoms of infection are a loss of smell and taste, fever, headache, shortness of breath, cough, muscle aches, and tiredness [6].
The coronaviral genome encodes four structural proteins, namely the envelope (E) protein, membrane (M) protein, nucleocapsid (N) protein, and spike (S) protein [5]. Most vaccine strategies have been focused on the spike protein, as it plays an essential role in viral entry to the host cell [5]. The E protein and M protein participate in the production and release of virus-like particles, while the N protein is a critical for viral replication and genome packaging [7,8]. During virus infection, the main proteases (Mpro) and cyclin-dependent kinases (CDKs) play a crucial role in the regulation and progression of cell division, which is essential in replication and viral transmissibility [9,10]. The main protease (Mpro) is essential for processing the polyproteins that are translated from the viral RNA, and SARS-CoV-2 modifies the CDK signaling pathway and regulates the cell division cycle in a way that leads to the enhancement of viral replication [11,12]. Consequently, MPro and CDK inhibitors are considered attractive targets for drugs that are used to treat the SARS-CoV-2 virus [13].
To speed up the treatment process, several antiviral drugs have been re-evaluated as an emerging antiviral option in the SARS-CoV-2 infection. Favipiravir has shown promising results in clinical studies in Japan, Russia, and China. Additionally, the oral antiviral drug nirmatrelvir/ritonavir (Paxlovid) has been approved by the FDA for emergency use [14].
Naturally occurring metabolites have proven to be promising sources of drug leads and structural motifs, based on the diversity of their structures and biological activities [15,16,17]. Several compounds, such as sesquiterpenes, diterpenes, steroids, alkaloids, and lipid derivatives, were isolated from soft corals [18,19,20]. Soft corals belonging to the family Xeniidea (Alcyonacea) are distributed throughout the oceans and seas [21,22], and the two genera that have been subjected to detailed chemical and biological investigation in the Red Sea are Xenia and Heteroxenia [23,24]. The genus Heteroxenia is distributed along the Egyptian Red Sea’s coast, with two common species, Heteroxenia fuscescens and Heteroxenia ghardaqensis [21]. Several secondary metabolites, such as steroids (with a high degree of oxygenation), sesquiterpenoids, ceramides, and glycerol derivatives, were isolated from both species. Many of these metabolites showed diverse biological activities, such as anticancer, anti-inflammatory, anti-diabetic, and analgesic activities [24,25,26,27,28,29,30].
Based on the above data, as well as the urgent necessity of developing and searching for natural drugs to treat the SARS-CoV-2 virus, the isolation of the secondary metabolites of the soft coral Heteroxenia fuscescens, followed by the in silico prediction of isolated compounds as potential inhibitors of Mpro and Cyclin-G-associated kinase (GAK) and a promising scaffold using molecular docking and modeling analysis, were carried out.
2. Results
2.1. Identification of the Isolated Compounds
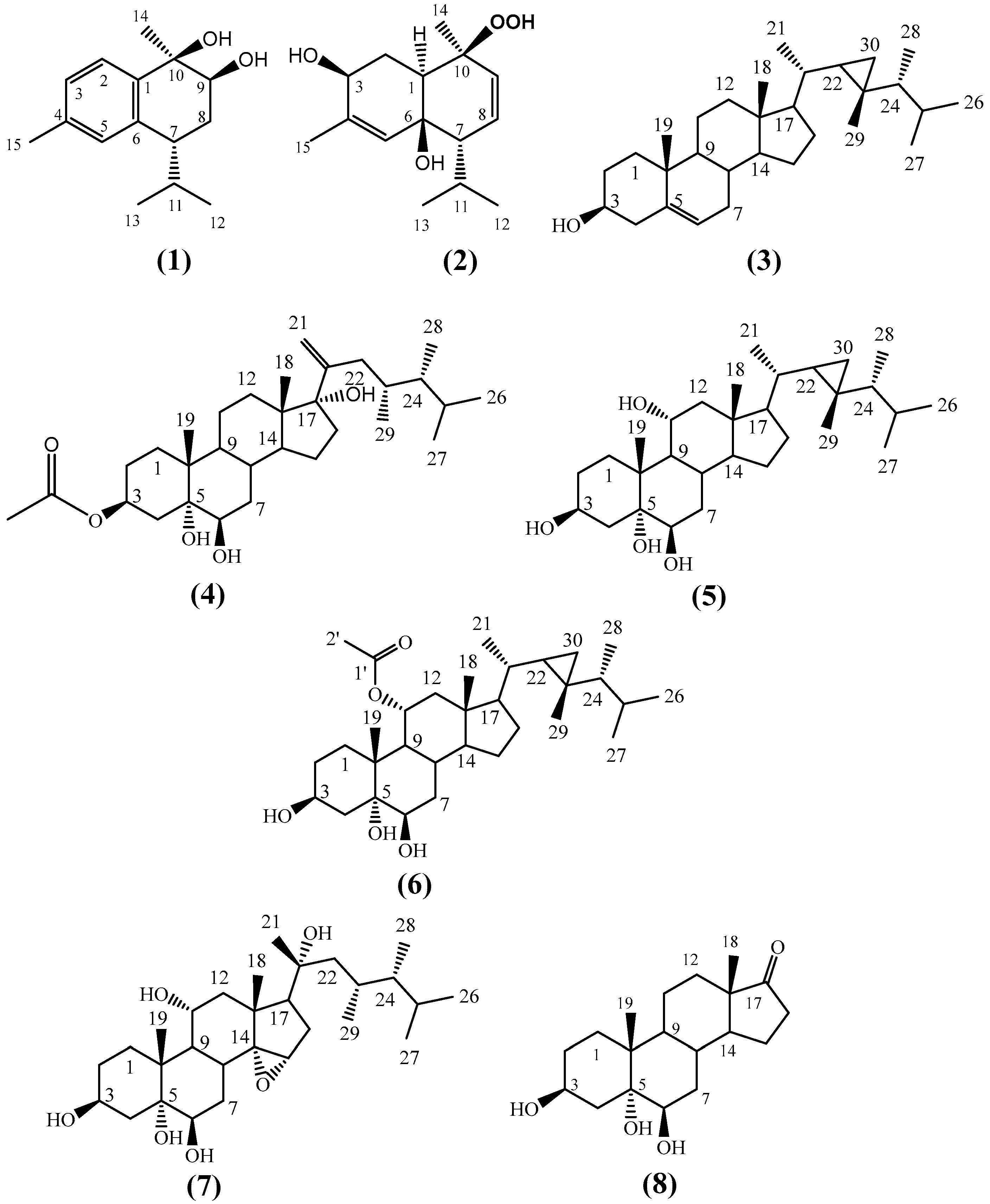
The crude extract of H. fuscescens was partitioned into fractions and isolated using several chromatographic techniques to yield eight compounds (1–8) (Figure 1). The structures of the known compounds were elucidated to be 3β,6β-dihydroxy-10β-hydroperoxycadin-4,8-diene, (2) [25], gorgost-5-(E)-ene-3-β-ol (3) [31,32], gorgost-3β,5α,6β,11α-tetrol (5) [33,34], 11α-acetoxy-gorgost-3β,5α,6β-triol (6) [30,35], 14α,15α-epoxy-23,24-dimethylcholesta,3β,5α,6β,11α,20α-pentol (7) [25], and 3β,5α,6β-trihydroxyandrosta-17-one (8) [25], based on comparisons of their 1D (1H and 13C) NMR spectroscopic data against the data in the literature (Tables S1–S3 and Figures S20–S31 in Supplementary Materials).
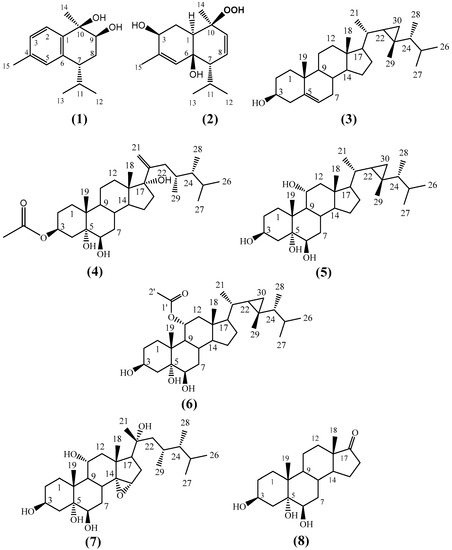
Figure 1.
Structure of the isolated compounds (1)–(8) from H. fuscescens.
Compound 1 was obtained as a colorless oil. The positive HR-FAB-MS showed a [M+H]+ at m/z 235.1701 (calcd for C15H23O2, 235.1698) and a base peak at [M-H2O+H]+ m/z 217.1605 (calcd for C15H21O, 217.1592); together with NMR spectroscopic data, the molecular formula was established to be C15H22O2, with five degrees of unsaturation (Figure S1).
The combined analysis of the 13C-NMR and HSQC spectra displayed 15 signals (Figures S4 and S5), which were categorized as four quaternary carbons (including three aromatic carbons at δC 139.5, 137.7, and 136.8, and one oxygenated at δC 74.5), six methines (including three aromatic carbons at δC 125.4, 127.5, and 128.8, and one oxygenated carbon at δC 73.8), one methylene at δC 27.3, and four methyls (including two quaternaries at δC 21.2, and 24.3, and two tertiaries at δC 18.0 and 21.6). The 1H-NMR spectrum (Figures S2 and S3) indicated the presence of three aromatic protons, including two ortho-coupling protons at δH 7.5 (d, J = 7.9 Hz, H-2) and δH 7.1 (d, J = 7.9 Hz, H-3), and one singlet proton at δH 7.0 (s, H-5). One oxygenated proton at δH 4.0 (dd, J = 11.7, 3.9 Hz, H-9), two quaternary methyls at [δH 1.4 (s, H3-14), 2.3 (s, H3-15)], and two tertiary methyls at [δH 0.78 (d, J = 7.3 Hz, H3-13), δH 1.06 (d, J = 7.1 Hz, H3-12)]. Three double bonds accounted for three degrees of unsaturation; consequently, the remaining degrees were due to the bicyclic structure of 1. From these data, compound 1 is assumed to be a bicyclic calamenene-type sesquiterpene [36,37] with two oxygenated carbons. The assignment of all carbons and protons, as well as the structure of compound 1, was elucidated based on the extensive analysis of 2D NMR (1H-1H COSY, HSQC, and HMBC).
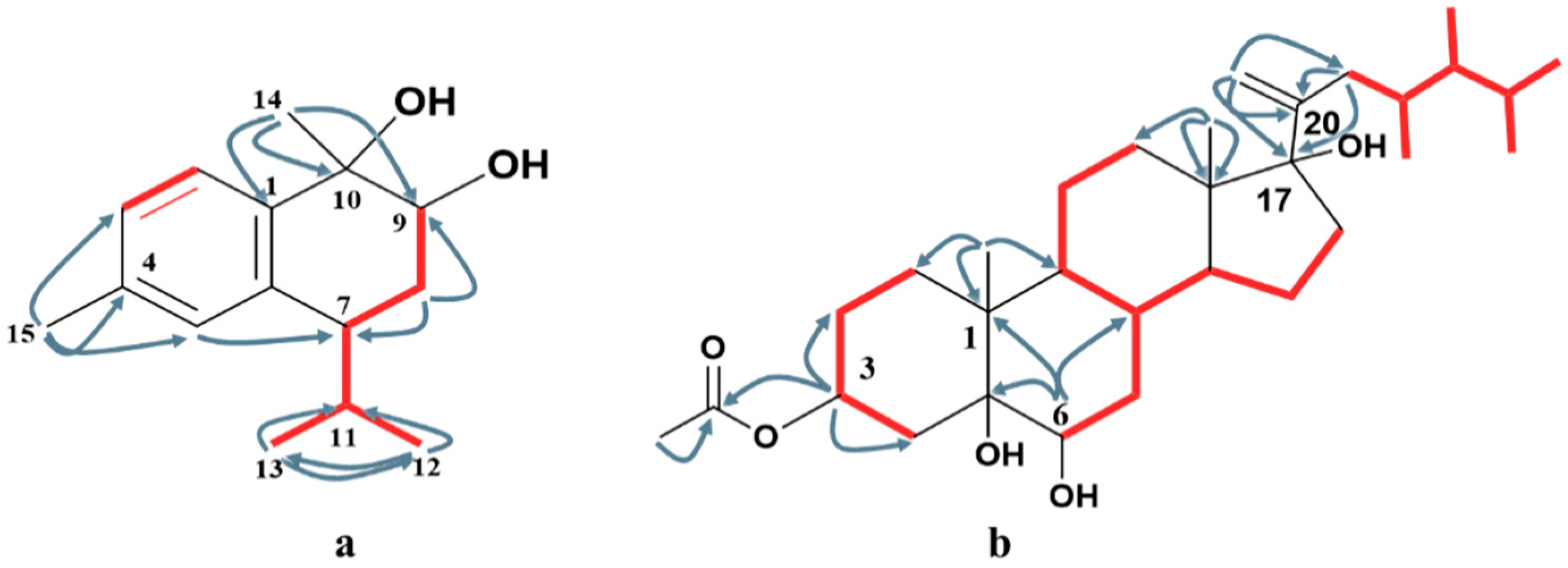
The HMBC spectrum (Figure 2 and Figure S6) clearly illustrated the attachment of H3-15 at C-4 via the presence of a correlation between the two ortho-coupling protons H-2 and H-3, and the observed correlations between H3-15 and C-3, C-4, and C-5. The second quaternary methyl H3-14 was attached to the oxygenated carbon C-10 based on the existence of cross-peaks between H3-14 and C-1, C-9, and C-10. The second oxygenated carbon was C-9, based on the downfield shift of H-9 δH 4.0 (dd, J = 11.7, 3.9 Hz, H-9), while the isopropyl moiety was attached to C-7 based on the HMBC correlations between H3-12 and C-7, C-11, and C-13; and between H3-13 and C-7, C-11, and C-12, respectively. The 1H-1H COSY spectrum (Figures S7 and S8) illustrated the presence of two discrete spins corresponding to H-2/H-3 in the aromatic ring and H-9/H2-8, H2-8/H-7, H-7/H-11, H-11/H3-12, and H-11/H3-13, which confirmed the overall structure of compound 1.
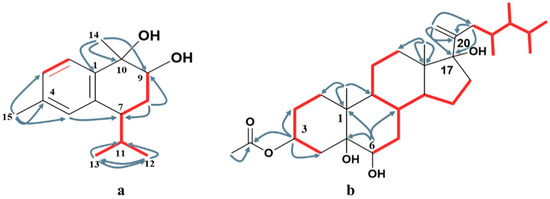
Figure 2.
Key HMBC ( ) and 1H-1H COSY (
) and 1H-1H COSY ( ) correlations of compounds 1 (a) and 4 (b).
) correlations of compounds 1 (a) and 4 (b).
 ) and 1H-1H COSY (
) and 1H-1H COSY ( ) correlations of compounds 1 (a) and 4 (b).
) correlations of compounds 1 (a) and 4 (b).
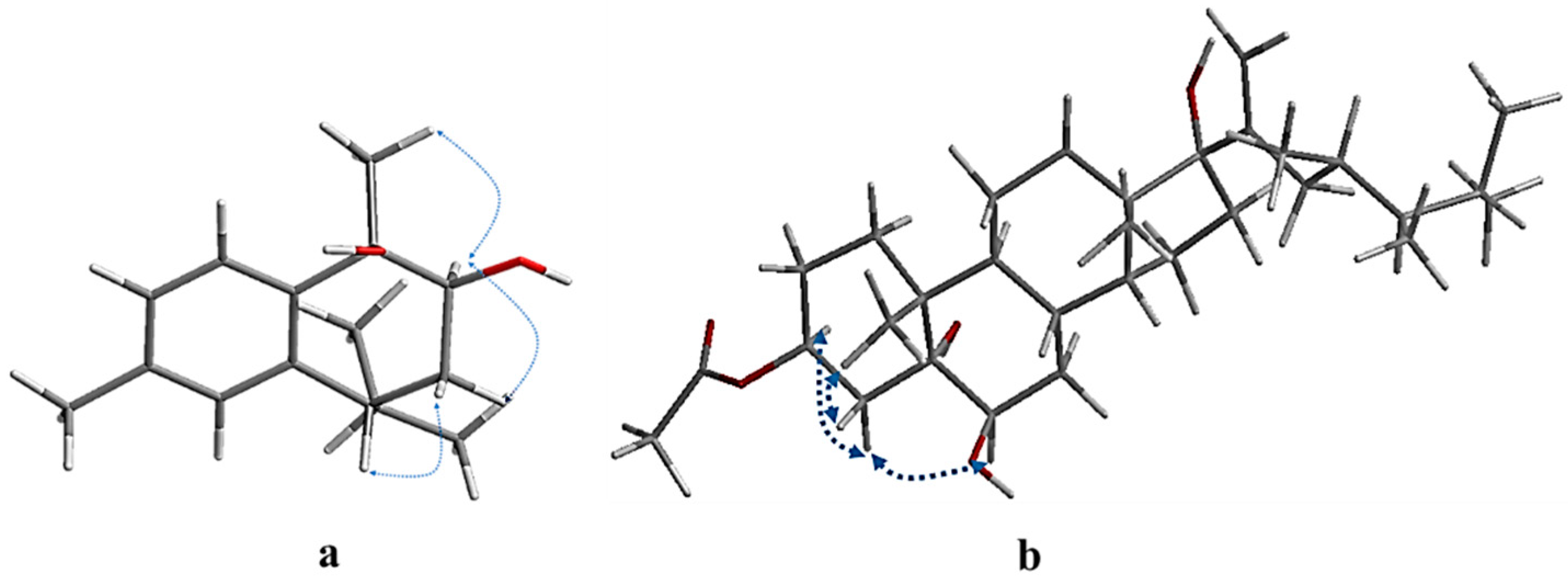
The relative configuration of compound 1 (7R*,9S*,10R*)-cis calamenene was deduced from the investigation via 1D and 2D NMR analyses (Figure 3, Figures S9 and S10), and via a comparison with the previously reported data of related compounds [38,39]. In particular, the main NOE correlations were observed between H-9 and H3-14. Additionally, the absence of an NOE correlation between H-9 or H3-14 and H-7 indicated that H-9 and H3-14 are in one direction, and H-7 is in the other direction. By comparing the chemical shifts of H-7 and C-7 (δH 2.84 m; δc 41.5) of compound 1 with relative metabolites as cis-muurola-4(14),5-diene with the 13C-NMR chemical shift of C-7 (δc 51.7) and 3-hydroxy calamenene with the 13C-NMR chemical shift of C-7 (δc 43.9) (Table 1) [38,39], we can conclude that the OH at C-9 and C-10 were β-oriented, while an isopropyl group at C-7 was α-oriented. From the above data, compound 1 is new, identified as (7R*,9S*,10R*) 9,10-dihydroxycalamenene and named fusceterpene A.
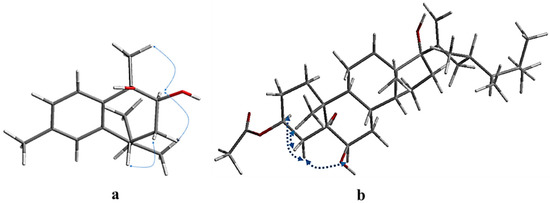
Figure 3.
Key NOESY correlations of compounds 1 (a) and 4 (b).

Table 1.
1H and 13C NMR data of compound 1.
Fuscesterol A 4 was isolated as a white powder. The HR-ESI-MS of compound 4 exhibited a [M+Na]+ peak at m/z 527.3673 corresponding to the molecular formula C31H52O5 and implying six degrees of unsaturation (Figure 1). These observations were in agreement with 31 signals in the 13C-NMR spectrum (Table 2 and Figure S13). They were clarified via an HSQC experiment (Figure S14) into six quaternary carbons (including one carbonyl at δC 172.8, 2 oxygenated at δC 87.7 and 76.6, and one olefinic at δC 152.3), eight methines (including two oxygenated at δC 76.4, 73.0), 10 methylenes (including one terminal olefinic at δC 112.0), and seven methyls. From the above data, compound 4 was a steroid in nature [40,41]. Four degrees of unsaturation were accounted for by the tetracyclic structure, one for vinylic methylene and one for carbonyl. The above data suggested that compound 4 was an acetoxylated dimethyl cholestane-type steroid with a terminal double bond on the side chain [42,43,44].

Table 2.
1H and 13C NMR data of compound 4.
1H-NMR data (Figure S12) showed signals for seven methyls, including four doublets at δH 0.93 (3H, J = 6.6 Hz), 0.89 (3H, J = 6.6 Hz), 0.80 (3H, J = 6.6 Hz), and 0.78 (3H, J = 6.6 Hz), which were assigned to H3-26, H3-27, H3-28, and H3-29, and two singlets at δH 0.65 and 1.18, assigned to H-18 and H-19, respectively. Two singlet signals at δH 5.14 s and 4.89 s were assigned to vinylic methylene at C-21. The location of the double bond was confirmed via the presence of cross-peaks between H2-21 and C-17, C-20, and C-22 in the HMBC spectrum (Figure S15). The presence of a hydroxylated group at C-17 (87.7) neighboring the vinylic methylene was confirmed from the HMBC correlation between H2-21, H3-18, and H2-16 with C-17. The downfield shift of the oxygenated methines at δH 5.17 (m) at C-3 was due to the attachment of an acetate moiety. The attachment was confirmed via the presence of cross-peaks between H-3 and C-2, C-4, and C-1′, respectively. The key 1H-1H COSY correlations were observed between H-2/H-3, H-3/H-4, H-7/H-8, H-14/H-15, and H-15/H-16 (Figures S16 and S17), which confirm the structure of compound 4.
The relative stereochemistry of compound 4 was deduced based on an NOE correlation in the NOESY spectrum (Figures S18 and S19) and comparing the 1H and 13C-NMR with the relative metabolites [42,44]. The key NOESY H-3/H-4a (δH 1.58), H-4a/H-6, and H3-19/H-4b (δH 2.16) indicated that OAc-3 and OH-6 were β-oriented. The α-orientation of the OH at C-5 and C-17 was clarified by comparing the 13C-NMR of C-5 and C-17 (δC 76.6 and δc 87.7), respectively, with relative metabolites as (23R) methyl-ergosta-20-ene-3β,5α,6β,17α-tetrol (δC 76.9 & δc 87.7), gorgostan-3β,5α,6β-triol-11α-acetate (C-5 δC 76.5), and klyflaccisteroid A (C-17 δC 86.7) [30,42,44]. Additionally, the relative configuration of the side chain was confirmed from the good agreement with the 1H and 13C-NMR data (23R, 24R)-23,24-dimethyl-cholest-20-ene side chain. Based on the above finding, compound 4 was identified as (23R*,24R*)-3β-acetoxy-5α,6β,17α-trihydroxy-23,24-dimethylcholesta-20-ene and named fuscesterol A.
2.2. Molecular Docking and Molecular Dynamic Simulation Results
To putatively determine the most probable molecular target for compounds 1–8, we virtually screened their structures against all currently available SARS-CoV-2 proteins that have been reported to be relevant to the viral life cycle or viral pathogenesis (https://swissmodel.expasy.org/repository/species/2697049; https://www.genome.jp/kegg-bin/show_pathway?hsa05171+H02398 (accessed on 1 September 2022); Table S1). Both Mpro and Cyclin-G-associated kinase (GAK) were the only proteins that were found to have considerable docking scores (<−8.0 kcal/mol) with compounds 4, 7, and 8 (for Mpro) and compounds 3–5 and 8 (for GAK).
The subsequent MDS validation experiments (50 ns long) revealed that the structures of compounds 4–7 and 8 were not good binders to GAK, because they were significantly unstable over the course of MDS, with an average RMSD ranging from 6Å to > 10 Å. Moreover, their calculated absolute binding free energy values (ΔGbinding) indicated a low affinity toward GAK’s binding site, and ranged from −2.5 to −3.2 kcal/mol.
In contrast, the structures of compounds 5, 7, and 8 have achieved stable binding over the course of 50 ns MDS runs, with average RMSDs ranging from 1.8 Å to 2.2 Å, and with ΔGbinding values of −8.6, −8.8, and −8.1 kcal/mol, respectively. Interestingly, the three steroids that shared the stable binding had a characteristic hydroxylation pattern at positions 3, 5, and 6 in rings A and B. In addition, we have reported that the triterpene class of compounds, which is structurally related to compounds 5, 7, and 8, is a promising scaffold to develop new Mpro inhibitors [45,46].
To investigate the binding mode of each compound inside the Mpro’s active site, the most populated binding poses for each compound structure was extracted from their 50-ns-long MDS trajectories.
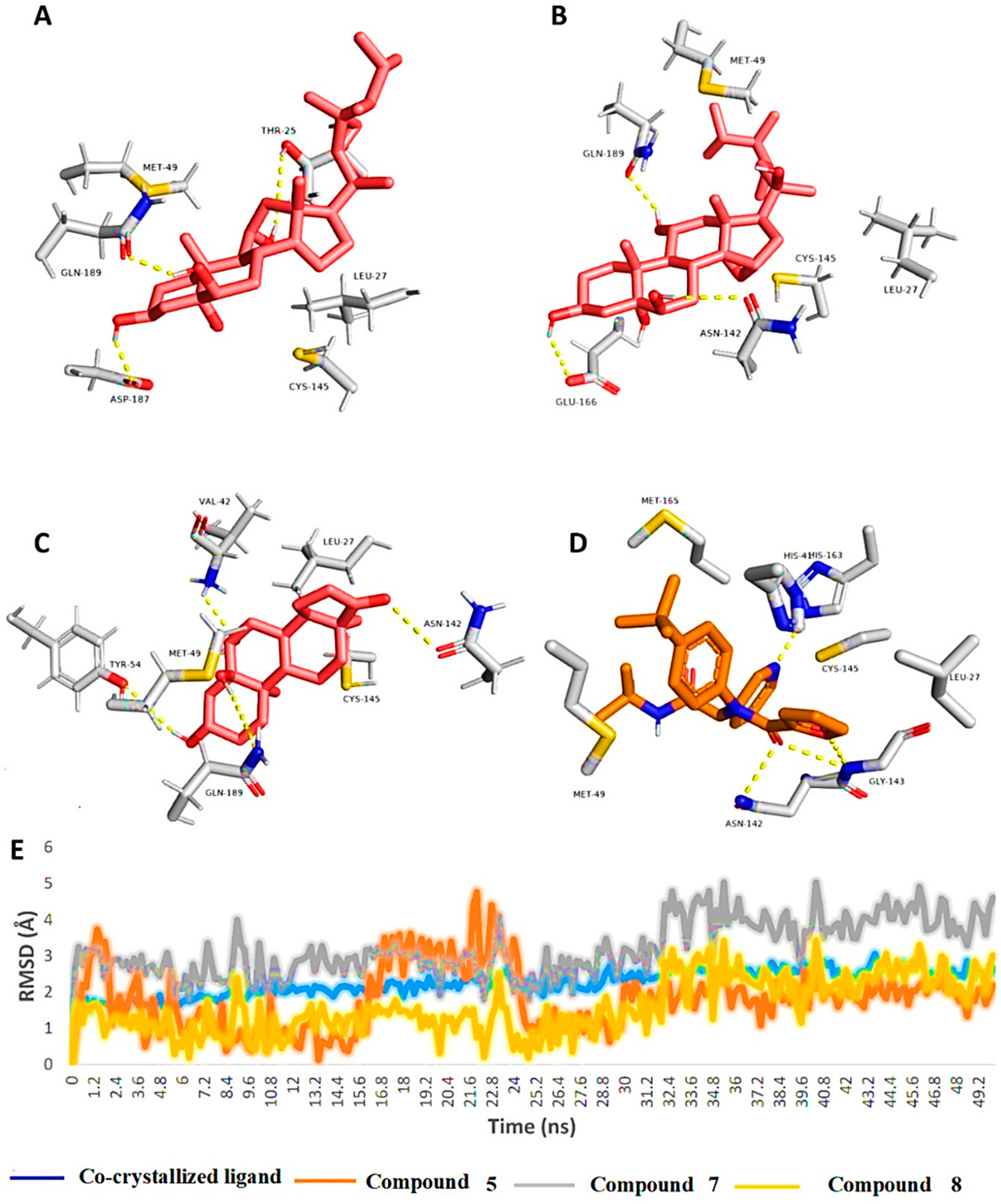
As shown in Figure 4, the binding modes of the investigated structures were different from each other. The structure of compound 5 established three stable H-bonds with THR-25, ASP-187, and GLN-189, in addition to two hydrophobic interactions with LEU-27 and MET-49. The structure of compound 7 also established three H-bonds, but with ASN-142, GLU-166, and GLN-189. Additionally, it interacted hydrophobically with LEU-27 and MET-49. Finally, the structure of compound 8 was able to form four stable H-bonds with VAL-42, TYR-54, ASN-142, and GLN-189, together with two hydrophobic interactions with LEU-27 and MET-49.
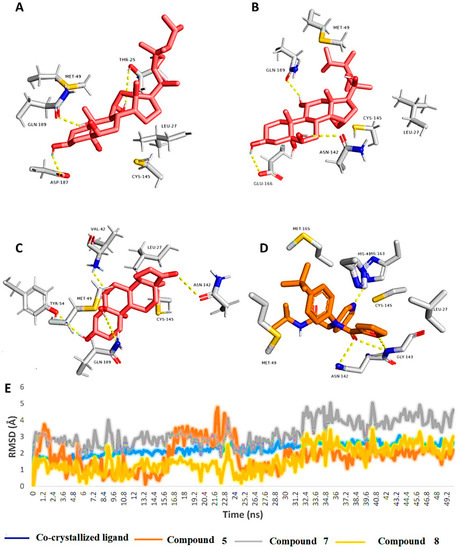
Figure 4.
Binding modes of compounds 5, 7, and 8 along with the co-crystalized inhibitor ML188 (A–D, respectively) inside the active site of SARS-CoV-2′s Mpro (PDB code: 7L0D). These binding modes were extracted from the 50-ns-long MDS trajectories as the most populated poses. (E): RMSDs of compounds 5, 7, and 8 along with the co-crystalized inhibitor ML188 inside the active site of SARS-CoV-2’s Mpro over the course of 50-ns-long MDS.
3. Discussion
In the previous literature, several bioactive metabolites were isolated from soft corals, which have the ability to inhibit the SARS-CoV-2 Mpro, as depresosterol, lopophytosterol, and cholest-5-ene-3β,7β-diol [47]. These compounds form stable hydrogen bonding through a polyhydroxy group in their structure with the amino acid in the active pocket of the SARS Mpro [47]. In the genus Heteroxenia (family Xeniidae), several polyoxygenated steroids were isolated and showed diverse biological activity [24,25,27,28,42].
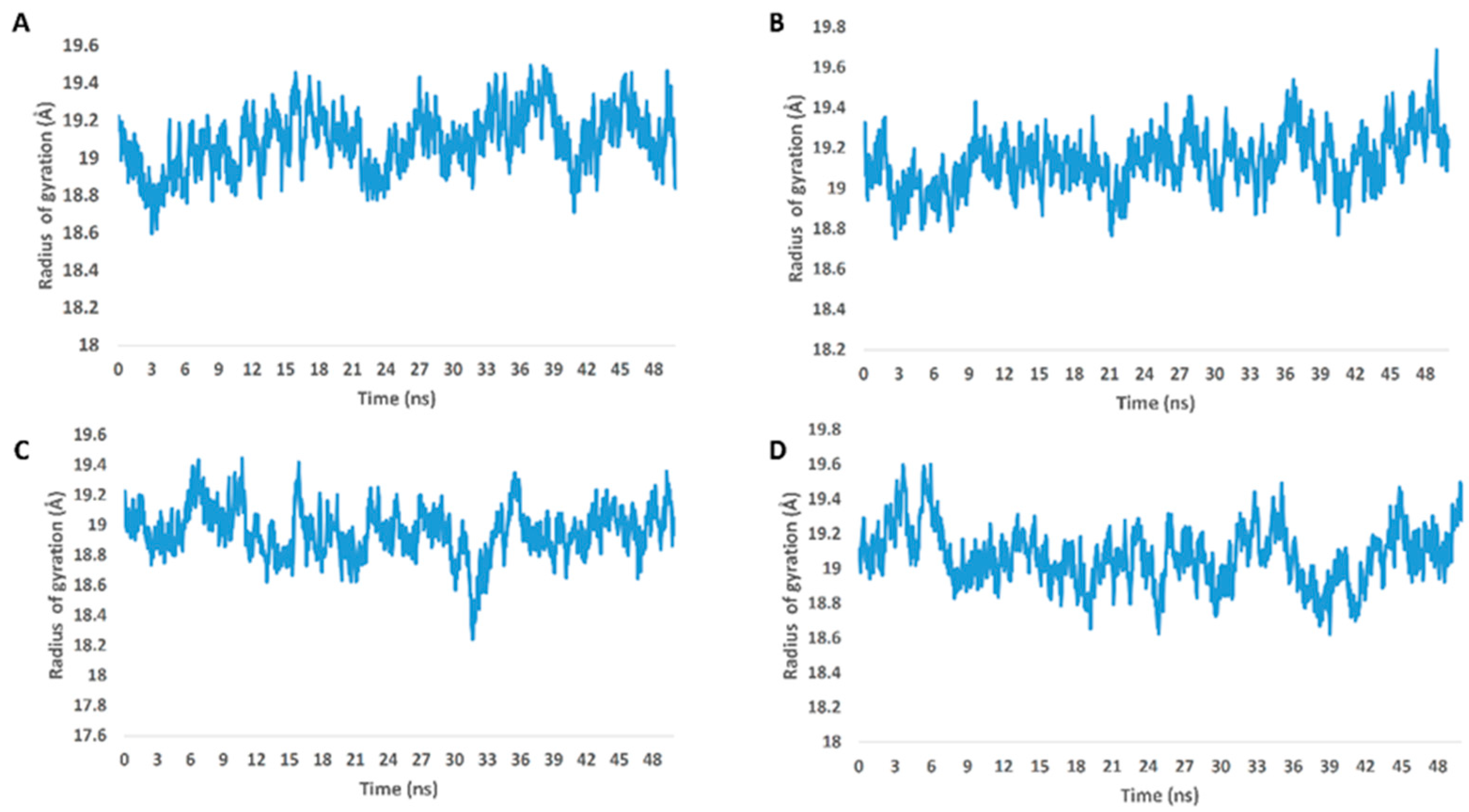
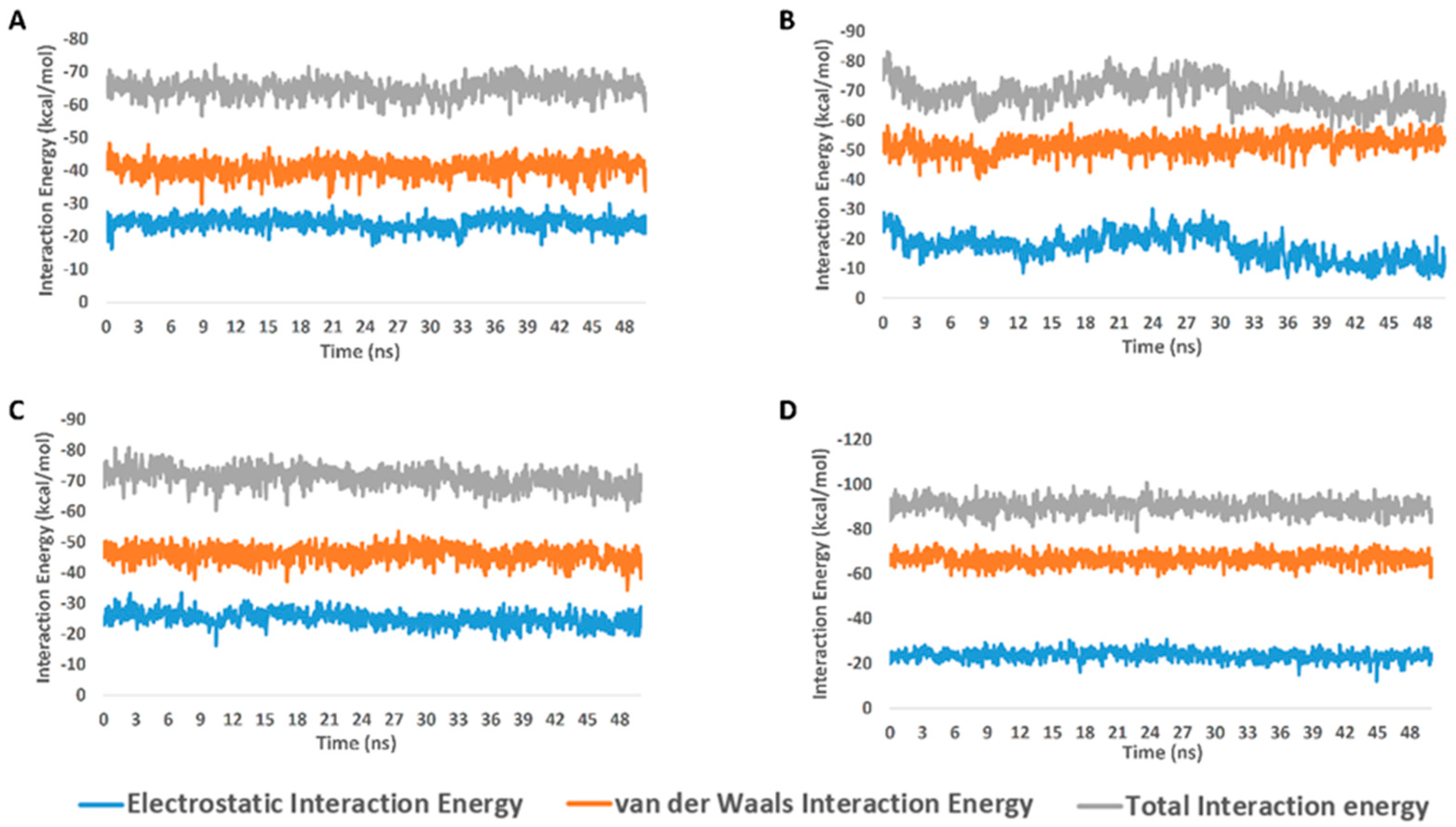
From the previous data, it can be concluded that GLN-189 was the key amino acid residue interacting with the three structures (i.e., compounds 5, 7, and 8) via H-bonding, while both the LEU-27 and MET-49 amino acid residues were the only residues hydrophobically interacting with them. The active steroids (5, 7, and 8) were interacting with three of the amino acids (GLN-189, LEU-27, and MET-49), which were included in the active site pockets of the SARS-CoV-2 Mpro [5], illustrating its possible activity. The dynamic behaviors of the three structures were convergent, where they were highly fluctuating over the course of the simulation, in comparison with the co-crystalized inhibitor (Figure 4). Their average RMSDs were 2.4 Å, 2.6 Å, and 1.3 Å for compounds 5, 7, and 8, respectively (Figure 4). The calculated radius of the gyration profile of Mpro in complex with each compound was also consistent, with an average of ~ 18.4 Å (Figure 5). With regard to the interaction energy of the three structures over the course of the simulation, they showed stable interaction energy profiles (i.e., electrostatic and van der Waals interaction energies), with an average of ~-80 kcal/mol (Figure 6). Accordingly, the calculated MM-PBSA binding energy of each compound (Table 3) was found to be convergent with or even higher than that of the co-crystalized inhibitor ML188, indicating the potential inhibitory activity of these compounds against the viral Mpro. Such structural information will be of great interest during the future development of novel Mpro inhibitors, depending on the scaffolds of these compounds (i.e., compounds 5, 7, and 8).
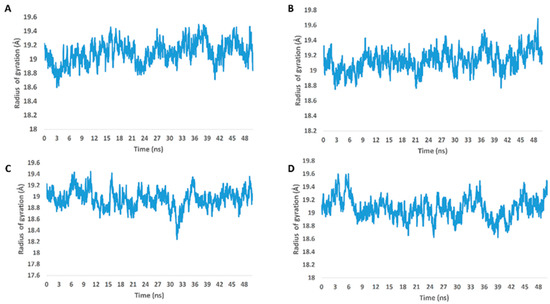
Figure 5.
Radius of gyration of Mpro in complex with compounds 5, 7, and 8 along with the co-crystalized inhibitor ML188 (A–D, respectively) over the course of 50-ns-long MDS.
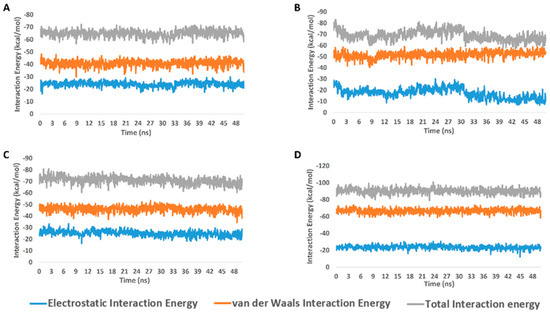
Figure 6.
Interaction energy profiles of compounds 5, 7, and 8 along with the co-crystalized inhibitor ML188 in complex with Mpro (A–D, respectively) over the course of 50-ns-long MDS.

Table 3.
MM-PBSA binding free energies of compounds 5, 7, and 8 along with the co-crystalized inhibitor ML188 in complex with Mpro.
With regard to the probable inhibitory activity SARS-CoV-2’s Mpro towards the isolated sesquiterpenes and sterols, docking, followed by MDS experiments, revealed that the polyhydroxylated sterols have good potential to bind with and to inhibit the enzyme’s catalytic activity. According to the dynamic binding mode analysis, the stability of such derivatives was achieved inside the enzyme’s active site through the formation of multiple H-bonds between the compounds’ hydroxyl groups and a number of hydrophilic amino acids. Hence, losing such essential hydroxyl groups or masking them via acetylation might lead to unstable binding (i.e., compounds 3, 4, and 6). Previously, a number of polyhydroxylated triterpenes (e.g., ursolic and maslinic acids) were found to significantly inhibit SARS-CoV’s Mpro [48,49]. Hence, such steroidal or triterpenoidal scaffolds may be promising in the future development of novel Mpro inhibitors.
4. Materials and Methods
4.1. General Experimental Procedures
Optical rotations were recorded on a Jasco DIP-370 polarimeter. The 1D (1H and 13C) and 2D (HSQC, HMBC, 1H-1H COSY, and NOESY) NMR experiments were recorded on a Bruker DRX 600 NMR spectrometer (Bruker, Billerica, MA, USA). HR-ESI-MS and HR-FAB-MS were measured on an LC-MS-Q-TOF (Agilent Tokyo, Japan) and a JMS-700 mass spectrometer (JEOL, Tokyo, Japan), respectively. Coupling constants are expressed in Hz, and chemical shifts in δ (ppm). Chromatographic separations were performed using column chromatography on a Merck silica gel (70–230), and Medium-Pressure Liquid Chromatography (MPLC) (Büchi Reveleris® Prep system, Flawil, Switzerland) was performed using a silica gel cartridge (40 μM, 12 g) and a C-18 cartridge (WP, 20 μM, 4 g) with a UV-ELSD detector. Thin-layer chromatography (TLC) was performed on glass pre-coated silica gel 60 F254 plates (Merck, Darmstadt, Germany) and reversed phase (RP-18 F254), which were visualized under UV light at (254 and 365 nm) and sprayed with 5% MeOH-H2SO4 reagent, followed by heating for 2–3 min.
4.2. Animal Material
Heteroxenia fuscescens soft coral was collected and identified by Dr. Aldoushy Mahdy (Faculty of Science, Al-Azhar University, Assiut branch, Egypt) from the Red Sea in front of the National Institute of Oceanography and Fisheries, Hurghada, Egypt. A voucher sample (HF 20) has been deposited at the Pharmacognosy Department, Faculty of Pharmacy, Al-Azhar University, Assiut Branch, Egypt.
4.3. Extraction and Isolation
The fresh material of H. fuscescens (370 g) was sliced and exhaustively extracted with 90% MeOH. The total MeOH extract (8.9 g) was fractionated using silica gel CC and eluted with 100% n-Hexane, n-Hexane:EtOAc, EtOAc, and MeOH (100%) to obtain five fractions of n-Hexane (1.6 g), n-Hexane:EtOAc (50:50, 1.4 g), EtOAc (100%, 2.3 g), EtOAc:MeOH (50:50, 2.7 g), and MeOH (100%, 4.2 g). The n-Hexane:EtOAc (50:50) (1.4 g) fraction was passed over a normal flash column to yield four sub-fractions (HE I-IV). Sub-fraction (HE-I) was separated on silica gel CC using n-Hexane:EtOAc in a gradient manner to afford pure compound 2 (2.0 mg) and compound 3 (11.3 mg), which were finally purified via NPTLC using n-Hexane:EtOAc (40:60). Sub-fraction HE-II was separated by RP-18 MPLC using a MeOH-H2O gradient from (30:70) to 100% MeOH to afford compound 1 (1.2 mg). Sub-fraction HE-IV was separated on NPTLC using n-Hexane:EtOAc (30:70) to obtain compound 4 (6.3 mg). The EtOAc fraction (2.3 g) was subjected to an RP-18 flash column using MeOH-H2O in a gradient manner (50:50 to 80:20) to afford six sub-fractions (Et I–VI). Compound 8 (2.2 mg) was obtained from sub-fraction Et II by using RP18-TLC and using (MeOH: H2O, 70:30) as a solvent system. Sub-fraction Et IV was passed over Sephadex (LH-20) and eluted with MeOH 100% to obtain compound 5 (10.2 mg) and compound 7 (3.1 mg). Compound 6 (4.2 mg) was obtained from sub-fraction VI, which passed over silica gel CC and was eluted with n-Hexane:EtOAc (85:15) to EtOAc:MeOH (90:10).
4.4. Spectral Data
4.4.1. Fusceterpene A (1)
Colorless oil; D20 + 62.2 (c 0.001, MeOH); for 1H and 13C-NMR data, see (Table 1). Positive HR-FAB-MS 235.1701 [M+H]+ (calcd for C15H23O2, 235.1698 m/z).
4.4.2. Fuscesterol A (4)
White amorphous powder; D20 + 25.4 (c 0.002, MeOH); for 1H and 13C-NMR data, see (Table 2). Positive HR-ESI-MS at m/z 527.3673 [M+Na]+ (calcd for C31H52NaO5, 527.3712 m/z).
4.5. Molecular Docking and Molecular Dynamic Simulation
(See Supplementary Materials).
5. Conclusions
This study highlighted the significance of identifying antiviral secondary metabolites from the Red Sea’s soft corals as naturally occurring SARS-CoV-2 inhibitors. A new steroid and a new sesquiterpene, along with six known compounds, were effectively isolated and structurally characterized, and they were essentially evaluated via in silico study against SARS-CoV-2 proteins. Through a stable hydrogen bond, the purified steroids containing a (3β,5α,6β-trihydroxy) moiety interact with the amino acid residue GLU-189, which highlights it as a perfect framework for the future creation of SARS-CoV-2 Mpro inhibitory medications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27217369/s1. Figure S1. Positive HRFAB-MS spectrum of compound 1; Figure S2. 1H-NMR spectrum of compound 1 (600 MHz, CDCl3); Figure S3. Expanded 1H-NMR spectrum (aromatic region) of compound 1 (600 MHz, CDCl3); Figure S4. 13C-NMR spectrum of compound 1 (150 MHz, CDCl3); Figure S5. HSQC spectrum of compound 1 (CDCl3); Figure S6. HMBC spectrum of compound 1 (CDCl3); Figure S7. 1H-1H COSY spectrum of compound 1 (CDCl3); Figure S8. Expanded 1H-1H COSY spectrum of compound 1 (CDCl3); Figure S9. NOESY spectrum of compound 1 (CDCl3); Figure S10. Expanded NOESY spectrum of compound 1 (CDCl3); Figure S11. Positive HR-ESI-MS spectrum of compound 4; Figure S12. 1H-NMR spectrum of compound 4 (600 MHz, CD3OD); Figure S13. 13C-NMR spectrum of compound 4 (150 MHz, CD3OD).; Figure S14. HSQC spectrum of compound 4 (CD3OD); Figure S15. HMBC spectrum of compound 4 (CD3OD); Figure S16. 1H-1H COSY spectrum of compound 4 (CD3OD); Figure S17. Expanded 1H-1H COSY spectrum of compound 4 (CD3OD); Figure S18. NOESY spectrum of compound 4 (CD3OD); Figure S19. Expanded NOESY spectrum of compound 4 (CD3OD); Figure S20. 1H-NMR spectrum of compound 2 (600 MHz, CDCl3); Figure S21. 13C-NMR spectrum of compound 2 (150 MHz, CDCl3); Figure S22. 1H-NMR spectrum of compound 3 (600 MHz, CDCl3); Figure S23. 13C-NMR spectrum of compound 3 (150 MHz, CDCl3); Figure S24. 1H-NMR spectrum of compound 5 (600 MHz, CD3OD); Figure S25. 13C-NMR spectrum of compound 5 (150 MHz, CD3OD); Figure S26. 1H-NMR spectrum of compound 6 (600 MHz, CD3OD); Figure S27. 13C-NMR spectrum of compound 6 (150 MHz, CD3OD); Figure S28. 1H-NMR spectrum of compound 7 (600 MHz, CD3OD); Figure S29. 13C-NMR spectrum of compound 7 (150 MHz, CD3OD); Figure S30. 1H-NMR spectrum of compound 8 (600 MHz, CD3OD); Figure S31. 13C-NMR spectrum of compound 8 (150 MHz, CD3OD); Table S1. 1H NMR data of compounds 2, 3 and 5; Table S2. 1H NMR data of compounds 6, 7 and 8; Table S3. 13C-NMR data of compounds 2, 3, 5, 6, 7 and 8; Table S4. All SARS-CoV-2 related proteins used in this study; Table S5. Docking scores of the isolated compounds (1–8) against the SARS CoV-2 targets ex-pressed in kcal/mol. References [50,51,52,53,54,55,56,57,58,59,60] are cited in the Supplementary materials.
Author Contributions
Conceptualization, K.S. and F.M.A.; validation, A.E.A., A.M.N., H.K.A. and M.R.K.; investigation, K.S., M.A.H.M., S.A.H.Z., R.A.A.H. and H.A.H.; methodology, F.M.A., A.M.S., S.A.H.Z. and A.E.A.; writing—original draft preparation, F.M.A., A.E.A. and M.A.H.M.; writing—review and editing, M.E.A.Z., A.M.N., H.K.A. and M.R.K.; visualization, R.A.A.H.; supervision, K.S.; project administration, K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank the Research and Education Support Center of the Faculty of Agriculture, Kyushu University, for the supporting facilities for NMR and mass analysis. Additionally, we thank the Deanship of Scientific Research, Imam Mohammad Ibn Saud Islamic University (IMSIU), Saudi Arabia, for supporting this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nair, R.; Soni, M.; Bajpai, B.; Dhiman, G.; Sagayam, K.M. Predicting the death rate around the world due to COVID-19 using regression analysis. Int. J. Swarm Intell. Res. 2022, 13, 1–13. [Google Scholar] [CrossRef]
- Allam, A.E.; Assaf, H.K.; Hassan, H.A.; Shimizu, K.; Elshaier, Y.A. An in silico perception for newly isolated flavonoids from peach fruit as privileged avenue for a countermeasure outbreak of COVID-19. RSC Adv. 2020, 10, 29983–29998. [Google Scholar] [CrossRef]
- Hwang, S.M.; Jung, Y.; Seo, H. Diagnostic Laboratory Characteristics of COVID-19 Patients Infected by Fomites: COVID-19 Outbreak in a South Korean Public Administrative Facility. Pathogens 2022, 11, 700. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, A.; Mohammad, T.; Anwar, S.; Amani, S.; Khan, M.S.; Husain, F.M.; Rehman, M.T.; Islam, A.; Hassan, M.I. Potential drug targets of SARS-CoV-2: From genomics to therapeutics. Int. J. Biol. Macromol. 2021, 177, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, A.; Mohammad, T.; Anwar, S.; AlAjmi, M.F.; Hussain, A.; Rehman, M.; Islam, A.; Hassan, M. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: Possible implication in COVID-19 therapy. Biosci. Rep. 2020, 40, BSR20201256. [Google Scholar] [CrossRef] [PubMed]
- Esakandari, H.; Nabi-Afjadi, M.; Fakkari-Afjadi, J.; Farahmandian, N.; Miresmaeili, S.-M.; Bahreini, E. A comprehensive review of COVID-19 characteristics. Biol. Proced. Online 2020, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Saadat, K.A. RNAi-mediated siRNA sequences to combat the COVID-19 pandemic with the inhibition of SARS-CoV-2. Gene Rep. 2022, 26, 101512. [Google Scholar] [CrossRef]
- Abouelela, M.E.; Assaf, H.K.; Abdelhamid, R.A.; Elkhyat, E.S.; Sayed, A.M.; Oszako, T.; Belbahri, L.; El Zowalaty, A.E.; Abdelkader, M.S.A. Identification of potential SARS-CoV-2 main protease and spike protein inhibitors from the genus aloe: An in silico study for drug development. Molecules 2021, 26, 1767. [Google Scholar] [CrossRef] [PubMed]
- Roe, K. High COVID-19 virus replication rates, the creation of antigen–antibody immune complexes and indirect haemagglutination resulting in thrombosis. Transbound. Emerg. Dis. 2020, 67, 1418–1421. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Chen, Y.; Qi, S.; Shi, D.; Feng, L.; Sun, D. A mini-review on cell cycle regulation of coronavirus infection. Front. Vet. Sci. 2020, 7, 586826. [Google Scholar] [CrossRef] [PubMed]
- Meijer, L. Cyclin-dependent kinases inhibitors as potential anticancer, antineurodegenerative, antiviral and antiparasitic agents. Drug Resist. Updat. 2000, 3, 83–88. [Google Scholar] [CrossRef]
- He, B.; Garmire, L. Prediction of repurposed drugs for treating lung injury in COVID-19. F1000Research 2020, 9, 609. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Parums, D.V. Current status of oral antiviral drug treatments for SARS-CoV-2 infection in non-hospitalized patients. Med. Sci. Monit. 2022, 28, e935952-1–e935952-4. [Google Scholar] [CrossRef] [PubMed]
- Veríssimo, A.C.; Pacheco, M.; Silva, A.M.; Pinto, D.C. Secondary metabolites from marine sources with potential use as leads for anticancer applications. Molecules 2021, 26, 4292. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Mahmoud, A.H.; Zidan, S.A.; Samy, M.N.; Alian, A.; Abdelmohsen, U.R.; Fouad, M.A.; Kamel, M.S.; Matsunami, K. Cytotoxicity and chemical profiling of the Red Sea soft corals Litophyton arboreum. Nat. Prod. Res. 2021, 36, 4261–4265. [Google Scholar] [CrossRef]
- Savić, M.P.; Sakač, M.N.; Kuzminac, I.Z.; Ajduković, J.J. Structural diversity of bioactive steroid compounds isolated from soft corals in the period 2015–2020. J. Steroid Biochem. Mol. Biol. 2022, 2022, 106061. [Google Scholar] [CrossRef] [PubMed]
- Abdelhafez, O.H.; Fahim, J.R.; Mustafa, M.; AboulMagd, A.M.; Desoukey, S.Y.; Hayallah, A.M.; Kamel, M.S.; Abdelmohsen, U.R. Natural metabolites from the soft coral Nephthea sp. as potential SARS-CoV-2 main protease inhibitors. Nat. Prod. Res. 2021, 36, 2893–2896. [Google Scholar] [CrossRef]
- Zidan, S.A.; Abdelhamid, R.A.; Al-Hammady, M.; Fouad, M.A.; Matsunami, K.; Orabi, M.A. Cytotoxic polyhydroxy sterols from the Egyptian Red Sea soft coral Sarcophyton acutum. Fitoterapia 2020, 147, 104765. [Google Scholar] [CrossRef]
- Ismail, H.A.; Hanafy, M.H.; Madkour, F.F.; Ahmed, M.I. Distribution of soft coral s in the Egyptian coasts of the Red Sea and Gulf of Aqaba. Int. J. Eng. Sci. 2017, 7, 14944. [Google Scholar]
- Koido, T.; Imahara, Y.; Fukami, H. High species diversity of the soft coral family Xeniidae (Octocorallia, Alcyonacea) in the temperate region of Japan revealed by morphological and molecular analyses. ZooKeys 2019, 862, 1. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Phan, C.S.; Ishii, T.; Kamada, T.; Hamada, T.; Vairappan, C.S. Terpenoids from Marine Soft Coral of the Genus Xenia in 1977 to 2019. Molecules 2020, 25, 5386. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Razik, A.F.; Nassar, M.I.; Elshamy, A.I.; Kubacy, T.M.; Hegazy, M.E.F.; Ibrahim, N.; Le Lamer, A.C.; Farrag, A.R.H. A new cytotoxic ceramide from Heteroxenia ghardaqensis and protective effect of chloroform extract against cadmium toxicity in rats. Arab. J. Chem. 2016, 9, 649–655. [Google Scholar] [CrossRef]
- Abdelkarem, F.M.; Desoky, E.E.K.; Nafady, A.M.; Allam, A.E.; Mahdy, A.; Nagata, M.; Miyamoto, T.; Shimizu, K. Isolation of new secondary metabolites from gorgonian soft coral Heteroxenia fuscescens collected from Red Sea. Phytochem. Lett. 2020, 36, 156–161. [Google Scholar] [CrossRef]
- Abdel-Lateff, A.; Alarif, W.M.; Al-Lihaibi, S.S.; Abdel-Naim, A.B. Antiproliferative effects of selected marine organisms collected from Red Sea. Pak. J. Pharm. Sci. 2017, 30, 381–386. [Google Scholar] [PubMed]
- Mohammed, R.; Seliem, M.A.E.; Mohammed, T.; Abed-ElFatah, A.; Abo-Youssef, A.; Thabet, M. Bioactive secondary metabolites from the Red Sea soft coral Heteroxenia fuscescens. Int. J. Appl. Res. Nat. Prod. 2011, 4, 15–27. [Google Scholar]
- Edrada, R.A.; Wray, V.; Witte, L.; van Ofwegen, L.; Proksch, P. Bioactive terpenes from the soft coral Heteroxenia sp. from Mindoro, Philippines. Z. Naturforsch C. 2000, 55, 82–86. [Google Scholar] [CrossRef]
- Elshamy, A.I.; Nassar, M.I.; Mohamed, T.K.; Madkour, H.A. A new hydroxymethyl diacylglycerol from methanol extract of Egyptian soft coral Heteroxenia ghardaqensis. J. Biol. Act. Prod. Nat. 2015, 5, 172–177. [Google Scholar]
- Elshamy, A.I.; Abdel-Razik, A.F.; Nassar, M.I.; Mohamed, T.K.; Ibrahim, M.A.; El-Kousy, S.M. A new gorgostane derivative from the Egyptian Red Sea soft coral Heteroxenia ghardaqensis. Nat. Prod. Res. 2013, 27, 1250–1254. [Google Scholar] [CrossRef]
- Zidan, S.A.; Abdelhamid, R.A.; Alian, A.; Fouad, M.A.; Matsunami, K.; Orabi, M.A. Diterpenes and sterols from the Red Sea soft coral Sarcophyton trocheliophorum and their cytotoxicity and anti-leishmanial activities. J. Asian Nat. Prod. Res. 2021, 24, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.E.F.; Mohamed, T.A.; Elshamy, A.I.; Hassanien, A.A.; Abdel-Azim, N.S.; Shreadah, M.A.; Abdelgawad, I.I.; Elkady, E.M.; Paré, P.W. A new steroid from the Red Sea soft coral Lobophytum lobophytum. Nat. Prod. Res. 2016, 30, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Umeyama, A.; Shoji, N.; Ozeki, M.; Arihara, S. Sarcoaldesterols A and B, two new polyhydroxylated sterols from the soft coral Sarcophyton sp. J. Nat. Prod. 1996, 59, 894–895. [Google Scholar] [CrossRef]
- Cheng, Z.B.; Xiao, H.; Fan, C.Q.; Lu, Y.N.; Zhang, G.; Yin, S. Bioactive polyhydroxylated sterols from the marine sponge Haliclona crassiloba. Steroids 2013, 78, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, H.; Wang, P.; Gong, W.; Xue, M.; Zhang, H.; Liu, T.; Liu, B.; Yi, Y.; Zhang, W. Bioactive polyoxygenated steroids from the South China sea soft coral, Sarcophyton sp. Mar. Drugs 2013, 11, 775–787. [Google Scholar] [CrossRef]
- Konečný, K.; Streibl, M.; Vašíčková, S.; Buděšínský, M.; Šaman, D.; Ubik, K.; Herout, V. Constituents of the liverwort Bazzania trilobata of Czech origin. Collect. Czechoslov. Chem. Commun. 1985, 50, 80–93. [Google Scholar] [CrossRef]
- Limna Mol, V.; Raveendran, T.; Naik, B.; Kunnath, R.; Parameswaran, P. Calamenenes–aromatic bicyclic sesquiterpenes–from the Indian gorgonian Subergorgia reticulata (Ellis and Solander, 1786). Nat. Prod. Res. 2011, 25, 169–174. [Google Scholar] [CrossRef]
- Salmoun, M.; Braekman, J.C.; Ranarivelo, Y.; Rasamoelisendra, R.; Ralambomanana, D.; Dewelle, J.; Darro, F.; Kiss, R. New calamenene sesquiterpenes from Tarenna madagascariensis. Nat. Prod. Res. 2007, 21, 111–120. [Google Scholar] [CrossRef]
- Young-Kyoon, K.; Cool, L.G.; Zavarin, E. cis-Calamenene-related sesquiterpenoids from Cupressus bakeri foliage. Phytochemistry 1994, 36, 961–965. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Z.; Han, X.; Li, X.L.; Lu, Z.Y.; Dou, B.B.; Zhang, W.Z.; Tang, X.L.; Li, P.L.; Li, G.Q. Four bioactive new steroids from the soft coral Lobophytum pauciflorum collected in South China Sea. Beilstein J. Org. Chem. 2022, 18, 374–380. [Google Scholar] [CrossRef]
- Ngoc, N.T.; Hanh, T.T.H.; Quang, T.H.; Cuong, N.X.; Nam, N.H.; Thung, D.C.; Van Kiem, P.; Van Minh, C. Polyhydroxylated steroids from the Vietnamese soft coral Sarcophyton ehrenbergi. Steroids 2021, 176, 108932. [Google Scholar] [CrossRef] [PubMed]
- Abdelkarem, F.M.; Desoky, E.E.K.; Nafady, A.M.; Allam, A.E.; Mahdy, A.; Ashour, A.; Mohamed, G.A.; Miyamoto, T.; Shimizu, K. Two new polyhydroxylated steroids from Egyptian soft coral Heteroxenia fuscescens (Fam.; Xeniidae). Nat. Prod. Res. 2021, 35, 236–243. [Google Scholar] [CrossRef]
- Tseng, W.R.; Huang, C.Y.; Tsai, Y.Y.; Lin, Y.S.; Hwang, T.L.; Su, J.H.; Sung, P.J.; Dai, C.F.; Sheu, J.H. New cytotoxic and anti-inflammatory steroids from the soft coral Klyxum flaccidum. Bioorg. Med. Chem. Lett. 2016, 26, 3253–3257. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.R.; Huang, C.Y.; Chen, B.W.; Tsai, Y.Y.; Shih, S.P.; Hwang, T.L.; Dai, C.F.; Wang, S.Y.; Sheu, J.H. New bioactive steroids from the soft coral Klyxum flaccidum. RSC Adv. 2015, 5, 12546–12554. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Burgio, G.; Thissera, B.; Orfali, R.; Jiffri, S.E.; Yaseen, M.; Sayed, A.M.; Rateb, M.E. Neoechinulin A as a promising SARS-CoV-2 Mpro inhibitor: In vitro and in silico study showing the ability of simulations in discerning active from inactive enzyme inhibitors. Mar. Drugs 2022, 20, 163. [Google Scholar] [CrossRef] [PubMed]
- Abd El Hafez, M.S.; AbdEl-Wahab, M.G.; Seadawy, M.G.; El-Hosseny, M.F.; Beskales, O.; Abdel-Hamid, A.S.A.; El Demellawy, M.A.; Ghareeb, D.A. Characterization, in-silico, and in-vitro study of a new steroid derivative from Ophiocoma dentata as a potential treatment for COVID-19. Sci. Rep. 2022, 12, 1–11. [Google Scholar] [CrossRef]
- Ibrahim, M.A.; Abdelrahman, A.H.; Mohamed, T.A.; Atia, M.A.; Al-Hammady, M.A.; Abdeljawaad, K.A.; Elkady, E.M.; Moustafa, M.F.; Alrumaihi, F.; Allemailem, K.S. In silico mining of terpenes from red-sea invertebrates for SARS-CoV-2 main protease (Mpro) inhibitors. Molecules 2021, 26, 2082. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Sayed, A.M.; Sharif, A.M.; Azhar, E.I.; Rateb, M.E. Olive-derived triterpenes suppress SARS-CoV-2 main protease: A promising scaffold for future therapeutics. Molecules 2021, 26, 2654. [Google Scholar] [CrossRef]
- Wen, C.C.; Kuo, Y.H.; Jan, J.T.; Liang, P.H.; Wang, S.Y.; Liu, H.G.; Lee, C.K.; Chang, S.T.; Kuo, C.J.; Lee, S.S. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007, 50, 4087–4095. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Clyde, A.; Galanie, S.; Kneller, D.W.; Ma, H.; Babuji, Y.; Blaiszik, B.; Brace, A.; Brettin, T.; Chard, K.; Chard, R.; et al. High Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Non-Covalent Inhibitor. J. Chem. Inf. Model. 2022, 62, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.M.; Alhadrami, H.A.; El-Gendy, A.O.; Shamikh, Y.I.; Belbahri, L.; Hassan, H.M.; Abdelmohsen, U.R.; Rateb, M.E. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (Mpro). Microorganisms 2020, 8, 970. [Google Scholar] [CrossRef] [PubMed]
- Amaro, R.E.; Baudry, J.; Chodera, J.; Demir, Ö.; McCammon, J.A.; Miao, Y.; Smith, J.C. Ensemble docking in drug discovery. Biophys. J. 2018, 114, 2271–2278. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the SC’06: 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; IEEE: New York, NY, USA, 2006; p. 43. [Google Scholar]
- Release, S. 3: Desmond Molecular Dynamics System, DE Shaw Research, New York, NY, 2017; Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2017. [Google Scholar]
- Schrodinger LLC Maestro. Version 9.0. Schrodinger LLC: New York, NY, USA, 2009.
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Kim, S.; Oshima, H.; Zhang, H.; Kern, N.R.; Re, S.; Lee, J.; Rous, B.; Sugita, Y.; Jiang, W.; Im, W. CHARMM-GUI free energy calculator for absolute and relative ligand solvation and binding free energy simulations. J. Chem. Theory Comput. 2020, 16, 7207–7218. [Google Scholar] [CrossRef]
- Ngo, S.T.; Tam, N.M.; Quan, P.M.; Nguyen, T.H. Benchmark of Popular Free Energy Approaches Revealing the Inhibitors Binding to SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021, 61, 2302–2312. [Google Scholar] [CrossRef]
- Miller, B.R., III; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA. py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).